Abstract
The entire genomes of 7 isolates of porcine circovirus (PCV) from pigs with congenital tremors (CT), type A2, or postweaning multisystemic wasting syndrome (PMWS) were cloned and sequenced. One isolate (CT-PCV-P7) originated from the late 1960s from a neonatal pig with CT, type A2. Two recent PCV isolates (CT-PCV-P5, CT-PCV-P6) were from 2 affected neonatal pigs, from different farms, with unrelated outbreaks of CT; type A2. Four isolates (PMWS-PCV-P1, PMWS-PCV-P2, PMWS-PCV-P3, PMWS-PCV-P4) originated from pigs with PMWS from 4 different farms. A comparative analysis of these PMWS and PCV isolates demonstrated 99% sequence identity with each other, and over 96% sequence identity with previously sequenced PCV2 isolates. The CT-PCV-P5 and CT-PCV-P6 isolates, however, shared 99% of the same identity with each other, and interestingly also with PMWS PCV isolates. There were no consistent genomic differences between PMWS and recent CT isolates. The CT-PCV-P7 showed 98% identity similarity to PK-15-derived PCV1 and demonstrated only 72% identity similarity to either CT-PCV-P5 or CT-PCV-P6. Phylogenetic analysis confirmed that the old isolate (CT-PCV-P7), and the new isolates (CT-PCV-P5, CT-PCV-P6, PMWS-PCV-P1, PMWS-PCV-P2, PMWS-PCV-P3, PMWS-PCV-P4) were correctly classified as PCV1 and PCV2, respectively.
Introduction
Circoviral genome consists of a single copy of circular single-stranded ambisense DNA genome (1). The size of the genome varies between 1.7 and 2.3 kb. Circoviruses are non-enveloped and have icosahedral symmetry. They have been isolated from chickens (chicken anemia virus) (2), pigs (porcine circovirus) (3), psittacines (psittacine beak and feather disease virus) (4), and pigeons (pigeon circovirus) (5). Porcine circovirus (PCV) type 1 (PCV1) was initially discovered as a noncytopathic contaminant of PK-15, a continuous porcine kidney cell line (6), and later characterized as a small icosahedral DNA virus with a circular genome (3). Porcine circovirus type 1 has been characterized by using an electron microscope (7), and its genome has been sequenced (8). The PK-15 PCV1 has never been associated with naturally occurring disease and the experimental inoculation of pigs did not result in clinical disease (9,10,11).
Phylogenetic analysis of PCV1, avian circoviruses, plant geminiviruses, and nanoviruses (previously known as plant circoviruses) classified PCV1 as most closely related to psittacine beak and feather disease virus. Both PCV1 and psittacine circovirus share features with, and were intermediate between the 2 plant viral groups (12). Additional analyses suggest that a predecessor to PCV1 and psittacine circovirus originated from a plant nanovirus that infected a vertebrate host and recombined with a vertebrate-infecting RNA virus, most likely a calicivirus (13).
Postweaning multisystemic wasting syndrome (PMWS) is clinically characterized by progressive weight loss, dyspnea, tachypnea, and icterus in postweaned pigs, and has been associated with infection by PCV2 (14,15,16). The complete genomic sequences of a number of PCV2 isolates associated with PMWS are available (17,18,19,20). Isolates of PCV2 differ from PCV1 antigenically and genetically (17,20,21). Isolates of PCV that are genetically similar to PK-15 cell PCV are referred to as “PK-15 PCV” (17) or “PCV1” (8). Also, isolates similar to those initially obtained from pigs with PMWS are referred to as “PMWS PCV” (17) or “PCV2” (8).
Congenital tremors (CT) in pigs are associated with demyelination of the brain and spinal cord. The most common form of CT in North America is transmissible and classified as type A2 (22). This form is characterized by clonic contractions of skeletal muscles of varying severity that decrease with time and usually resolve by 4 wk of age. Recently, using indirect fluorescent assay (IFA) in-situ hybridization and polymerase chain reaction (PCR), we demonstrated the association between PCV2 in neonatal pigs and naturally occurring CT type A2 from 4 farms in the Midwestern U.S. (23). Abortions associated with PCV2 have been demonstrated in experimentally (24), and naturally (25), infected sows. Intrauterine inoculation of fetuses with PCV2 resulted in virus replication in the fetuses (26). These findings implicate PCV2 as a pathogen in syndromes other than PMWS and suggest that PCV2 can cross the placenta and congenitally infect pigs.
