Abstract
The aim of this study was to compare, by randomly amplified polymorphic DNA (RAPD), the diversity of Streptococcus suis serotypes 1/2 and 2 isolates recovered at slaughter houses from the tonsils of clinically healthy pigs. The pigs belonged to herds with or without clinical signs of S. suis disease.
Overall, a low diversity was observed among isolates of serotype 1/2. A representative isolate recovered from a diseased animal presented a relatively high similarity (85%), with most isolates recovered from carrier pigs, from herds either with or without clinical signs of S. suis disease. For serotype 2 isolates, a relatively high degree of heterogeneity was observed in the whole population. Two subpopulations were observed for serotype 2 isolates, which arose from herds with clinical signs. Interestingly, the representative isolate coming from the diseased pig was included in a small closed cluster, with 2 isolates recovered from carrier pigs belonging to the same herd. On the other hand, most of the S. suis serotype 2 isolates originating from herds with no history of S. suis disease, were closely related (90% similarity). Furthermore, they presented different RAPD patterns from those originating from animals from the herd presenting S. suis clinical signs due to this serotype. Results suggest that, in the herds studied, clinical manifestations due to serotype 2 are probably related to the virulence of a specific isolate. Conversely, for the herd affected with serotype 1/2, clinical manifestations of the disease were more likely to be the result of inherent herd factors than the virulence of the specific isolate.
Introduction
Streptococcus suis infection has been considered a major worldwide problem in the swine industry, particularly in the past 10 y. This pathogen is responsible for pathological conditions, such as, meningitis, endocarditis, arthritis, pneumonia, and septicemia followed by sudden death (1). Thirty-five capsular types of S. suis have been described, with types 2 and 1/2 being the most prevalent serotypes recovered from diseased animals in Canada (2). The S. suis serotype 2 is also recognized as an agent of zoonosis since it has been identified as a cause meningitis; septicemia; and endocarditis in humans, particularly those who are occupationally exposed to pigs or pig products (3,4,5).
The primary natural habitat of S. suis is the upper respiratory tract of pigs, particularly the tonsils and nasal cavities. Since most healthy pigs are carriers of multiple serotypes of S. suis, as well as untypable strains in those sites, isolation of a specific serotype is difficult (1,6). An immunomagnetic separation technique (IMS) for the selective isolation of S. suis serotypes 2 and 1/2 from tonsils of carrier animals has recently been standardized in our laboratory (7). An IMS was used to isolate a representative number of isolates of S. suis serotypes 1/2 and 2, originating from carrier pigs belonging to herds with or without manifestations of S. suis disease.
Since a carrier state in convalescent animals and asymptomatic infection of pigs may create reservoirs of S. suis, genetic studies are required to better characterize an S. suis population potentially responsible for the transmission of infection within a closed herd (8). Furthermore, intensive swine production in developed countries is likely to be, at least in part, at the origin of the spread of particular S. suis clones worldwide (9).
Genetic differences between and within serotypes of S. suis have been detected with different typing methods such as; multilocus enzyme electrophoresis (10,11), restriction endonuclease analysis (12,13), ribotyping (14,15,16,17), macrorestriction analysis (18,19), and randomly amplified polymorphic DNA (RAPD) (9). Isolates from different geographic origins were studied in each case, and a few studies on genetic variability within a given serotype, in a closed population, are available (8,20,21). The latter studies were carried out with a limited number of isolates that originated from clinically healthy animals, and most were restricted to S. suis serotype 2.
In recent years, RAPD has become a very powerful technique for the detection of diversity among isolates (22,23,24). This method has the advantage of being independent of gene expression, thereby allowing for the study of DNA polymorphism between organisms without requiring knowledge of their molecular biology. Furthermore, Chatellier et al (9) found that fingerprinting by RAPD analysis may be used successfully for the genetic characterization and epidemical identification of S. suis serotype 2 isolates. Randomly amplified polymorphic DNA, as well as other molecular typing methods, revealed the existence of a correlation between the genetic background of S. suis and the expression of certain putative virulence markers, such as muramidase-released protein (MRP), extracellular factor (EF), and hemolysin (11,16,17,18). Although the expression of these proteins is, in general, associated with pathogenicity, their role as virulence factors is still unclear (25).
