Abstract
The peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-dependent transcription factor that has been demonstrated to regulate fat cell development and glucose homeostasis. PPARγ is also expressed in a subset of macrophages and negatively regulates the expression of several proinflammatory genes in response to natural and synthetic ligands. We here demonstrate that PPARγ is expressed in macrophage foam cells of human atherosclerotic lesions, in a pattern that is highly correlated with that of oxidation-specific epitopes. Oxidized low density lipoprotein (oxLDL) and macrophage colony-stimulating factor, which are known to be present in atherosclerotic lesions, stimulated PPARγ expression in primary macrophages and monocytic cell lines. PPARγ mRNA expression was also induced in primary macrophages and THP-1 monocytic leukemia cells by the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA). Inhibition of protein kinase C blocked the induction of PPARγ expression by TPA, but not by oxLDL, suggesting that more than one signaling pathway regulates PPARγ expression in macrophages. TPA induced the expression of PPARγ in RAW 264.7 macrophages by increasing transcription from the PPARγ1 and PPARγ3 promoters. In concert, these observations provide insights into the regulation of PPARγ expression in activated macrophages and raise the possibility that PPARγ ligands may influence the progression of atherosclerosis.
The peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear receptor superfamily of transcription factors that regulate patterns of gene expression in response to the binding of small molecular weight ligands (1–3). PPARγ mRNA is most highly expressed in adipose tissue, the adrenal gland, spleen, and large colon (4–7). Several lines of evidence indicate that PPARγ plays an important role in regulating adipocyte differentiation and glucose homeostasis. PPARγ and the retinoid X receptor (RXR) form heterodimers on regulatory elements in a number of adipose-specific promoters that stimulate transcription in response to PPARγ or RXR-specific ligands (3, 5, 8, 9). Furthermore, forced expression of PPARγ in certain fibroblast cell lines induces adipocyte differentiation in a manner that is strongly potentiated by PPARγ- and RXR-specific ligands (8–11). Although the identities of the ligands that regulate PPARγ activity in vivo remain to be established with certainty, 15-deoxy-Δ12,14-prostaglandin J2 (15ΔPGJ2) and certain polyunsaturated fatty acids have been demonstrated to stimulate PPARγ-dependent transcription (9, 10, 12, 13). In addition, synthetic ligands such as troglitazone and BRL49653 have been identified that are specific PPARγ activators (14). Troglitazone and structurally related thiazolidinediones significantly reduce peripheral resistance to insulin in obesity and type 2 diabetes mellitus in both animals and humans and have recently been instituted as adjunctive therapy in diabetic patients (15–18).
The roles of PPARγ in other tissues are poorly understood. Recent studies indicate that PPARγ is expressed in cells of the monocyte/macrophage lineage (19–21). Several lines of evidence suggest that PPARγ may exert anti-inflammatory effects by negatively regulating the expression of pro-inflammatory genes. Treatment of peritoneal macrophages with 15ΔPGJ2 or several synthetic PPARγ ligands reduced the expression of inducible nitric oxide synthase by interferon γ (IFN-γ) and inhibited induction of gelatinase B and scavenger receptor A gene transcription in response to phorbol ester stimulation (20). Similarly, treatment of primary human monocytes with PPARγ-specific ligands blocked phorbol ester induction of interleukin 6 (IL-6), tumor necrosis factor α, and IL-1β (21). Anti-inflammatory effects of PPARγ ligands have not as yet been established in vivo, however, and it is possible that PPARγ exerts complex effects on macrophage function that are not strictly related to inflammation.
Macrophages are thought to play critical pathogenic roles in several chronic inflammatory diseases, including atherosclerosis (reviewed in refs. 22 and 23). Fatty streaks, the earliest visible lesions of atherosclerosis, contain large numbers of macrophage foam cells derived from circulating monocytes that adhere to activated endothelium and migrate into the artery wall (reviewed in ref. 24). These cells subsequently differentiate into macrophages that express the scavenger receptor A gene, as well as other scavenger receptors that mediate the uptake of oxidized low density lipoprotein (oxLDL) (25). Because these receptors are not subject to negative regulation by high levels of intracellular cholesterol, massive accumulation of cholesterol esters can occur in macrophages, resulting in foam cell formation. In addition to their uptake of oxLDL, macrophage foam cells are thought to influence the progression of atherosclerosis by several additional mechanisms, including promoting LDL oxidation (24), secretion of pro-inflammatory cytokines and other humoral factors that exert paracrine and autocrine effects in the artery wall (22, 23), and secretion of matrix metalloproteinases that have been suggested to remodel extracellular matrix proteins in arterial lesions, increasing the risk of plaque rupture, thrombus formation, and the clinical sequelae of myocardial infarction and stroke (26).