Although complete genomic sequences of a number of PMWS-associated PCV2 are available, genetic analysis of PCV isolates associated with CT have not yet been reported. We sequenced and analyzed genomes of a PCV1 isolate associated with CT in the late 1960s, 2 PCV2 strains associated with CT in the late 1990s, and 4 PCV2 strains from different geographical regions of Indiana, USA, that were obtained from pigs showing signs of PMWS. The purpose of this study was to compare genome sequences of PCV1 and PCV2 associated with CT, with those of PCV2 involved with PMWS.
Materials and methods
Tissue samples and a cell line infected with PCV
Pigs infected with PCV2 were initially identified by signs of either PMWS or CT, followed by microscopic examination of tissue sections. Presence of PCV2 in tissue was further confirmed by in-situ hybridization using a PCV-specific oligonucleotide probe. The IFA was done using antiserum against PCV2 and PCR, using primers specific to PCV2. Four PCV2 isolates collected from pigs showing signs of PMWS were named PMWS-PCV-P1, PMWS-PCV-P2, PMWS-PCV-P3, and PMWS-PCV-P4. Two PCV2 isolates collected from pigs showing signs of CT were named CT-PCV-P5 and CT-PCV-P6. Pigs with CT did not have any gross or microscopic lesions, and they tested negative for porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus and other cytopathic viruses.
A pregnant sow was experimentally inoculated with the cell culture supernatant from CT-pig kidney cells (27). A cell line, porcine central nervous system (PCNS), was subsequently derived from the brain of a piglet showing signs of CT. For this study, PCNS cells (stored in the early 1970s) were revived in Eagle's minimum essential medium (EMEM) (Life Technologies, Incorporated) containing 10% reconstituted fetal bovine serum (FetalClone III; HyClone Incorporated, Logan, Utah, USA). Cells were harvested and tested for PCV by in-situ hybridization using a PCV-specific oligonucleotide probe, electron microscopy (EM), and by PCR using PCV1-specific primers. This PCV1 isolate was named “CT-PCV-P7.”
DNA isolation and PCR
The PCNS cells grown in EMEM were harvested when cells started floating in the medium. The cell pellet was lysed by sodium dodecyl sulfate (SDS) pronase (500 μg/mL pronase in 10 mM Tris, pH7.4, 10 mM EDTA, and 0.5% SDS) and incubated at 37°C overnight. The total cellular DNA was isolated by phenol extraction followed by ethanol precipitation.
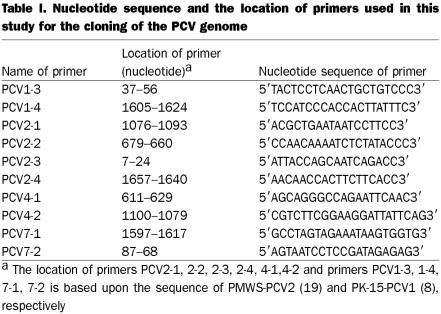
Lymph nodes for PMWS-PCV-P1, -P2, -P3, -P4, and CT-PCV-P6, as well as liver for CT-PCV-P5, were homogenized in EMEM using a tissumizer (Fisher Scientific, Ottawa, Ontario). This was followed by sonication using a sonicator (Misonix Incorporated, Farmingdale, New York, USA). Tissue homogenates were incubated with equal volumes of SDS-pronase (1 mg/mL pronase in 20 mM Tris, pH7.4, 20 mM EDTA, and 1% SDS) at 37°C overnight. Total cellular DNA was obtained by phenol extraction and ethanol precipitation. The DNA was used as a template for polymerase chain reaction (PCR) using Vent DNA polymerase (New England BioLab, Mississauga, Ontario) with 2 pairs of primers to amplify the entire PCV genome. We were not able to amplify the entire genomes of all virus isolates with the same sets of primers, therefore, we used PCV2-1/PCV2-2 and PCV2-3/PCV2-4 sets of primers for P1, P2, P3, P5, and P6 virus strains; PCV2-1/PCV2-2 and PCV4-1/PCV4-2 for P4 virus strain; and PCV1-3/PCV1-4 and PCV7-1/PCV7-2 for P7 virus strain (Table I). The PCR products were analyzed in 1% agarose gel and visualized with an ultra-violet (UV) transilluminator. Proper precautions were taken during PCV amplification and cloning to avoid the possibility of cross contamination.