The objective of the present study was to compare, by RAPD, the diversity of S. suis serotypes 1/2 and 2 isolates recovered at slaughter houses from tonsils of clinically healthy pigs belonging to herds with or without clinical signs of S. suis disease.
Materials and methods
Herds
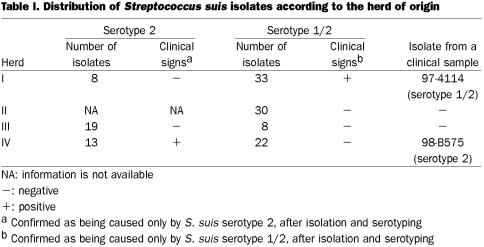
Herd I was considered an endemically affected herd for S. suis serotype 1/2, because recurrent clinical disease (sudden death, septicemia, meningitis), were diagnosed to be caused by this serotype. In fact, isolation of S. suis type 1/2 in pure culture, or as a predominant bacteria from clinical specimens (mainly brains), during at least 6 mo was recorded. This herd was considered an endemically affected herd for S. suis serotype 1/2 but not for serotype 2 (Table I), since no disease associated with this serotype was reported in the herd. Isolates of serotype 2 recovered from healthy animals in this herd were considered “isolates from a herd nonendemically affected by S. suis serotype 2.” Herd II did not present any signs of endemic infection due to S. suis, and only occasional isolation of this microorganism; mainly from lungs, were reported. Serotype 1/2 was never reported from clinical cases in this herd. No signs of endemic infection with any serotype of S. suis could be detected in herd III. On the other hand, clinical infection was regularly detected in herd IV primarily as meningitis caused by S. suis serotype 2, without any indication of isolation of serotype 1/2 strains from diseased animals. Thus, isolates of serotype 2 from this herd were considered “isolates from a serotype 2 clinically affected herd,” whereas isolates of serotype 1/2 from clinically healthy animals were considered “isolates from a herd nonendemically affected by S. suis serotype 1/2” (Table I). All herds were closed farrow to finish herds, where replacement gilts were selected from their own finishing barns. In endemically affected herds, most clinical cases were observed in piglets between 5 and 7 wk of age.
Table I.
Bacteria
Strains of S. suis serotype 1/2 from different geographical origins and the reference strain (2651), used to standardize the RAPD technique for this serotype, came from our collection. The 133 S. suis field isolates used in this study are indicated in Table I. The isolates were recovered at slaughter houses from tonsils of clinically healthy pigs belonging to herds where clinical manifestations of S. suis disease of a given serotype were or were not present. Isolation of S. suis was carried out using the IMS technique (7). Furthermore, 2 representative clinical isolates of serotypes 1/2 and 2 recovered from herds I and IV, respectively, were included in this study (Table I). These isolates, recovered from the brains of 5 affected animals at different time intervals, were considered to be representative of the disease population since they were found to be genotypically identical to each other. All isolates underwent coagglutination testing (26) using sera against serotypes 1 and 2, as well as negative serum (negative control). Isolates of serotype 2 or 1/2 were confirmed biochemically as being S. suis using standard procedure (27).
Detection of MRP, EF, and hemolysin
Production of MRP, EF, and hemolysin were detected on selected S. suis serotype 2 isolates (28).