In the present studies, we present evidence that PPARγ is highly expressed in macrophage-derived foam cells of both early and advanced atherosclerotic lesions in a pattern that is highly correlated with the presence of oxidation-derived epitopes. Factors found to induce PPARγ expression in macrophages include oxLDL, macrophage colony-stimulating factor (M-CSF) and granulocyte/macrophage colony-stimulating factor (GM-CSF), which have been documented to be present in atherosclerotic lesions (reviewed in (23)). PPARγ could also be induced by the phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) in a manner that was dependent on new protein synthesis and was specifically inhibited by a protein kinase C (PKC) inhibitor. Induction of PPARγ expression by TPA was mediated by the PPARγ1 and PPARγ3 promoters, whereas PPARγ2 was not detected.
METHODS
Cell Culture.
Murine bone marrow progenitor cells and primary peritoneal macrophages were isolated and cultured as described (27). EML cells and MPRO cells were generously provided by S. Collins (Fred Hutchinson Cancer Institute) and cultured as described (28). Treatment with all trans retinoic acid was at 10 μM. THP-1 (American Type Culture Collection) cells were cultured in RPMI 1640 (GIBCO) supplemented with 10% heat inactivated fetal calf serum (Gemini Biological Products, Calabasas, CA), 100 units/ml penicillin, and 100 mg/ml streptomycin. Recombinant cytokines (human and murine) were obtained from R&D Systems or Endogen (Cambridge, MA) and used at the following concentrations, unless otherwise indicated: human M-CSF, 20 ng/ml; murine GM-CSF, 5 ng/ml; IFN-γ, 100 units/ml; lipopolysaccharide (LPS), 5 μg/ml; TPA, 0.1 μM; oxLDL, 50 μg/ml; and cycloheximide, 20 μg/ml. PKC inhibitor bisindolylmaleimide II and protein kinase A inhibitor H-89 were obtained from Calbiochem.
Western Blot Analysis.
Western blot analysis was performed by using standard procedures (29). Incubation with primary antibody (rabbit IgG anti-human PPARγ), developed against the N-terminal aa 20–104 (7) recognizing the 55-kDa PPARγ protein was performed at 1:500 dilution overnight at 4°C. The secondary antibody (goat anti-rabbit from Dako) was diluted at 1:4,000 and incubated at room temperature for 1 h. Detection was performed by using chemiluminescence (NEN Renaissance). Protein levels were determined to be similar in each sample by using an antibody against β-actin (Sigma).
RNase Protection.
The full-length PPARγ2 coding region plus 33 bp of the 5′ untranslated region was inserted in the inverted orientation (3′ to 5′ in front of the T7 promoter) into the EcoRI site of the expression vector pSG5 (Stratagene). The resulting plasmid pSG5-PPARγ2-inv was digested with EcoRV and religated, yielding the vector pSG5-PPARγ2-RPA that was used as a template for the synthesis of the antisense RNA probe, allowing specific measurement of PPARγ2 mRNA. For the specific analysis of the PPARγ3 relative to PPARγ1 mRNA, a template for probe synthesis was constructed starting from a reverse transcription–PCR product, using human adipose tissue RNA as template, with the oligonucleotides LF-44 (5′-GTCGGCCTCGAGGACACCGGAGAG-3′, which binds sense at exon A1) and LF-21 (5′-GGCTCTTCATGAGGCTTATTGTAGAGCTGA-3′, which binds antisense at the exon 2). The amplified fragment was inserted blunt into the EcoRV site of pBluescript SK+ (Stratagene). The resulting plasmid pBS-PPARγ3-RPA, which contains part of the exon A1, the full-length exon A2, exon 1, and part of exon 2, was used as a template for the probe synthesis. Common PPARγ probes corresponding to nt 800–1,093 of the murine and human cDNAs, respectively, were subcloned into pBluescript SK+. Isolation of total RNA, preparation of antisense probes, and RNase protection experiments were done following standard protocols (29). A β-actin antisense probe was used to verify equivalent amounts of total mRNA.
Transfection Assays.
RAW 264.7 cells were transiently transfected by using 2 μl Lipofectamin (GIBCO/BRL) with 0.5 μg of luciferase reporter plasmid and 0.1 μg of β-actin-lacZ reporter plasmid as internal transfection control (20). After transfection, cells were treated with 0.1 μM TPA in 0.5% fetal bovine serum for 16 h. THP-1 cells were transfected by electroporation (30). Luciferase and β-galactosidase enzymatic activities were determined, and luciferase activity was normalized to the β-galactosidase standard as described (31). The PPARγ1 promoter construct, pGL3γ1p3000, containing 3 kb of 5′ flanking information has been described (7). The human PPARγ3 promoter reporter construct contained ≈800 bp of 5′ flanking information upstream of the PPARγ3 promoter. This fragment was isolated by PCR using the oligonucleotide pair 5′-CGTTAAAGGCTGACTCTCGTTTGA-3′, binding in the PPARγ3 exon A2, and 5′-TCATGTAGGTAAGACTGTGTAGAA-3′, binding sense at position −800 of the PPARγ3 promoter, and the PAC clone 8,856 as template (7). The PCR product was sequenced and cloned into the reporter vector pGL3 (Promega) creating the expression vector pGL3γ3p800.