Table I.
Transfection with complete PCV2 clones and detection of PCV2
Various PCR products were initially cloned into the SmaI site of pUC18 by blunt-end ligation. Plasmids containing PCV2 genomes from nucleotide (nt) 1067–1768 and 1–1657 (pPCV-P1, -P2, and -P3) were constructed and digested with SacII (at 1474 nt) to yield a linear-form of full PCV genomes. Semi-confluent monolayers of PCV-free PK-15 cells in 6-well plates were transfected with 1 μg of ligated PCV2 genome using the Lipofectin-mediated transfection protocol (Life Technologies Incorporated) and then the cells were passaged 3 times. After the third passage, cells were harvested, cytospined, and fixed with acetone. Polyclonal antibody against PMWS-PCV2 raised in a rabbit (purchased from Morozov and Paul, Iowa State University, Ames, Iowa, USA) was used for IFA. For EM, water was added to cell pellets and the cell contents were centrifuged at 10 000 rotations per minute (RPM) for 5 min. Supernatants were collected and centrifuged at 20 000 RPM for 40 min. The pellet was resuspended in water containing 3% phosphotungstic acid and 1% bovine serum albumin. Samples were nebulized onto the carbon-coated grid and examined with an electron microscope (Philips 201; Philips Electronics, Markham, Ontario).
DNA sequencing and sequence analysis
Plasmids containing PCV DNA were sequenced using universal forward and reverse primers. Subsequently, both strands of DNA were sequenced by primer walking using an Applied Biosystems 373A automated sequencer. The entire genomes of 7 PCV isolates (PMWS-PCV-P1; -P2; -P3; -P4; and CT-PCV-P5, -P6, and -P7) were analyzed using the GCG sequence analysis software (Wisconsin package).
Phylogenetic calculations
The sequence alignments were gained by the ClustalW program. The phylogenetic calculations were performed by the PHYLIP program package version 3.572c (28). For the parsimony analysis, the programs Protpars or DNApars were used. For the distance analysis, the Protdist (Dayhoff's PAM 001 matrix) or DNAdist (Kimura 2-parameter) followed by Fitch (with global rearrangements) were applied. During the bootstrap analysis, the above calculations were preceded by the Seqboot (100 datasets) and followed by the Consense program to get the consensus tree. Finally, the results were visualized by the TreeView program (29). A more detailed description of the applied methods has been published elsewhere (30).
Results
Generation of infectious PCV2 clones
To test whether the cloned PCV2 genomes could produce infectious virions, the religated or unligated form of SacII-digested pPCV-P1, -P2, and -P3 were used to transfect PCV-free PK-15 cells. Cells were harvested after third passage and analyzed for the presence of PCV2 antigen by IFA. A number of cells transfected with SacII-digested religated PCV2 DNA were positive, whereas cells transfected with SacII-digested unligated PCV2 DNA were negative for PCV2 antigen by IFA (data not shown). To determine whether transfection with PCV2 DNA resulted in the production of PCV2 virion, PK-15 cells transfected with PCV2 DNA were analyzed by EM. Small spherical viruses, approximately 17 nm in diameter, were observed in cells transfected with SacII-digested religated PCV2 DNA (data not shown). These results confirmed that the full-length PCV2 genomes amplified by PCR were infectious, suggesting that PCR amplification using Vent polymerase (having proof-reading ability) did not introduce any lethal mutation in PCV2 genomes. Therefore, we used Vent polymerase for PCR amplification of complete genomes of other PCV field strains.