RAPD fingerprinting
The RAPD analysis for S. suis serotype 2 was performed as described by Chatellier et al (9). Since primers used for serotype 2 strains (9) did not clearly differentiate serotype 1/2 strains (unpublished observations), the standardization of RAPD for S. suis serotype 1/2 was carried out in this study with 9 strains from different origins. Four of them were from different geographical origins: 95–9225 (Argentina); 98–09 (Mexico); 98–475 (Canada); and 2651 (The Netherlands, reference strain), whereas the 5 remaining strains were from different herds in Quebec. After testing 10 primers, 10 nucleotides in length, with a G + C content of 40 to 70% (not shown), 3 were chosen to amplify the DNA of S. suis serotype 1/2 strains: OPB07 (GGTGACGCAG), OPB10 (CTGCTGGGAC), and CLAU (AAAGGATGCT). The PCR mixture consisted of buffer (10 mM Tris-HCl, pH 8.3; 50 mM KCl; 2.5 mM Mg Cl2), 100 μM each of the 4 deoxynucleotide triphosphates (Pharmacia Biotech Incorporated, Baie d'Urfé, Quebec), 0.4 μM primer, 50 ng of DNA extracted and purified (29), and 2.5 U of Taq DNA polymerase (Pharmacia) in a total volume of 25 μL. Primers used were synthesized (Custom Primers; Gibco BRL, Burlington, Ontario). Each sample was subjected to an initial cycle of denaturation (5 min at 94°C) (DNA Thermal Cycler 480; Perkin-Elmer Applied Biosystems, Foster City, California, USA). Each of the 35 subsequent cycles consisted of denaturing at 94°C for 30 s, annealing at 35°C for 30 s, and extension at 72°C for 1 min. The last cycle, included an extension at 72°C for 5 min. Amplified products were separated by electrophoresis in a 1.2% agarose gel (Sigma-Aldrich, Oakville, Ontario), and were visualized as white bands on the black background by UV transillumination following ethidium bromide staining. A 1 kb DNA ladder (Gibco) was used in each gel as a molecular size standard. A negative control consisting of the same reaction mixture substituting water for the template DNA, was included in each run. A positive control, containing the same reaction mixture and template DNA from the well-characterized reference strain was also included. Individual isolates were tested under identical conditions at least 3 times with the selected primers.
Pattern analysis
Photographs of each gel were digitized with a video camera connected to a microcomputer (Alpha ease TM; Alpha Innotech Corporation, San Leandro, California, USA). After conversion, the data were normalized and analyzed (Molecular Analyst Software, Fingerprinting, version 1.12; Bio-Rad Laboratories, Mississauga, Ontario). Degrees of homology were determined by Dice comparisons. Clustering correlation coefficients were calculated by the unweighted pair group method with arithmetic averages. Once completed, a dendrogram was drawn showing the hierarchical representation of linkage levels between isolates. The definition of 2 strains as “different” was automatically carried out by the software.
The genetic diversity level was calculated as the ratio between the number of different patterns and the total population number. Statistical analysis of data was performed to analyze the difference of diversity level between isolates originating from herds with or without clinical signs of S. suis disease. Chi-square values were then calculated (Statistical Analysis Systems (SAS), version 8.0; Cary, North Carolina, USA). Differences between the groups were considered significant at P ≤ 0.05.