Immunohistochemistry.
Immunohistochemistry studies were performed by using human coronary arteries obtained from recipients of heart transplants. Arteries were immediately removed from the heart and placed into fixative (4% paraformaldehyde, 5% sucrose) containing antioxidants (1 mM EDTA and 50 μM butylated hydroxytoluene) to prevent oxidative artifacts that may affect lipid-rich tissues obtained postmortem. After paraffin embedding, 7-μm-thick serial sections of 42 arterial segments containing a broad spectrum of atherosclerotic lesions were immunostained with an avidin-biotin-alkaline phosphatase method (32). The primary antibodies and dilutions used to detect PPARγ, oxidation-specific epitopes such as malondialdehyde (MDA)-lysine, macrophages, and smooth muscle cells are listed in Table 1 (7, 33–38). Some of the tissues were counterstained with methyl green. Controls consisted of parallel sections stained without the primary antibody and were devoid of specific staining. Specificity of staining with the P2-20 antiserum against PPARγ was verified by competitive immunostaining. A 1:50 dilution of the antibody was incubated for 1 h at room temperature with an equal volume of PBS containing 10 μg/ml of a peptide (PPARγ aa 2–20). Staining after 10 min of substrate exposure was compared with that obtained with the same antibody incubated with PBS.
Table 1.
Characteristics of antibodies used for immunohistochemistry
| Target epitope | Antibody | Type | Dilution (substrate time) | Source | Ref. |
|---|---|---|---|---|---|
| Human PPARγ | P20-104 | Rabbit IgG | 1:50 (30 min) | J.A. and J.N. | 7 |
| Human PPARγ | P2-20 | Goat IgG | 1:100 (30 min) | Santa Cruz Biotechnology | |
| MDA-lysine | MDA2 | Mouse IgG1 (Mab) | 1:100 (10 min) | W.P. and J.L.W. | 33, 34 |
| Macrophages | HAM56 | Mouse IgM (Mab) | 1:200 (30 min) | Axel Accurate (Westbury, NY) | 35 |
| CD68 (macrophages) | Clone KP1 | Mouse IgG1 (Mab) | 1:100 (10 min) | Dako | 36 |
| Actin (smooth muscle cells) | HHF35 | Mouse IgG1 (Mab) | 1:1,000 (30 min) | Enzo Diagnostics | 37 |
RESULTS
Expression of PPARγ in Human Atherosclerotic Lesions.
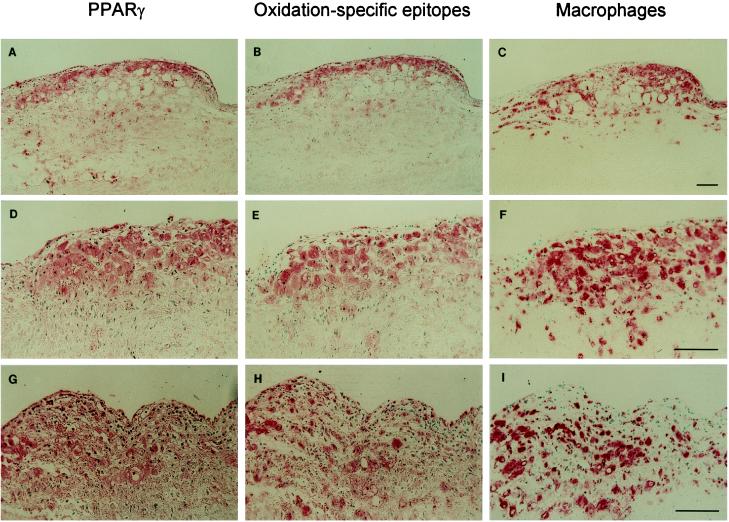
To determine whether PPARγ is expressed in vivo in atherosclerotic lesions, immunohistochemistry studies of human coronary arteries obtained from cardiac transplant recipients were performed. Staining with a rabbit anti-PPARγ IgG directed against PPARγ1 aa 20–104, which are common to all PPARγ isoforms, showed that PPARγ was highly expressed in early and intermediate atherosclerotic lesions (Fig. 1 A, D, and G). A very similar pattern of staining was observed by using a mAb directed against aa 2–20 and could be abolished by preincubation of the antibody with a peptide of this sequence, indicating that it was specific (data not shown). In early atherosclerotic lesions, staining was predominantly found in large foam cells and generally colocalized with staining for the macrophage-specific markers CD68 (Fig. 1 C, F, and I) and HAM56 (data not shown), although some staining clearly occurred in areas that were negative for either of these macrophage-specific markers. No significant staining for PPARγ 20–104 was observed in the underlying media or unaffected sections of the aorta, and the fatty streak lesions shown in Fig. 1 did not stain for a marker of smooth muscle cells [HHF-35 (37)]. PPARγ staining was strikingly similar to that of oxidation-specific epitopes detected by using the mAb MDA2 (Fig. 1 B, E, and H). Staining of transitional lesions and atheromas also showed some colocalization of PPARγ and oxidation-specific epitopes. However, at the edges of the necrotic core and in the core itself increasing dissociation between PPARγ and oxidation epitopes on one side and macrophage epitopes on the other was evident (data not shown). In these areas, progressive loss of staining for CD68 and HAM56 was noted, and staining with the anti-PPARγ antibody became more diffuse and not strictly cell associated. This finding may reflect differential loss of epitopes during progressive apoptosis/necrosis of PPARγ-expressing cells or the generation of neoepitopes that are recognized by the PPARγ antibodies.