Sequence comparison
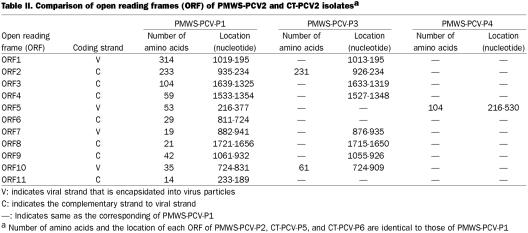
We sequenced the entire genomes of 4 PCV2 isolates associated with PMWS (PMWS-PCV-P1, -P2, -P3, and -P4), 2 PCV2 isolates associated with CT in the late 1990s (CT-PCV-P5 and -P6), and 1 PCV1 isolate associated with CT in the late 1960s (CT-PCV-P7). Sequence information for all 7 isolates has been submitted to Gene Bank. The accession numbers are as follows: PMWS-PCV-P1, #AY09945; PMWS-PCV-P2, #AY099496; PMWS-PCV-P3, #AY099497; PMWS-PCV-P4, #AY099498; CT-PCV-P5, #AY099499; CT-PCV-P6, #AY099500; and CT-PCV-P7, #AY099501. Sequences of these isolates were compared with that of the previously described PCV2 isolates (17) and PCV1 isolate (8). Genomes of PMWS-PCV-P1, -P2, and -P4 were 1768 nt long, whereas PMWS-PCV-P3 was 6 nt shorter than the rest of PMWS-PCV2 isolates. This is due to a 6-nt deletion between 820 to 825 nt. All 4 PMWS-PCV2 isolates had an overall 99% nt sequence identity with each other. The coding strand, number of amino acids, and the location of each open reading frame (ORF) in the genomes of PMWS-PCV2 are listed (Table II). The amino acid sequence of ORF1 was highly homologous (approximately 99% homology at the amino acid level) among all PMWS-PCV2 isolates. Observed changes in amino acid residues in various ORFs of PMWS-PCV2 isolates are listed (Table III). The ORF2 had more amino acid changes than ORF1 among PMWS-PCV2, but still had an approximately 97% homology. Open reading frames 3, 4, 7, and 8 were identical among PMWS-PCV2 isolates, and there were only few changes in the rest of ORFs (Table III). The ORF5 of PMWS-PCV-P4 and ORF10 of PMWS-PCV-P3 were 54 and 26 amino acids longer respectively than their counterpart in the rest of our PMWS-PCV2 isolates.
Table II.
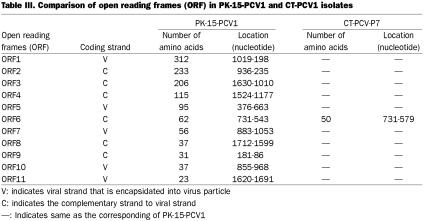
Table III.
Two CT-PCV2 isolates (CT-PCV-P5 and -P6), which were isolated in the late 1990s, were 1768 nt long. These CT-PCV2 isolates had approximately 99% nt sequence identity. Interestingly, CT-PCV2 isolates also demonstrated approximately 99% nt sequence identity with PMWS-PCV2 isolates. The genomes of PMWS-PCV-P3 and CT-PCV-P5 were identical. Both PMWS-PCV2 and CT-PCV2 genomes encode 11 potential ORFs. The amino acid changes in the various ORFs of CT-PCV2 compared to those of PMWS-PCV-P1 are listed (Table III).
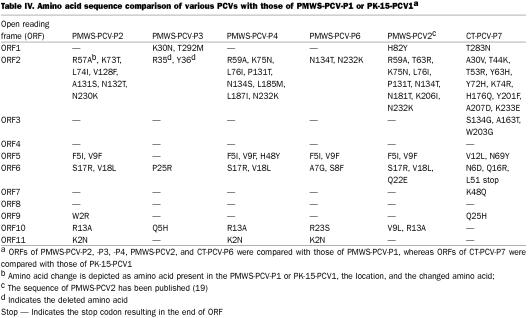
The genome of CT-PCV-P7 was 1759 nt long and has the potential to code for 11 ORFs (Table IV). The CT-PCV-P7 genome only had approximately 72% nt sequence identity with PMWS-PCV2 or CT-PCV2, but shared a surprising approximately 98% nt sequence identity with PCV1. Amino acid sequences of all ORFs of CT-PCV-P7 were also highly homologous to those of PCV1. Amino acid changes in various ORFs of CT-PCV-P7 compared to their counterpart in PCV1 are listed (Table IV).
Table IV.