Results
Standardization of RAPD for serotype 1/2 isolates
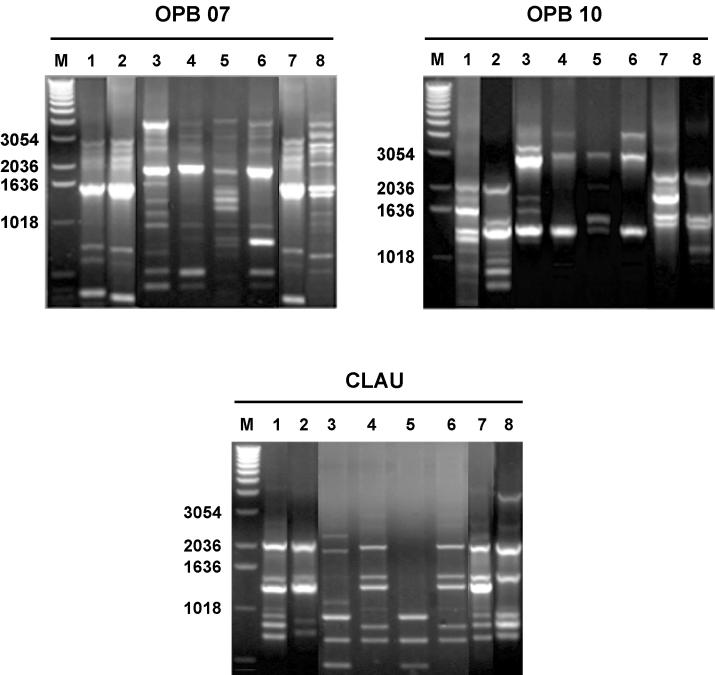
To standardize RAPD for S. suis serotype 1/2, strains from very different geographical origins were included. The 3 primers used yielded heterogeneous band patterns, allowing for differentiation of the strains analyzed (Figure 1). Reproducible strain-specific RAPD patterns were obtained when isolates were tested at least 3 times. However, slight variations between different repetitions were observed in the faint bands. Low-intensity bands were therefore not included in the analysis.
Figure 1. Illustration of RAPD patterns generated with primers OPB07, OPB10, and CLAU from different strains of S. suis serotype 1/2. Lanes 1: isolate from herd II; lanes 2: isolate from herd III; lanes 3: strain 98–09 (Mexico); lanes 4: strain 98–475 (Canada); lanes 5: serotype 1/2 reference strain 2651 (The Netherlands); lanes 6: strain 95–9225 (Argentina); lanes 7: isolate from herd I; and lanes 8: isolate from herd IV.
M: 1kb DNA ladder.
General genetic diversity of isolates of S. suis serotypes 1/2 and 2 as defined by RAPD fingerprint
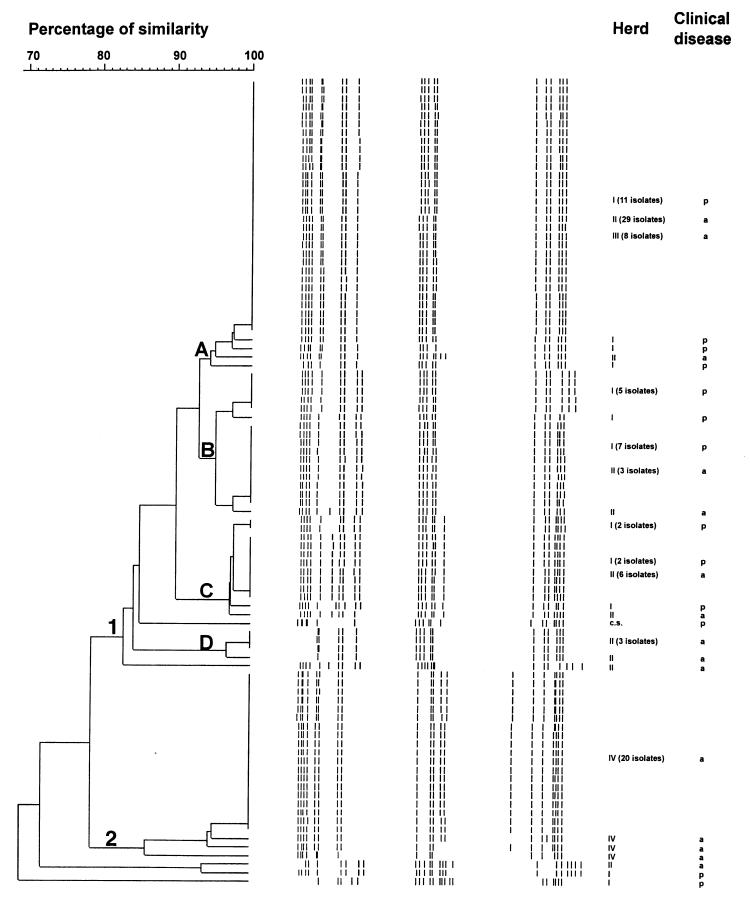
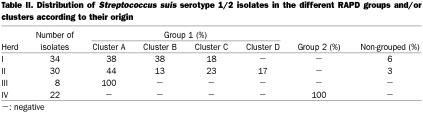
The genetic relationship among all isolates of S. suis serotypes 1/2 and 2 are presented in dendrograms shown in Figures 2 and 3, respectively. The 2 isolates recovered from diseased animals were also included for comparative purposes. No significant difference in the dendograms could be observed when the 2 isolates were omitted (not shown). Overall, 69% similarity was detected among isolates of serotype 1/2. In the present study, 97% of the isolates, were clustered into 2 groups (1 and 2), revealing a 79% similarity between them. Four clusters with 94 to 97% similarity were identified within group 1. The other group did not present any clusters. Interestingly, some isolates of S. suis serotype 1/2 originating from different herds shared the same RAPD pattern.