Figure 1.
Immunohistochemistry with antibodies to PPARγ, oxidation-specific epitopes, and macrophages. Sections shown are serial, but not always consecutive sections of human coronary arteries immunostained with an antiserum to PPARγ (aa 20–104) (A, D, and G), the oxidation-specific mAb MDA2 (B, E, and H), and a macrophage-specific mAb, anti-CD68 (C, F, and I), as described. (A–C) Early atherosclerotic lesion showing a striking colocalization between PPARγ and oxidation-specific epitopes. (×58.) (D–F) Higher magnification (×116) of the shoulder area of a lesion rich in macrophage/foam cells, demonstrating the cellular nature of the PPARγ staining. (G–I) Higher magnification (×116) of a fatty streak demonstrating that PPARγ staining generally but not exclusively colocalized with macrophages. Lesions A–I were virtually free of smooth muscle cell-derived cells staining for actin (data not shown).
Regulation of PPARγ Expression During Macrophage Differentiation.
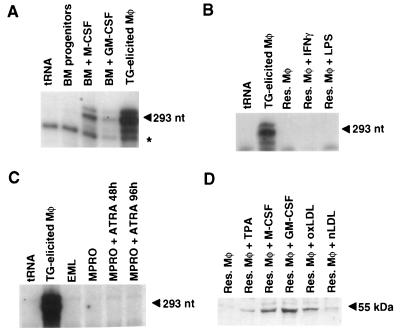
To identify factors that might regulate PPARγ expression in macrophages, murine bone marrow progenitor cells were cultured in the presence of M-CSF or GM-CSF for 3 days and PPARγ mRNA levels were quantitated by RNase protection assay. No PPARγ expression was observed in the undifferentiated bone marrow population, whereas expression was detected in the adherent macrophage population that was induced by M-CSF (Fig. 2A). GM-CSF also induced PPARγ mRNA expression in the adherent macrophage population, although less strongly than M-CSF (Fig. 2A). Intriguingly, although very high levels of PPARγ mRNA were present in thioglycolate-elicited peritoneal macrophages, almost no PPARγ mRNA could be detected in resident peritoneal macrophages (Fig. 2 A and B).
Figure 2.
Regulation of PPARγ expression during macrophage differentiation. (A) PPARγ mRNA is up-regulated during differentiation of bone marrow progenitor cells in response to M-CSF and GM-CSF and is highly expressed in thioglycolate-elicited peritoneal macrophages. Total RNA was prepared from undifferentiated bone marrow progenitor cells, from the adherent macrophage population arising from bone marrow (BM) progenitor cells cultured for 3 days in M-CSF (BM + M-CSF), GM-CSF (BM + GM-CSF), and from thioglycolate (TG)-elicited macrophages. RNase protection analysis was performed by using an antisense PPARγ probe, as described. The specific 293-nt PPARγ protection product is indicated by an arrow. A nonspecific band is indicated by an asterisk. (B) PPARγ mRNA is expressed at very low levels in resident peritoneal macrophages and is not induced by treatment with IFN-γ or LPS. Total RNA was prepared from thioglycolate-elicited and resident macrophages exposed to the indicated stimuli and analyzed for expression of PPARγ as in A. (C) PPARγ is not up-regulated during granulocyte differentiation. Total RNA was isolated from EML and MPRO cells before and after granulocyte differentiation in response to all trans retinoic acid (ATRA). By 96 h, more than 90% of the MPRO cells exhibited segmented nuclei characteristics of fully differentiated neutrophils. (D) PPARγ protein levels are induced in resting peritoneal macrophages by TPA, M-CSF, GM-CSF, and oxLDL, but not native LDL (nLDL). Resident peritoneal macrophages were treated with the indicated compounds for 72 h, as described. Cells were harvested in SDS sample buffer, and soluble proteins were resolved by SDS/PAGE. The 55-kDa PPARγ protein was detected by using a specific antibody recognizing the A/B domain common to the PPARγ1 and PPARγ2 proteins. For the RNase protection experiments in A–C, equivalent amounts of total RNA were verified by the use of an antisense β-actin probe in each sample. For the Western blot experiment in D, equivalent amounts of proteins were verified by parallel Western blots by using an antibody to β-actin.