Meehan et al (8) observed the presence of a nonanucleotide sequence at the apex of the stem-loop structure of PCV1, similar to that described of nanoviruses and geminiviruses of plants. All our PCV isolates had conserved stem-loop structure and nonanucleotide (A/TAGTATTAC), representing the origin of rolling-circle DNA replication (31,32). The potential glycosylation sites (N-X-T or N-X-S, where ‘X’ is any amino acid), which were previously reported by Hamel et al, (17), were conserved in all our PCV isolates except ORF6 of CT-PCV-P7 where the ‘N’ amino acid residue at number 6 was replaced with ‘D.’
Phylogenetic analysis
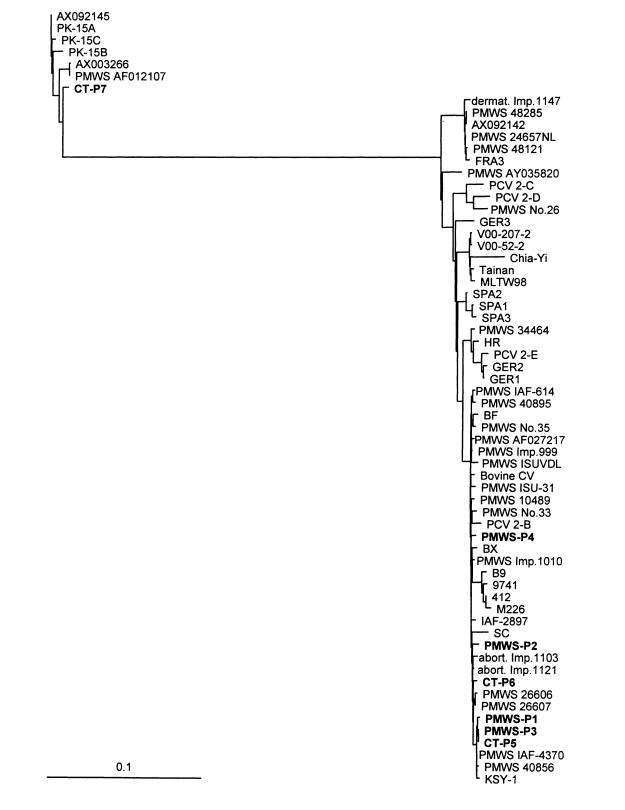
The main objective of phylogenetic analysis was to classify PCV isolates of this study with some of the already known PCV1 and PCV2 isolates. When either the deduced amino acid sequences of individual proteins, such as the replication-associated protein (ORF1/replicase/rep/P35.8 protein) and the capsid protein (ORF2/P27.9/P27.8 protein), or the nt sequence of the full genome, were used for phylogenetic analysis, both the distance matrix and the parsimony analyses yielded 2 distinct clusters of fairly similar topology. Since the differences between the strains were moderate, and the distance matrix analysis seemed to yield more consistent data (30), we chose to present our findings using the distance matrix analysis of full genomes (Figure 1) (the full alignment and the other trees are available upon request).
Figure 1. Distance matrix analysis of the full genome sequences of porcine and bovine circovirus isolates. Gaps and neighboring regions not being obviously homologous were removed from the alignment; the final length of the edited alignment was 1718 nt. Branch length is proportional to the phylogenetic distance of the isolates. The bar represents a 10% difference between the 2 sequences. Unrooted tree. The names of the sequences described in this paper are in bold. Strains (other than ours) and their GenBank accession numbers are as follows. The applied names show also the disease if known. If strain name is not available, the accession number is shown or the described restriction enzyme fragmentation type. PK-15 cell line derived strains: PK-15A (U49186), PK-15B (Y09921), PK-15C (AF071879); PMWS strains: AF012107, Imp.1011 48285 (AF055394), 24657NL (AF201897), Imp.1011 48121 (AF055393), AY035820, No.26 (AB072302), 34464 (AF264043), IAF-614 (AF118097), 40895 (AF264042), No.35 (AB072303), AF027217, Imp.999 (AF055391), ISUVDL 98-15237 (AF147751), ISU-31 (AJ223185), 10489 (AF264040), No.33 (AB072301), Imp.1010-Stoon (AF055392), 26606 (AF264038), 26607 (AF264039), IAF-4370 (AF118097), 40856 (AF264041); strains from abortions (abort.): Imp.1103 (AJ293868), Imp.1121 (AJ293867), porcine dermatitis and nephropathy (dermat.): Imp.1147 (AJ293869); strains without described disease: AX092145, AX003266, AX092142, FRA3 (AF201311), GER3 (AF201307), V00-207-2 (AF305533), V00-52-2 (AF305532), Chia-Yi (AF364094), Tainan (AF166528), MLTW98 (AF154679), SPA2 (AF201309), SPA1 (AF201308), SPA3 (AF201310), HR (AF381176), GER2 (AF201306), GER1 (AF201305), BF (AF381175), BX (AF381177), B9 (AF086834), 9741 (AF086835), 412 (AF085695), M226 (AF086836), IAF-2897 (AF408635), SC (AF465211), KSY-1 (AF454546); strains with type names referring to restriction enzyme fragmentation pattern: PCV 2-C (AF109398), PCV 2-D (AF117753), PCV 2-E (AF109399), PCV 2-B (AF112862); bovine circovirus: Bovine CV (AF109397).
