Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is an RNA virus in the order Nidovirales, family Arteriviridae, genus Arterivirus. The virus induces a prolonged viremia, replicates in macrophages, and produces persistent infection. The purpose of this study was to determine if PRRSV could persist for 90 d or more in a large population of breeding-age gilts housed under environmental conditions typical of commercial swine production and to determine if experimentally infected gilts could shed virus to naïve sentinel gilts beyond 90 d postinfection. Using the intranasal route, we inoculated 120 PRRSV-naïve gilts, 4 mo of age, with 5 mL of cell culture fluid containing a total dose of 102.4 TCID50 of a field isolate (MN-30100) of PRRSV. The index gilts were organized into 3 groups (A, B, and C), 40 gilts per group. To assess the dynamics of the experimental infection, a monitor group of 30 index gilts was blood-tested on days 0, 3, 7, 14, 30, 60, 90, 120, 150, and 180 postinfection. PRRSV viremia was detected with the polymerase chain reaction (PCR) on days 3, 7, and 14 and by virus isolation (VI) on days 7 and 14. PRRSV antibodies were detected from day 14 by enzyme-linked immunosorbent assay (ELISA). To assess shedding, 30 PRRSV-naïve sentinel gilts were commingled with the index gilts on day 90 postinfection and tested by PCR, VI, and ELISA every 15 d until 180 d postinfection; all samples were negative. To assess persistence, 40 index and 10 sentinel gilts were slaughtered at 120 (group A), 150 (group B), or 180 (group C) d postinfection. Evidence of PRRSV was not detected by PCR or VI in any tissue samples from the 120 index gilts. These results indicate that persistence and shedding of PRRSV are of short duration in breeding-age gilts.
Introduction
Infection with porcine reproductive and respiratory syndrome virus (PRRSV) is a major problem in the swine industry (1). Estimated losses of $228 per inventoried sow per year, due to increased mortality rate, reduced growth rate, and elevated costs of medication and vaccination, have been reported (2). PRRSV is an RNA virus in the order Nidovirales, family Arteriviridae, genus Arterivirus (3). Other members of the Arterivirus group include lactate-dehydrogenase-elevating virus of mice, equine arteritis virus, and simian hemorrhagic fever virus (4). These viruses can induce a prolonged viremia in the presence of antibodies, replicate in macrophages, and produce persistent infection (5). Persistent infection is defined as “the continued presence of a virus in a host for extended periods of time post infection” (6,7). RNA viruses such as PRRSV do not revert to inactive states after infection but, rather, continue to replicate at some level within certain sites in the body (5,6,7). Experimental studies have recovered viable PRRSV from tonsil tissue of growing pigs 157 d postinfection, and PRRSV nucleic acid has been detected in boar semen 92 d postinfection (8,9).
In endemically infected populations, PRRSV may be transmitted in the breeding herd, resulting in recurrent episodes of PRRS-related reproductive disease, as well as infection of weaned pig populations secondary to vertical or horizontal transmission from sow to pig prior to weaning (10,11). Evidence of PRRSV transmission in endemically infected breeding herds has been reported (11,12). Within such populations, PRRSV-infected and naïve subpopulations of sows coexist, infected animals appear to cluster in small groups or exist as singletons, and naïve sows can produce PRRSV antibodies following exposure to virus (12,13).
However, few data are available regarding the duration of PRRSV persistence within a large experimental population of breeding-age swine housed under commercial conditions. Bierk and colleagues (14) recently investigated chronic PRRSV infection in an endemically infected field population. Diagnostic data from 60 adult breeding swine (45 sows and 15 boars) indicated that approximately 2% of the sampled population harbored PRRSV. No conclusions could be drawn regarding whether the PRRSV-positive animals were persistently infected, nor could the investigators determine the duration of persistence, owing to the inability to identify the exact time of infection of individual animals. The same group demonstrated persistent infection and shedding of PRRSV from experimentally infected sows to contact controls among non-pregnant sows from 49 to 86 d postinfection (15). Limitations of this study included the use of small groups of animals, the use of facilities that were not representative of commercial swine systems, and the inability to assess PRRSV persistence beyond 90 d postinfection. Therefore, the purpose of our study was to determine if PRRSV could persist in a population of breeding-age gilts housed under commercial conditions for 120 d or more and if experimentally infected gilts could shed virus beyond 90 d postinfection.