PPARγ expression during granulocyte differentiation was evaluated in EML and MPRO cells, which were derived from bone marrow progenitor cells by retroviral insertion of a dominant-negative retinoic acid receptor (28). EML cells represent a very early multipotent hematopoietic progenitor cell, whereas MPRO cells exhibit a promyelocytic phenotype and can be induced to differentiate into granulocytes by treatment with high concentrations of all-trans retinoic acid (28). PPARγ mRNA could not be detected in EML or MPRO cells either before or after differentiation, suggesting that the induction of PPARγ in myeloid cells is macrophage-specific (Fig. 2C).
The presence of low levels of PPARγ mRNA in resident peritoneal macrophages and much higher levels of expression in atherosclerotic lesions and thioglycolate-elicited macrophages suggested that PPARγ expression is induced by humoral factors that are produced during inflammatory response. PPARγ expression was therefore evaluated in resident peritoneal macrophages following treatment with a panel of factors that influence macrophage function. Consistent with the results obtained with bone marrow progenitor cells, treatment of resident peritoneal macrophages with M-CSF and GM-CSF led to a marked increase in PPARγ protein levels, as detected by Western blot analysis (Fig. 2D). In addition, TPA and oxLDL induced PPARγ protein levels (Fig. 2D). In contrast, treatment of resident peritoneal macrophages with IFN-γ or LPS did not stimulate PPARγ expression (Fig. 2B).
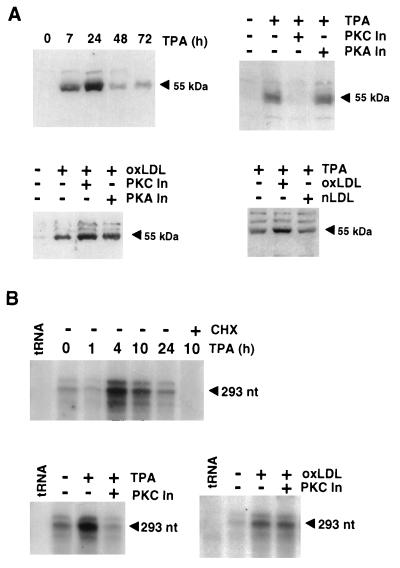
To further investigate the mechanisms that may regulate PPARγ expression in vivo, experiments were performed in the THP-1 monocytic leukemia cell line. Undifferentiated THP-1 cells express low levels of PPARγ under basal conditions (Fig. 3). Treatment of THP-1 cells with TPA and oxLDL resulted in significant increases in PPARγ expression at both the protein (Fig. 3A) and mRNA (Fig. 3B) levels, consistent with observations with resident peritoneal macrophages. Induction of PPARγ mRNA by TPA was maximal 4–10 h after treatment (Fig. 3B), with protein levels peaking at 24 h and then returning to near baseline levels by 48 h (Fig. 3A). Induction of PPARγ mRNA was blocked by cycloheximide treatment, indicating a requirement for new protein synthesis (Fig. 3B). Intriguingly, the PKC inhibitor bisindolylmaleimide II blocked induction of PPARγ mRNA and protein expression by TPA, but not by oxLDL (Fig. 3 A and B), whereas the protein kinase A inhibitor H89 had no effect on the induction of PPARγ expression by either oxLDL or TPA (Fig. 3A). OxLDL treatment potentiated the effects of a maximal activating concentration of TPA on PPARγ expression, consistent with the possibility that they control PPARγ expression by different mechanisms (Fig. 3A).
Figure 3.