The most evident result of the distance matrix analysis of various porcine and bovine circovirus genomes, was the clear separation of isolates into 2 clusters (Figure 1). No intermediate genotypes were found, though the number of examined genomes (including our 7 new sequences) reached 64, and the origin of the isolates covered geographically distant regions.
The smaller cluster (type 1), contained isolates from the different lineages of the PK-15 cell line and 2 circovirus strains (PMWS accession number AF012107 and our CT-PCV-P7), isolated from different pathological entities (PMWS and CT). The other fairly large cluster (type 2), contained the remaining 57 isolates, including 24 from pigs with PMWS; 2 strains from abortions; 1 strain from porcine dermatitis and nephropathy syndrome; our 2 CT-PCV2 isolates (CT-PCV-P5 & -P6); and a bovine isolate (AF109397).
Discussion
The goal of this study was to determine genetic variability in PCV associated with PMWS or CT. Our PMWS-PCV2 isolates yielded, approximately, a 99% nt sequence identity with each other and also, approximately, a 95% nt identity with PMWS-associated PCV2 strains isolated in the United Kingdom, Canada, France, Germany, Spain, Netherlands, China, South Korea, Japan, and the United States (17,18,19,20,21,33) indicating that various PMWS-PCV2 isolates are highly homologous regardless of their place of origin.
Although 2 CT-PCV2 isolates and 1 CT-PCV1 isolate originated from neonatal pigs with CT type A2, they shared only 72% nt sequence identity. The genomes of the 2 CT-PCV2 isolates were similar to recent isolates of PMWS-PCV2, whereas the CT-PCV1 isolate was very close to PCV1 variants. Based on phylogenetic calculations, different PCV isolates can be divided into 2 groups. The PCV1 variants, our CT-PCV-P7, and a single uncharacterized PMWS isolate (AF012107) comprise PCV1. The remaining 20 recent PMWS-PCV2 and our 2 CT-PCV2 isolates (CT-PCV-P5 & -P6) comprise PCV2. Phylogenetic analyses of a smaller number of PCV isolates and their clustering into PCV1 and PCV2, have been performed earlier (20,22).
The PK-15-PCV (a PCV1 isolate) was clinically nonpathogenic in inoculation studies in weaned pigs (9,10,11), whereas our CT-PCV-P7 isolate (also a PCV1), was derived from a neonatal pig with CT in the late 1960s and seemingly caused congenital tremors in progeny when inoculated into a pregnant sow at 70 d of gestation (27). It is unclear whether PK-15-PCV could also cause CT or if CT-PCV-P7 is pathogenic in weaned pigs. The age, route of infection, and/or some other factors may determine the pathogenicity and clinical manifestations of PCV1 and PCV2. The presence of, approximately, a 99% nt sequence identity among the CT-PCV2 and PMWS-PCV2 strains indicates that recent outbreaks of PMWS and CT are associated with the same type of PCV, such as, PCV2. The reported age of pigs with naturally occurring PMWS is 6 to 12 wk (14,15,34), whereas CT is a disease of newborn pigs (23). Porcine circovirus requires cell division for its replication (35). Neural cell division occurs exclusively during fetal development, and may be the only period when PCV could replicate in nervous tissues leading to signs of CT. Recently it has been demonstrated that experimental inoculation of cesarean-derived, colostrum-deprived piglets with PCV2 resulted in clinical signs of PMWS (36). However, coinfection of PCV2 with porcine parvovirus or PRRSV causes an enhancement in the severity of PMWS (11,37,38,39). Other viruses might enhance PCV replication by directly or indirectly causing division of PCV target cells. It still remains to be proven whether the presence of PCV (PCV1 or PCV2), in pigs with CT is coincidental, or if PCV plays a major role in developing CT.