Materials and methods
Source of animals and housing
We obtained 120 PRRSV-naïve gilts, 4 mo of age, from a source known to be negative for PRRSV on the basis of 5 years of diagnostic data and the absence of clinical signs of PRRS in all phases of production (2,10,11). The gilts were housed at the research farm of the University of Minnesota Swine Disease Eradication Center, in a 10-pen, mechanically ventilated finishing building; the pens were 10 × 2.5 m in size and had partially slatted floors. The animals were placed 12 per pen and provided with 1 m2 of space. During the study, animals were cared for under the guidelines of the University of Minnesota Institutional Animal Care Committee policies.
Infection model
Upon arrival at the farm, all gilts were individually identified with numbered ear tags. On day 0, they were infected intranasally with 5 mL of cell culture fluid containing a total dose of 102.4 TCID50 of a field isolate (MN-30100) of PRRSV (14). To assess the PRRSV status of the population over the course of the study, a monitor group of 30 index gilts was organized by randomly selecting 3 animals from each pen. This sample size was sufficient to estimate prevalence when the true expected prevalence was ≤ 10% or ≥ 90% with ± 10% accuracy and 95% confidence. On day 0, ten 8-wk-old PRRSV-naïve gilts from the same source were housed in a separate facility 30 m from the experimental facility to serve as negative controls; thus, lateral introduction of PRRSV was monitored.
Assessment of persistence and shedding
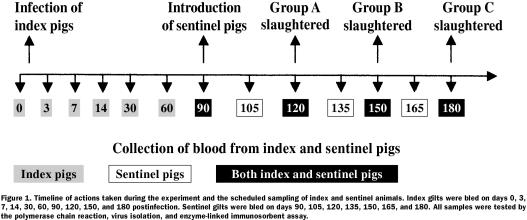
Following experimental infection, the 120 index gilts were organized into 3 groups (A, B, and C), 40 gilts per group. To determine whether PRRSV could persist within the experimentally infected gilts for 120 to 180 d postinfection, group A would be marketed at 120 d postinfection, group B at 150 d, and group C at 180 d; selected tissues would be collected at slaughter and tested for PRRSV. The sample size of 40 gilts per slaughter group was capable of detecting at least 1 PRRSV-infected gilt, assuming an estimated prevalence of 2%, with 95% confidence (14). To determine whether the experimentally infected population could shed PRRSV during the period of 90 to 180 d postinfection, 30 PRRSV-naïve, age-matched sentinel gilts were introduced at day 90 after infection of the index population. The sentinel gilts were individually tagged, mixed directly with the index gilts (3 sentinels per pen), and bled every 15 d to determine their PRRSV status. Figure 1 provides a timeline that summarizes the events during the study period.
Figure 1. Timeline of actions taken during the experiment and the scheduled sampling of index and sentinel animals. Index gilts were bled on days 0, 3, 7, 14, 30, 60, 90, 120, 150, and 180 postinfection. Sentinel gilts were bled on days 90, 105, 120, 135, 150, 165, and 180. All samples were tested by the polymerase chain reaction, virus isolation, and enzyme-linked immunosorbent assay.
Sampling methods and diagnostic testing
The PRRSV-infected monitor gilts were blood-tested on days 0, 3, 7, 14, 30, 60, 90, 120, 150, and 180 postinfection. All 30 sentinel gilts were blood-tested upon arrival at the facility (on day 90 after infection of the index animals) and then every 15 d until day 180 after infection of the index gilts, for a total of 7 samplings (on days 90, 105, 120, 135, 150, 165, and 180). The negative-control gilts were blood-tested on arrival at the farm and at the end of the study. Serum was tested for PRRSV nucleic acid by polymerase chain reaction (PCR) (TaqMan PCR; Perkin-Elmer Applied Biosystems, Foster City, California, USA), for viable PRRSV by virus isolation (VI), using MARC-145 continuous-cell lines and porcine alveolar macrophages (16,17,18), and for PRRSV antibodies by enzyme-linked immunosorbent assay (ELISA) (IDEXX ELISA; IDEXX Laboratories, Westbrook, Maine, USA).