Regulation of PPARγ expression in THP-1 cells by TPA and oxLDL is mediated by PKC-dependent and independent mechanisms, respectively. (A) Regulation of PPARγ expression in THP-1 monocytic leukemia cells as detected by Western blot analysis. (Upper Left) THP-1 cells were cultured in the presence of 0.1 μM TPA for the indicated times prior to analysis by Western blot. The arrowhead indicates the specific 55-kDa PPARγ band. (Upper Right) THP-1 cells were treated with 1 nM TPA in the presence or absence of the PKC inhibitor (PKC In) bisindolylmaleimide II (1 μM) or the protein kinase A inhibitor (PKA In) H89 (10 μM). (Lower Left) THP-1 cells were treated with 25 μg/ml oxLDL in the presence or absence of the same concentration of PKC and PKA inhibitors. (Lower Right) THP-1 cells were treated with a maximal activating concentration of TPA (0.1 μM) and 50 μg/ml of either oxLDL or native LDL. Equivalent loading of protein in each sample was verified by simultaneous Western blots for β-actin (data not shown). (B) Regulation of PPARγ mRNA expression in THP-1 monocytic leukemia cells as detected by RNase protection assays. (Upper) THP-1 cells were treated with 0.1 μM TPA for the indicated times prior to isolation of total RNA. The arrowhead indicates the specific 293 nt protected fragment for PPARγ. (Lower Left) THP-1 cells were treated with 10 nM TPA and the PKC inhibitor as indicated at the same concentration used in A. (Lower Right) THP-1 cells were treated with 25 μg oxLDL and the indicated PKC or PKA inhibitors at the same concentrations. Equivalent amounts of RNA were verified by using an antisense β-actin probe (data not shown).
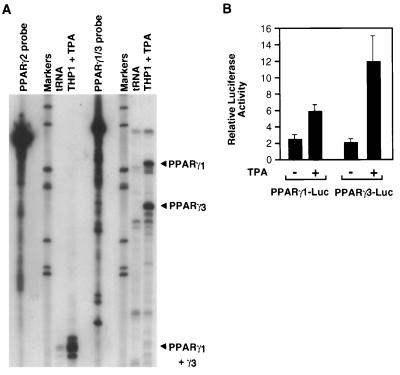
PPARγ has been demonstrated to be under the control of at least three different promoters that direct expression of PPARγ1, PPARγ2, and PPARγ3 mRNAs, which may serve to permit differential regulation of PPARγ expression in response to tissue-specific transcription factors and the local environment of hormones and other signaling molecules (L.F. and J.W., unpublished data). RNase protection experiments were therefore performed by using two different antisense probes that permitted specific detection of the different PPARγ mRNA species generated by each promoter. These experiments demonstrated that PPARγ1 and -γ3 isoforms were expressed in TPA-treated THP-1 cells, but not PPARγ2 (Fig. 4A). Consistent with these findings, both the PPARγ1 and -γ3 promoters were induced by TPA in the RAW 264.7 macrophage cell line (Fig. 4B). Similar results were obtained in TPA-treated THP-1 cells (data not shown).
Figure 4.
Regulation of PPARγ expression in THP-1 cells is mediated by the PPARγ1 and PPARγ3 promoters. (A) RNase protection analysis of PPARγ mRNA in TPA-treated THP-1 cells by using antisense mRNA probes that distinguish PPARγ1, PPARγ2, and PPARγ3 transcripts. The PPARγ2 probe is predicted to give protected fragments of 216 nt for PPARγ2 and a common fragment of 104 nt for PPARγ1 and PPARγ3. The PPARγ1 probe is predicted to give protected fragments of 218 nt for PPARγ1 and 174 nt for PPARγ3. (B) Expression of the PPARγ1 and PPARγ3 promoters in RAW 264.7 cells and regulation by 0.1 μM TPA. The PPARγ1-luciferase reporter gene contains 3.0 kb of 5′ flanking information, whereas the PPARγ3-luciferase reporter gene contains 800 bp of 5′ flanking information.
DISCUSSION
Recent studies have demonstrated that PPARγ is expressed in monocytes and macrophages and that natural and synthetic PPARγ ligands inhibit the production of a number of inflammatory mediators by these cells (20, 21). These observations have raised the possibility that PPARγ may play a physiologic role in modulating the magnitude and duration of inflammatory responses in which macrophages play prominent roles. The presence of large numbers of macrophages and a broad spectrum of inflammatory mediators in atherosclerotic lesions raised the question whether PPARγ is expressed in lesion macrophages, whether it is specific to particular stages of lesion development, and whether specific ligands may, in theory, activate this pathway and thus influence the inflammatory process. In the present studies, PPARγ was found to be highly expressed in foam cells in atherosclerotic lesions and in thioglycolate-elicited macrophages, whereas very little PPARγ was found in bone marrow progenitor cells, resting peritoneal macrophages, or undifferentiated myeloid cell lines. Several factors were found to induce PPARγ expression in differentiated macrophages, including M-CSF, GM-CSF, and oxLDL, suggesting that these factors may be of importance in regulating expression of PPARγ in macrophages in vivo.
PPARγ expression is directed by three distinct promoters, with the PPARγ1 and PPARγ2 promoters primarily used to drive PPARγ expression in adipose tissue. In the present studies, PPARγ expression in TPA-treated THP-1 cells was entirely accounted for by the PPARγ1 and PPARγ3 promoters. Consistent with this observation, both the PPARγ1 and PPARγ3 promoters were activated by TPA in transiently transfected RAW 264.7 cells. It will therefore be of considerable interest to determine whether other inducing factors, such as M-CSF, GM-CSF, and oxLDL, differentially regulate the PPARγ1, PPARγ2, and PPARγ3 promoters. OxLDL and TPA exerted additive effects on PPARγ expression. Furthermore, although TPA-dependent activation of PPARγ could be blocked by a PKC inhibitor, oxLDL activation of PPARγ expression could not, suggesting that PPARγ expression in the macrophage can be regulated by more than one pathway.