Porcine circovirus type 1 was identified during the 1960s and 1970s (6,8), whereas PCV2 was identified in the late 1990s (17,18,20). On the basis of sequence analysis, it appears that PCV2 may be derived from PCV1. However, the large phylogenetic distance between the 2 types and the seeming total lack of intermediates contradicts a direct and recent connection. These findings do not support the role of the PK-15 cell line as an origin of widespread PCV infection, such as, vaccine borne disease. The PCV2 cluster includes a single bovine-origin circovirus isolate. It is unknown how widespread circoviral infection is in bovids. Based on the high similarity between this single bovine isolate and PCV2s, the tempting, but contradictory, speculation is that the 2 different PCV lineages evolved previously and simultaneously in porcine and bovine hosts, as seen in the case of adenoviruses (40). However, such PCV1 and PCV2 strain evolution may have occurred in 2 or more unidentified host species.
Footnotes
Acknowledgments
The authors thank Rick Westerman for helping with the GCG program, Marry Woodruff for her expertise in IFA and EM, and Jane Kovach for secretarial assistance. This work was partially funded by the National Pork Producers Council and an Indiana Value-added Grant.
Dr. Kuipel's present address is Animal Health Diagnostic Laboratory, Department of Pathobiology and Veterinary, Diagnostic Investigations, College of Veterinary Medicine, Michigan State University, G300 Veterinary Medical Center, East Lansing, Michigan 48824 USA.
Dr. Anothayanontha's present address is Antech Diagnostics, North Cave Creek Road, Phoenix, Arizona 85022 USA.
Address all correspondence and reprint requests to Dr. S.K. Mittal; telephone: (765) 496-2894; fax: (765) 494-9830; e-mail: mittal@purdue.edu
Received January 17, 2002. Accepted May 23, 2002.
References
- 1.Todd D, McNulty MS, Mankertz A, et al. Circoviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, et al. eds. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press, 2000:299–303.
- 2.Yuasa N, Taniguchi T, Yoshida I. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis 1979;23:366.
- 3.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature 1982;295:64–66. [DOI] [PubMed]
- 4.Pass DA, Perry RA. The pathology of psittacine beak and feather disease. Aust Vet J 1984;61:69–74. [DOI] [PubMed]
- 5.Woods LW, Latimer KS, Barr BC, et al. Circovirus-like infection in a pigeon. J Vet Diagn Invest 1993;5:609–612. [DOI] [PubMed]
- 6.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene — Erste Abteilung Originale — Reihe A: Medizinische Mikrobiologie und Parasitologie 1974;226:153–167. [PubMed]
- 7.Stevenson GW, Kiupel M, Mittal SK, Kanitz CL. Ultrastructure of porcine circovirus in persistently infected PK-15 cells. Vet Pathol 1999;36:368–378. [DOI] [PubMed]
- 8.Meehan BM, Creelan JL, McNulty MS, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol 1997;78:221–227. [DOI] [PubMed]
- 9.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. ArchVirol 1986;91:271–276. [DOI] [PubMed]
- 10.Allan G, McNeilly F, Cassidy J. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol 1995;44: 49–64. [DOI] [PubMed]
- 11.Krakowka S, Ellis JA, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet Pathol 2000;37:254–263. [DOI] [PubMed]
- 12.Niagro FD, Forsthoefel AN, Lawther RP, et al. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol 1998;143:1723–1744. [DOI] [PubMed]
- 13.Gibbs MJ, Weiller GF. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci USA 1999;96: 8022–8027. [DOI] [PMC free article] [PubMed]
- 14.Kiupel M, Stevenson GW, Mittal SK, Clark EG, Haines DM. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet Pathol 1998;35:303–307. [DOI] [PubMed]
- 15.Ellis J, Hassard L, Clark E, et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J 1998;39:44–51. [PMC free article] [PubMed]
- 16.Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest 2000;12:3–14. [DOI] [PubMed]
- 17.Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol 1998;72:5262–5267. [DOI] [PMC free article] [PubMed]
- 18.Meehan BM, McNeilly F, Todd D, et al. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol 1998;79:2171–2179. [DOI] [PubMed]
- 19.Morozov I, Sirinarumitr T, Sorden SD, et al. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol 1998;36: 2535–2541. [DOI] [PMC free article] [PubMed]
- 20.Mankertz A, Domingo M, Folch JM, et al. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res 2000;66:65–77. [DOI] [PubMed]
- 21.Allan G, McNeilly F, Kennedy S. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest 1998;10:3–10. [DOI] [PubMed]
- 22.Done B. The congenital tremor in pigs. Vet Annu 1976;16:98–102.