Figure 2. Genetic relationship among 94 isolates of S. suis serotype 1/2 recovered from carrier pigs belonging to herds with or without clinical signs. The dendogram was generated by unweighted pair group method with arithmetic averages (UPGMA). There are 2 groups (1 and 2) and 4 clusters (A, B, C and D).
c.s. — clinical sample.
a — absent.
p — present.
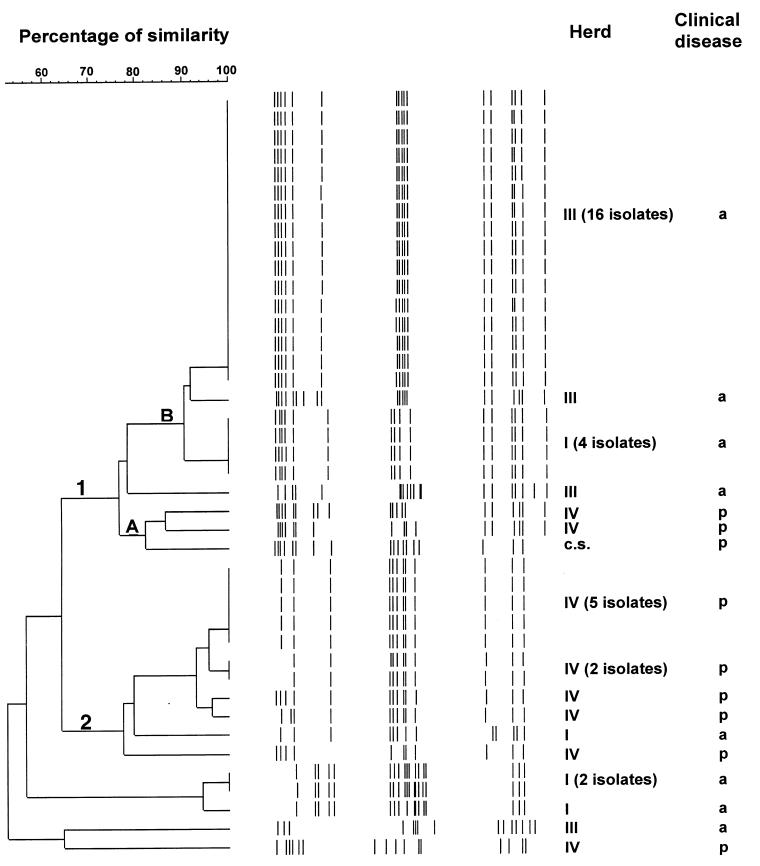
Figure 3. Genetic relationship among 41 isolates of S. suis serotype 2 recovered from carrier pigs belonging to herds with or without clinical signs. The dendogram was generated by unweighted pair group method with arithmetic averages (UPGMA). There are 2 groups (1 and 2) and 2 clusters (A and B).
c.s. — clinical sample.
a — absent.
p — present.