Tissues collected at slaughter always included tonsil and lymph nodes from at least 2 sites from each gilt. Specifically, the sternal and superficial inguinal lymph nodes were required, because of the ease of accessing these sites at slaughter and because of data from a previous study that demonstrated frequent detection of PRRSV nucleic acid by PCR in these sites (15). Additional lymphoid tissue (tracheobronchial, medial iliac, and, or, lateral retropharyngeal) was collected when it could clearly be identified. Along with each group of 40 index gilts, 10 sentinel gilts were processed in a similar manner. Large pieces of tissue were collected during evisceration on the kill floor or as carcasses were chilling at 4°C. The samples were confirmed to be of lymphoid origin by microscopic examination and then were pooled by individual animal, transported on ice to the University of Minnesota Veterinary Diagnostic Laboratory, homogenized (so that all tissues would be evenly represented during testing and even small amounts of persisting PRRSV would not be missed), tested for PRRSV by PCR and VI, and evaluated for the presence of lesions suggestive of PRRSV infection (17,18,19). The open reading frame 5 (ORF 5) region of the nucleic acid of representative PRRSV isolates recovered from the index and sentinel gilts was sequenced to determine the degree of homology with the isolate used for the experimental infection (20). Serum from the negative controls was tested by ELISA; these gilts were not included in any of the 3 slaughter groups.
Results
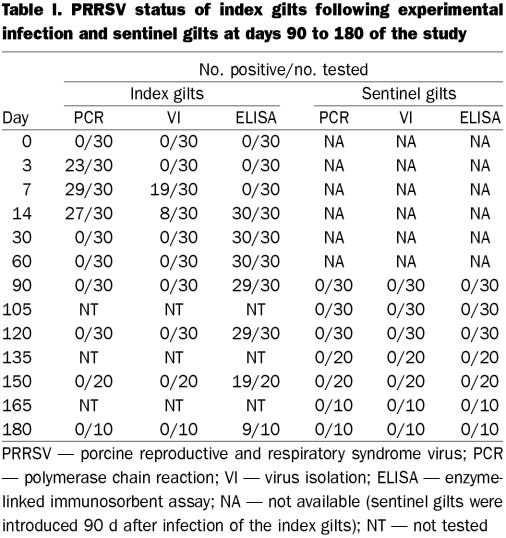
Table I summarizes the diagnostic and test data for the index and sentinel groups, respectively. All 30 index gilts in the monitor group were PRRSV negative on arrival, as verified by PCR, VI, and ELISA. Serial testing of the monitor gilts indicated successful experimental infection. On day 3 postinfection, 23 were PCR positive but none were VI positive. They were depressed, anorexic, and feverish (temperature 40 to 41.5°C) for approximately 48 to 72 h postinfection. No deaths occurred. ORF 5 molecular sequencing of a randomly selected PRRSV isolate recovered from an index pig indicated 100% homology with the isolate used for the experimental infection. On day 7 postinfection, 29 of the monitor gilts were PCR positive and 19 were VI positive; however, all were ELISA negative (sample-to-positive ratio < 0.4). On day 14 postinfection, all 30 monitor gilts were ELISA positive, but only 27 and 8 were PCR and VI positive, respectively. The numbers of ELISA-positive monitor gilts from day 30 to 180 postinfection were as follows: 30/30 (days 30 and 60), 29/30 (days 90 and 120), 19/20 (day 150) and 9/10 (day 180). All samples collected on days 30 to 180 were PCR and VI negative. All sentinel gilts were PRRSV negative by PCR, VI, and ELISA on arrival (day 90 after infection of the index population) and remained so throughout days 90 to 180 postinfection.
Table I.