- 23.Stevenson GW, Kiupel M, Mittal SK, et al. Tissue distribution and genetic typing of porcine circoviruses in pigs with naturally occurring congenital tremors. J Vet Diagn Invest 2001;13:57–62. [DOI] [PubMed]
- 24.West KH, Bystrom JM, Wojnarowicz C, et al. Myocarditis and abortion associated with intrauterine infection of sows with porcine circovirus 2. J Vet Diagn Invest 1999;11:530–532. [DOI] [PubMed]
- 25.O'Connor B, Gauvreau H, West K, et al. Multiple porcine circovirus 2-associated abortions and reproductive failure in a multisite swine production unit. Can Vet J 2001;42:551–553. [PMC free article] [PubMed]
- 26.Sanchez RE, Jr., Nauwynck HJ, McNeilly F, Allan GM, Pensaert MB. Porcine circovirus 2 infection in swine foetuses inoculated at different stages of gestation. Vet Microbiol 2001;83: 169–176. [DOI] [PubMed]
- 27.Gustafson DP, Kaintz CL. Experimental transmission of congenital tremors in swine. Proc Annu Meet US Anim Health Assoc 1974:338–345. [PubMed]
- 28.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2), 2001. Cladistics. 1989;5:164–166.
- 29.Page RDM. TREEVIEW-an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. [DOI] [PubMed]
- 30.Harrach B, Benkö M. Phylogenetic analysis of adenovirus sequences. Proof of necessity of establishing a third genus in the Adenoviridae family. In: Wold WSM, ed. Adenovirus Methods and Protocols. Methods in Molecular Medicine. Totowa: Humana Press, 1998:309–339.
- 31.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol 1997;71:2562–2566. [DOI] [PMC free article] [PubMed]
- 32.Mankertz A, Mankertz J, Wolf K, Buhk HJ. Identification of a protein essential for replication of porcine circovirus. J Gen Virol 1998;79:381–384. [DOI] [PubMed]
- 33.LeCann P, Albina E, Madec F, Cariolet R, Jestin A. Piglet wasting disease. Vet Res 1997;141:660. [PubMed]
- 34.Rosell C, Segales J, Plana-Duran J, et al. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol 1999;120:59–78. [DOI] [PubMed]
- 35.Tischer I, Peters D, Rasch R, Pociuli S. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch Virol 1987;96:39–57. [DOI] [PubMed]
- 36.Bolin SR, Stoffregen WC, Nayar GP, Hamel AL. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J Vet Diagn Invest 2001;13: 185–194. [DOI] [PubMed]
- 37.Allan G, Kennedy S, McNeilly F. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol 1999;121:1–11. [DOI] [PubMed]
- 38.Allan GM, McNeilly F, Ellis J, et al. Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol 2000;145:2421–2429. [DOI] [PubMed]
- 39.Harms PA, Sorden SD, Halbur PG, et al. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol 2001;38:528–539. [DOI] [PubMed]
- 40.Russell WC, Benkö M. Adenoviruses (Adenoviridae): animal viruses. In: Webster RG, Granoff A, eds. Encyclopedia of Virology. London: Academic Press, 1999:14–21.