In vivo, oxLDL is believed to be generated within the artery wall as the consequence of reaction with pro-oxidant molecules generated by activated macrophages and other vascular cells. Extensive evidence has been provided for the occurrence of lipid peroxidation, and the presence of oxLDL in atherosclerotic lesions of humans and animal models (32, 33, 38–41). Notably, in early, macrophage-rich lesions, oxidation-specific epitopes were mostly observed within, or in close vicinity to, macrophages. The remarkable colocalization between oxidation-specific epitopes and PPARγ, in particular in the early lesions, suggests that oxLDL, or oxidant stress itself, may be an important regulatory factor of PPARγ expression in lesions.
The observation that PPARγ is highly expressed in macrophage foam cells of atherosclerotic lesions underscores the importance of determining the biological role of this transcription factor in the regulation of macrophage gene expression. This issue is especially relevant given the frequency of cardiovascular complications in subjects with type 2 diabetes mellitus, who now have the option to be treated with thiazolidinediones. Although current information would suggest a potential protective role for PPARγ, based on inhibition of inflammatory cytokines, gelatinase B, and the scavenger receptor A gene, the development of atherosclerosis is a complex phenomenon involving many gene products. Indirect evidence from human patients treated with troglitazone and studies of a balloon injury model of atherosclerosis in rats suggest protective effects of PPARγ ligands on lesion development (42, 43). Intervention studies in appropriate animal models and prospective clinical studies of patients treated with synthetic PPARγ ligands will be required to determine whether these substances positively or negatively influence the development of atherosclerosis or its complications.
Acknowledgments
We thank Florencia Casanada and Mercedes Silvestre for assistance with immunohistochemistry, Elizabeth Miller for preparation of oxLDL, Brian Smith for assistance with macrophage culture, and Tanya Schneiderman for assistance with manuscript preparation. J.L.W., W.P., and C.K.G. were supported by the National Institutes of Health (Specialized Center of Research on Molecular Medicine and Atherosclerosis Grants HL14197-25 and HL59694). C.K.G. is an Established Investigator of the American Heart Association. J.H. was supported by a postdoctoral training grant from the National Institutes of Health, J.L.W. was supported by a National Institutes of Health Medical Scientist Training Program grant to the University of California at San Diego. A.L. was supported by a National Institutes of Health Clinician Investigator Award. L.F. and J.A. acknowledge support from the Association pour la Recherche contre le Cancer, from the Institut National de la Santé et de la Recherche Médicale, from the Institut Pasteur de Lille, and from Ligand Pharmaceuticals. J.A. is a research director of the Centre National de la Recherche Scientifique.
ABBREVIATIONS
- PPARγ
peroxisome proliferator-activated receptor γ
- oxLDL
oxidized low density lipoprotein
- 15ΔPGJ2
15-deoxy-Δ12,14-prostaglandin J2
- M-CSF
macrophage colony-stimulating factor
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- IFN-γ
interferon γ
- TPA
12-O-tetradecanoylphorbol 13-acetate
- LPS
lipopolysaccharide
- PKC
protein kinase C
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemberger T, Desvergne B, Wahli W. Annu Rev Cell Dev Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- 3.Schoonjans K, Staels B, Auwerx J. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 4.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 6.Chawla A, Schwartz E J, Dimaculangan D D, Lazar M A. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 7.Fajas L, Auboeuff D, Raspé E, Schoonjans K, Lefebvre A-M, Saladin R, Najib J, Laville M, Fruchart J-C, Deeb S, et al. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 8.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 9.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 10.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 11.Reginato M J, Krakow S L, Bailey S T, Lazar M A. J Biol Chem. 1998;273:1855–1858. doi: 10.1074/jbc.273.4.1855. [DOI] [PubMed] [Google Scholar]