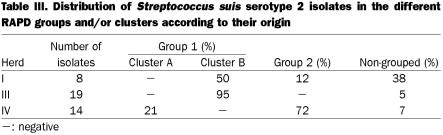
On the other hand, 41 isolates of serotype 2 revealed a similarity of 52%, and most were also assigned to the 2 groups (1 and 2). Clusters (A and B) could only be identified in group 1. Cluster similarity varied between 82 and 90%, while group similarity was 76% for group 1 and 77% for group 2. Five isolates did not fall into any cluster in any group. Isolates of S. suis serotype 2 from different herds did not share the same RAPD pattern.
Genetic variation of isolates originating from a given herd
Genetic variation depended on the serotype and herd studied. For serotype 1/2, within herd clustering was observed for isolates originating from herds III and IV. However, these isolates represented only 32% of the S. suis serotype 1/2 population studied. In the case of serotype 2, most S. suis isolates originating from a given herd tended to be more closely related, especially in herds with no clinical signs of S. suis infection.
Genetic variation of isolates originating from pigs belonging to herds without clinical signs of S. suis disease
Similar diversity levels, calculated as the ratio between the number of different patterns and the total population number, were observed for serotypes 2 and 1/2 isolates (0.26 and 0.23, respectively). Overall, clustering within herds could not be established for isolates of serotype 1/2 (Figure 2). Interestingly, group 2 is formed only by isolates originating from herd IV, which represent 37% of all isolates collected from herds with no clinical signs of S. suis serotype 1/2 disease. In the case of serotype 2, 81% of isolates originating from herds with no history of S. suis disease due to this serotype, are genetically related and are grouped in cluster B of group 1, as depicted in Figure 3.
Genetic variation of isolates originating from pigs belonging to herds with clinical signs of S. suis disease
Clustering could not be established for isolates of S. suis serotype 1/2 (Table II). The representative isolate of S. suis serotype 1/2, recovered from a diseased pig, was genetically related (85% similarity) to most isolates originating from both types of herds, with (herd I) or without (herd II and III) clinical signs of S. suis disease (Figure 2). On the other hand, 2 clearly distinct subpopulations were observed among S. suis serotype 2 isolates. Seventy-two percent of the isolates were included in group 2 and a few others in cluster A of group 1 (Table III). Despite this apparent homogeneity, 9 different RAPD patterns were revealed when the 13 isolates included in this population were tested. The representative isolate of S. suis serotype 2, recovered from a diseased pig, was included in a small closed cluster (cluster A group 1) with 2 isolates from unaffected carrier pigs belonging to the same herd with clinical signs of S. suis disease (Figure 3).
Table II.
Table III.
Presence of MRP, EF, and suilysin in serotype 2 isolates
All isolates of S. suis serotype 2, including isolates recovered from clinical disease, presented a typical Canadian phenotype EF−, suilysin−, and MRP variable (28).
Discussion
Differences between S. suis isolates at the genetic level have been previously studied. In general, considerably high levels of genetic diversity were observed within and between S. suis serotypes. However, most studies were conducted with isolates from different geographic origins (9,10,11,12,14,16,17,18). Furthermore, limited information exists regarding the diversity between isolates of a given S. suis serotype recovered from carrier pigs belonging to non-endemically affected herds, since all studies were conducted with isolates from herds with clinical manifestations of S. suis infection (8,20,21). Results obtained in this study may be applied only to the herds studied and no generalizations should be made. In fact, the effect of different herd managements (1 site, 3 sites, multisites), as well as the presence of other infections, such as porcine respiratory and reproductive syndrome virus (PRRSV), may have an influence on the transmission of different S. suis strains in a given herd.