At each slaughter date (120, 150, and 180 d postinfection), 50 pools of tissue (from 40 index and 10 sentinel animals) were collected, for a total of 150 tissue pools. The approximate ages of the gilts at slaughter were 240, 270, and 300 d for groups A, B, and C, respectively. Samples of tonsil and sternal and superficial inguinal lymph nodes were collected from all index and sentinel gilts. In addition, samples of tracheobronchial, medial iliac, and lateral retropharyngeal lymph nodes were collected from 66 of the 120 index gilts; the total number of lymph node sites was 5 for 10 gilts, 4 for 23 gilts, and 3 for 33 gilts. Microscopic evidence of PRRSV infection was not detected, and all tissue pools were negative by PCR and VI. The negative-control gilts remained ELISA negative throughout the study.
Discussion
The primary objective of this study was to determine if PRRSV could persist in a population of breeding-age gilts for 120 to 180 d postinfection and whether infected gilts could shed the virus to naïve sentinels beyond 90 d postinfection. Although diagnostic data from the monitor gilts indicated successful experimental infection, with viremia detected for up to 14 d postinfection, PRRSV was not detected in any of the tissue pools from the index gilts slaughtered at 120, 150, or 180 d postinfection. Serum samples collected on days 3, 7, and 14 were positive for most of the index gilts by PCR (23, 29, and 27, respectively, out of 30); however, fewer of the samples were VI positive (0, 19, and 8, respectively, out of 30), which indicates that PCR is more sensitive than VI. The sensitivity of the TaqMan PCR, used in this study, has been reported to be 0.1 TCID50/mL (17).
Shedding of PRRSV from index animals to commingled sentinels was not observed during the period of 90 to 180 d postinfection. Therefore, under the conditions of our study, PRRSV persistence and shedding within a large population of experimentally infected breeding-age gilts appear to be of short duration (less than 120 and 90 d, respectively). Different results have recently been reported for PRRSV persistence in experimentally infected nursery pigs: Horter and associates (21) indicated that PRRSV was detectable in 100% of 60 experimentally infected 3-wk-old pigs at 63 d postinfection and in 90% of the pigs 105 d postinfection. One possible explanation for the difference between the studies may be that the duration of PRRSV persistence is related to the age of the host. It is well documented that the viremic period in adult swine is shorter than that in younger animals (22,23). In our study, the index gilts were infected at 4 mo of age and slaughtered at 240 to 300 d of age. Therefore, one could speculate that the immune response of the adult is different following infection with PRRSV.
Another reason may be the difference in the diagnostic methods used in the 2 studies. Horter and associates used a sequential testing scheme that involved a series of tests, including PCR, VI, and swine bioassay (21,24). Although a large percentage (47% to 81%) of the samples tested were positive by PCR or VI, samples negative in both tests were tested by swine bioassay. We debated whether to include swine bioassay; however, in contrast to the study of Horter and associates, 100% of the tissue pools from the 120 index animals in our study were both PCR and VI negative. We recently tested 869 tissue samples from breeding-age female swine by PCR, VI, and swine bioassay: the samples that were PCR and VI negative (n = 868) were also negative by swine bioassay, and the single sample that was PCR or VI positive was also positive by swine bioassay (25). Therefore, because of the results of this study, the results of previous studies evaluating PRRSV persistence in breeding-age female swine (15), and the potential costs of testing 120 homogenates by swine bioassay, we did not include this test. Finally, the 2 studies used different isolates of PRRSV. Horter and associates selected VR-2332, a cell-culture-adapted strain previously recovered from pulmonary tissue from an infected nursery-aged pig. In contrast, we used a field strain (MN-30100), isolated from lymphoid tissue of an asymptomatic, seropositive, aviremic sow (14). Our study isolate was capable of inducing fetal death following the inoculation of 2 PRRSV-naïve pregnant sows at 95 d of gestation. The sows were necropsied 14 d later, and samples of maternal and fetal tissue and blood were collected. One sow had 10 fresh, 6 partially autolyzed, and 2 mummified fetuses; the other sow had 6 fresh and viable fetuses devoid of microscopic lesions of PRRSV infection. Fetal samples from both litters were positive for PRRSV (14).