- 12.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Willson T M, Lenhard J M, Lehman J M. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee R, Davies P J A, Crombie D L, Bischoff E D, Cesario R M, Jow L, Hamann L G, Boehm M F, Mondon C E, Nadzan A M, et al. Nature (London) 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- 16.Nolan J J, Ludvik B, Beerdsen P, Joyce M, Olefsky J. N Engl J Med. 1994;331:1188–1193. doi: 10.1056/NEJM199411033311803. [DOI] [PubMed] [Google Scholar]
- 17.Willson T M, Cobb J E, Cowan D J, Wiethe R W, Correa I D, Prakash S R, Beck K D, Moore L B, Kliewer S A, Lehmann J M. J Med Chem. 1996;39:665–8. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 18.Saltiel A R, Olefsky J M. Diabetes. 1996;45:1661–1669. doi: 10.2337/diab.45.12.1661. [DOI] [PubMed] [Google Scholar]
- 19.Greene M E, Blumberg B, McBride O W, Yi H F, Kronquist K, Kwan K, Hsieh L, Greene G, Nimer S D. Gene Expression. 1995;4:281–299. [PMC free article] [PubMed] [Google Scholar]
- 20.Ricote M, Li A C, Willson T M, Kelly C J, Glass C K. Nature (London) 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 21.Jiang C, Ting A T, Seed B. Nature (London) 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 22.Berliner J A, Navab M, Fogelman A M, Frank J S, Demer L L, Edwards P A, Watson A D, Lusis A J. Circulation. 1995;91:2488–2496. doi: 10.1161/01.cir.91.9.2488. [DOI] [PubMed] [Google Scholar]
- 23.Ross R. Nature (London) 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 24.Steinberg D, Parthasarathy S, Carew T E, Khoo J C, Witztum J L. N Engl J Med. 1989;320:915–924. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- 25.Krieger M, Herz J. Annu Rev Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- 26.Lee R T, Libby P. Arterioscler Thromb Vasc Biol. 1997;17:1859–1867. doi: 10.1161/01.atv.17.10.1859. [DOI] [PubMed] [Google Scholar]
- 27.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai S, Bartelmez S, Sitnicka E, Collins S. Genes Dev. 1995;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F M. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 30.Zhu Y, Pless M, Inhorn R, Mathey-Prevot B, D’Andrea A D. Mol Cell Biol. 1996;16:4808–4817. doi: 10.1128/mcb.16.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H, Moulton K, Horvai A, Parik S, Glass C K. Mol Cell Biol. 1994;14:2129–2139. doi: 10.1128/mcb.14.3.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palinski W, Hörkkö S, Miller E, Steinbrecher U P, Powell H C, Curtiss L K, Witztum J L. J Clin Invest. 1996;98:800–814. doi: 10.1172/JCI118853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palinski W, Rosenfeld M E, Ylä-Herttuala S, Gurtner G C, Socher S A, Butler S, Parthasarathy S, Carew T E, Steinberg D, Witztum J L. Proc Natl Acad Sci USA. 1989;86:1372–1376. doi: 10.1073/pnas.86.4.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palinski W, Ylä-Herttuala S, Rosenfeld M E, Butler S, Socher S A, Parthasarathy S, Curtiss L, K, Witztum J L. Arteriosclerosis. 1990;10:325–335. doi: 10.1161/01.atv.10.3.325. [DOI] [PubMed] [Google Scholar]
- 35.Gown A M, Tsukada T, Ross R. Am J Pathol. 1986;125:191–207. [PMC free article] [PubMed] [Google Scholar]
- 36.Micklem K, Rigney E, Cordell J, Simmons D, Stross P, Turley H, Seed B, Mason D. Br J Haematol. 1989;73:6–11. doi: 10.1111/j.1365-2141.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 37.Tsukada T, Tippens D, Mar H, Gordon D, Ross R, Gown A M. Am J Pathol. 1986;126:51–60. [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenfeld M E, Palinski W, Ylä-Herttuala S, Butler S, Witztum J L. Arteriosclerosis. 1990;10:336–349. doi: 10.1161/01.atv.10.3.336. [DOI] [PubMed] [Google Scholar]
- 39.Ylä-Herttuala S, Palinski W, Rosenfeld M E, Parthasarathy S, Carew T E, Butler S, Witztum J L, Steinberg D. J Clin Invest. 1989;84:1086–1095. doi: 10.1172/JCI114271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palinski W, Ord V A, Plump A S, Breslow J L, Steinberg D, Witztum J L. Arterioscler Thromb. 1994;14:605–616. doi: 10.1161/01.atv.14.4.605. [DOI] [PubMed] [Google Scholar]
- 41.Hammer A, Kager G, Dohr G, Rabl H, Ghassempur I, Jürgens G. Arterioscler Thromb Vasc Biol. 1995;15:704–713. doi: 10.1161/01.atv.15.5.704. [DOI] [PubMed] [Google Scholar]
- 42.Minamikawa J, Yamauchi M, Inoue D, Koshiyama H. J Clin Endocrinol Metab. 1998;83:1041. doi: 10.1210/jcem.83.3.4668-1. [DOI] [PubMed] [Google Scholar]
- 43.Law R E, Meehan W P, Xi X-P, Graf K, Wuthrich D A, Coats W, Faxon D, Hsueh W A. J Clin Invest. 1996;98:1897–1905. doi: 10.1172/JCI118991. [DOI] [PMC free article] [PubMed] [Google Scholar]