The combination of 3 primers allows RAPD to successfully detect genotypic differences between strains of S. suis serotype 1/2, as reported for serotype 2 by Chatellier et al (9), although primers are not exactly the same for both serotypes. Ideally, exactly the same primers should have been used for both serotypes. However, the primers previously described as highly discriminatory for serotype 2 (9), did not clearly differentiate strains of serotype 1/2 from different origins. This suggests that a standardization may be required when changing the target serotype.
The genetic diversity among field isolates of serotype 1/2, using the RAPD proposed in this study, was lower than anticipated from previous reports (10,15,17,18). Interestingly, certain RAPD patterns were highly conserved between herds. For example, the same RAPD pattern was found in 3 out of 4 herds studied. This may suggest herd-to-herd contamination, as reported for Staphylococcus aureus. In fact, Rivas et al (30) analyzed S. aureus originating from 12 dairy herds in New York State and also found high inter-farm conservation of some S. aureus ribotypes. On the other hand, more than one RAPD pattern was found within each of the herds analyzed in the present study. This finding suggests that the same herd can be contaminated by different strains, as previously reported (8,31).
Although the entire population of S. suis serotype 1/2 presented a low level of diversity, clustering was not observed for isolates recovered from a given herd endemically infected by this serotype. The level of genetic diversity for S. suis isolates originating from a herd presenting serious clinical signs due to this serotype was similar to that obtained with isolates recovered from carrier animals in 3 unaffected herds. In addition, a representative isolate recovered from a diseased animal has a relatively high similarity (85%) with most isolates recovered from carrier pigs belonging to both categories of herds. In this study, as was the case in other previous reports (8,20), a single pattern was observed when at least 5 isolates from different diseased animals were compared. These observations suggest that the disease caused by this particular isolate within the studied herd was related more to inherent factors affecting the herd such as stress, overpopulation, and lowered immunity, than to the virulence of a specific isolate. In fact, differences in the virulence of S. suis serotype 1/2 isolates are thus far unknown. A similar epidemiological pattern was described by Müiler-Graf et al (32), who sug- gested that there is no obvious distinction between “carried” and “invasive” Streptococcus pneumoniae.
The situation seems to be different for field isolates of serotype 2. A relatively high degree of heterogeneity was observed among all S. suis isolates. Evidence for this genetic heterogeneity is supported by previous reports on the genetic diversity of S. suis serotype 2 isolates (9,11,12,14,16,17,19). All these studies were performed using a relatively large number of isolates originating from diseased pigs. Interestingly, the present study was carried out using a large majority of isolates originating from carrier pigs. This high level of heterogeneity was evident when isolates originating from different herds, or those from the endemically affected herd, were compared. In fact, a highly significant difference (P = 0.0088) was recorded when the genetic diversity level of isolates of S. suis serotype 2, originating from a herd presenting serious clinical signs due to this serotype, was compared to that of non-clinically affected herds (0.69 and 0.26, respectively). Previous studies have also shown large degrees of variability in genetic patterns of S. suis serotype 2 isolates recovered from tonsils of clinically healthy pigs from closed, endemically affected herds (8,20,21).
The representative clonal isolate of serotype 2 recovered from diseased pigs was closely related genetically to only 2 isolates originating from carrier pigs belonging to the same herd. This fact may indicate that there is a tendency for only 1 strain to cause mortality in a single herd (8,21). Indeed, detection of potentially virulent strains in asymptomatic pigs may only be a chance occurrence. It is possible that these animals were in the incubation period and would have developed clinical signs later. Since the animals were not individually monitored, it was not possible to ascertain whether a true asymptomatic carrier state existed. As expected, no significant differences regarding the virulence markers MRP, EF, and hemolysin were observed between isolates recovered from affected or healthy animals in this study. Virulence factors or markers for Canadian strains are still unknown (28).