This study brought forth new information on the dynamics of PRRSV persistence in a large population of experimentally infected breeding-age gilts. At the time of writing, this is the largest PRRSV persistence study ever conducted in this age group, housed under these conditions. Other strengths of this study include the use of a field isolate previously recovered from an aviremic sow that harbored PRRSV in lymphoid tissues, the use of commercial facilities to mimic field conditions, and the introduction of PRRSV-naïve sentinels directly commingled with index animals, which maximized animal-to-animal interaction. However, this study had several limitations. Although we did fulfill our sample selection criteria, collecting samples of tonsil and sternal and superficial inguinal lymph nodes from all 120 index gilts, we were unable to collect a broader spectrum of samples, because of the inherent difficulty of collecting samples at a slaughterhouse. Therefore, we cannot rule out the possibility that certain gilts harbored PRRSV in lymphoid sites that were not sampled. Definitive visual confirmation of the supplementary sites sampled was difficult in some gilts. These lymph nodes were frequently small and often covered with adipose tissue. Furthermore, inadequate lighting in the refrigeration room, time limitations on the kill floor, and inverted suspension of the carcasses made collection from all sites in all gilts impossible. Visual confirmation of the supplementary sites was possible in 66 of the 120 index gilts, and from these gilts we exceeded our minimum sampling criteria in an attempt to strengthen our conclusions. However, although we cannot ignore the fact that our detection of persistently infected gilts might have been improved with better sampling methods, the fact that the sentinel gilts remained negative throughout the study indicated either the absence of carriers or the inability of these gilts to shed PRRSV.
We did not attempt to document PRRSV persistence before 120 d after infection. From our previous work (15), we hypothesized that PRRSV would persist for up to 86 d, particularly when we used the same isolate, concentration, route of infection, and genetic source of pigs. Therefore, the gilts in this study were not slaughtered until 120 d postinfection. In retrospect, the use of a positive control to document PRRSV persistence from days 60 to 90 postinfection would have enhanced the study design. A proposed strength of our study was the size of the test group; however, breeding-herd inventories in the commercial swine industry are much larger, frequently 1000 to 5000 sows. Therefore, although we based our group size on published data (14), if future studies are conducted in large breeding herds, larger samples may be required to increase the sensitivity of detecting carriers should they exist at levels of less than 2%. Finally, owing to the sheer size, time requirements, and expense of the study, we were able to conduct only 1 replicate.
To conclude, our results indicate that PRRSV transmission may be limited to less than 90 d postinfection and persistence to less than 120 d postinfection in a large population of breeding-age gilts. These results demonstrate the value of eliminating PRRSV-naïve subpopulations, enhancing viral clearance, and preventing the shedding of PRRSV through the exposure of an isolated, static population of non-pregnant breeding-age gilts to a defined concentration of PRRSV. Future studies should focus on repeating the study using larger samples and tissue sets, as well as assessing the key immunologic responses that bring about elimination of virus from the persistently infected gilts between days 90 and 120 postinfection. Answers to these issues could prove to be very helpful in controlling PRRS throughout the global swine industry.
Footnotes
Acknowledgments
We thank the National Pork Board, the Minnesota Pork Producers Association, the Pig Improvement Company, and Genetiporc for financial support and animal resources during the study.
Address correspondence and reprint requests to Dr. S.A. Dee, tel: 612-625-8781, fax: 612-625-1210, e-mail: deexx004@tc.umn.edu
Received December 18, 2001. Accepted March 21, 2002.
References
- 1.Loula TJ. Mystery pig disease. Agri Prac 1991;12:23–34.
- 2.Dee SA, Joo HS, Polson DD, Marsh WE. Evaluation of the effects of nursery depopulation on the profitability of 34 pig farms. Vet Rec 1997;140:498–500. [DOI] [PubMed]
- 3.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 1997;142:629–633. [PubMed]
- 4.Plagemann PGW, Moennig V. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv Virus Res 1992;41:99–192. [DOI] [PMC free article] [PubMed]
- 5.Collins JE, Benfield DA, Christianson WT, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 1992;4:117–126. [DOI] [PubMed]
- 6.Ahmed R, Morrison LA, Knipe DM. Persistence of viruses. In: Fields BN, Knipe DM, Howley PM, eds. Field's Virology. Philadelphia: Lippincott-Raven, 1996:219–249.
- 7.Ahmed R, Morrison LA, Knipe DM. Viral persistence. In: Nathanson N, Ahmed R, Gonzalez-Scarano F, et al, eds. Viral Pathogenesis. Philadelphia: Lippincott-Raven, 1997: 181–206.
- 8.Wills RW, Zimmerman JJ, Yoon KJ, et al. Porcine reproductive and respiratory syndrome virus: a persistent infection. Vet Microbiol 1997;55:231–240. [DOI] [PubMed]
- 9.Christopher-Hennings J, Nelson EA, Hines RJ, et al. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J Vet Diagn Invest 1995;7: 456–464. [DOI] [PubMed]
- 10.Dee SA, Joo HS. Clinical investigation of recurrent reproductive failure with PRRS virus in a swineherd. J Am Vet Med Assoc 1994;204:1017–1018. [PubMed]
- 11.Dee SA, Philips RE. Use of polymerase chain reaction to detect vertical transmission of PRRS virus in piglets from gilt litters. Swine Health Prod 1999;7:237–239.
- 12.Dee SA, Joo HS, Henry S, et al. Detecting subpopulations after porcine reproductive and respiratory syndrome virus infection in large breeding herds using multiple serologic tests. Swine Health Prod 1996;4:181–184.
- 13.Dee SA, Molitor TW, Rossow KD. Epidemiological and diagnostic observations following elimination of PRRS virus from a breeding herd of pigs by the test and removal protocol. Vet Rec 2000;146:211–213. [DOI] [PubMed]
- 14.Bierk MD, Dee SA, Rossow KD, et al. Diagnostic investigation of chronic porcine reproductive and respiratory syndrome virus in a breeding herd of pigs. Vet Rec 2001;148:687–690. [DOI] [PubMed]
- 15.Bierk MD, Dee SA, Rossow KD, Collins JE, Otake S, Molitor TW. Transmission of PRRS virus from persistently infected sows to contact controls. Can J Vet Res 2001;65:261–266. [PMC free article] [PubMed]
- 16.Snyder ML, Mermer B, Anderson PR, Wensvoort G, Hill HT. Evaluative data for an immunodiagnostic ELISA for PRRS. Proc 2nd Int Symp PRRS 1995:15.
- 17.Molitor TW, Tune KA, Shin J, Collins J, Kapur V. Applications of TaqManTM PCR in the detection of PRRS virus. Proc Allen D. Leman Swine Conf 1997:173–175.
- 18.Bautista EM, Goyal S, Yoon IJ, Joo HS, Collins J. Comparison of porcine alveolar macrophages and CL 2621 for the detection of porcine reproductive and respiratory syndrome virus and anti-PRRS antibody. J Vet Diagn Invest 1993;5:163–165. [DOI] [PubMed]
- 19.Rossow KD, Morrison RB, Goyal SM, Singh GS, Collins JE. Lymph node lesions in neonatal pigs congenitally exposed to porcine reproductive and respiratory syndrome virus. J Vet Diagn Invest 1994;6:368–371. [DOI] [PubMed]
- 20.Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Virol 1995;140:1451–1460. [DOI] [PMC free article] [PubMed]
- 21.Horter D, Pogranichnyy R, Chang C-C, Yoon K-J, Zimmerman J. Persistence of porcine reproductive and respiratory syndrome virus in pigs. Proc Allen D. Leman Swine Conf 2001:27. [DOI] [PubMed]
- 22.Yoon IJ, Joo HS, Christianson WT, Morrison RB, Dial GD. Persistent and contact infection in nursery pigs experimentally infected with porcine reproductive and respiratory syndrome virus. Swine Health Prod 1993;1:5–8.
- 23.Christianson WT, Choi CS, Collins JE, et al. Pathogenesis of PRRS virus infection in mid-gestation sows and fetuses. Can J Vet Res 1993;57:262–268. [PMC free article] [PubMed]
- 24.Swenson SL, Hill HT, Zimmerman JJ. Excretion of porcine reproductive and respiratory syndrome virus after experimentally induced infection in boars. J Am Vet Med Assoc 1994;204: 1943–1948. [PubMed]
- 25.Dee SA, Torremorell M, Rossow K, Otake S, Faaberg K. Identification of genetically diverse sequences of PRRSV in a swineherd. Can J Vet Res 2001;65:254–260. [PMC free article] [PubMed]