The RAPD patterns showed that most of the S. suis serotype 2 isolates originating from the herds studied, without a history of S. suis disease, were closely related (90% similarity). Furthermore, they presented different RAPD patterns from those originating from diseased animals from the herd where clinical signs of infection with S. suis serotype 2 were manifested. These observations may suggest the existence of some non-virulent strains that are well adapted to survive in pigs, allowing for the establishment of a non-harmful relationship with the host.
Although several differences in the molecular epidemiology of serotypes 1/2 and 2 appear to exist, in the present study these serotypes are apparently transmitted in a similar mode. Horizontal transmission appears to occur in both herds, as the same RAPD pattern was shared between several isolates recovered from a given herd. In fact, it is generally accepted that S. suis is only transmitted between animals with “nose to nose” contact. However, experimental airborne transmission of S. suis serotype 2 was recently demonstrated by Berthelot-Hérault et al (33). Important to note is that, to the best of our knowledge, there is no information about possible changes in the S. suis population with age. In fact, the isolates included in this study came from tonsils at slaughter whereas clinical signs were observed at younger age.
In conclusion, the RAPD test used in this study has the potential to reveal information about the epidemiology of S. suis infections, and may allow for the study of routes of transmission and sources of S. suis within a given population. Results suggest that, for the herds studied, clinical manifestations due to serotype 2 are probably related to the virulence of a specific isolate. Conversely, for the serotype 1/2 studied, clinical manifestations of the disease were more likely to be the result of inherent herd factors than the virulence of a specific isolate.
Footnotes
Acknowledgments
The authors thank Julie-Melanie Trudel for technical assistance. This study was supported by Conseil de Recherche en Pêche et en Agroalimentaire de Quebec # 4778 and by the Canadian Network on Bacterial Pathogens of Swine.
Address correspondence and reprint requests to Dr. Marcelo Gottschalk; telephone: (450) 773-8521 ext. 8374; fax: (450) 778-8108; e-mail: gottschm@medvet.umontreal.ca
Received February 11, 2002. Accepted May 9, 2002.
References
- 1.Higgins R, Gottschalk M. Streptococcal disease. In: Straw B, D'Allaire S, Mengeling W, Taylor D, eds. Diseases of swine. 8th ed. Iowa State University Press, Ames, 1999:563–578.
- 2.Higgins R, Gottschalk M. Distribution of Streptococcus suis capsular types in 1999. Can Vet J 2000;41:414. [PMC free article] [PubMed]
- 3.Peetermans WEC, Moffie BG, Thompson J. Bacterial endocarditis caused by Streptococcus suis type 2. J Infect Dis 1989;159:595–596. [DOI] [PubMed]
- 4.Büngener W, Bialek R. Fatal Streptococcus suis septicemia in an abattoir worker. Eur J Clin Microbiol Infect Dis 1989;8: 306–308. [DOI] [PubMed]
- 5.Arends JP, Zanen HC. Meningitis caused by Streptococcus suis in humans. J Infect Dis 1988;10:131–137. [DOI] [PubMed]
- 6.Monter Flores JL, Higgins R, D'Allaire S, Charette R, Boudreau M, Gottschalk M. Distribution of the different capsular types of Streptococcus suis in 19 swine nurseries. Can Vet J 1993;34: 170–171. [PMC free article] [PubMed]
- 7.Gottschalk M, Lacouture S, Odierno L. Immunomagnetic isolation of Streptococcus suis serotypes 2 and 1/2 from swine tonsils. J Clin Microbiol 1999;37:2877–2881. [DOI] [PMC free article] [PubMed]
- 8.Mogollon JD, Pijoan C, Murtaugh MP, Collins JE, Cleary PP. Identification of epidemic strains of Streptococcus suis by genomic fingerprinting. J Clin Microbiol 1991;29:782–787. [DOI] [PMC free article] [PubMed]
- 9.Chatellier S, Gottschalk M, Higgins R, Brousseau R, Harel J. Relatedness of Streptococcus suis serotype 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J Clin Microbiol 1999;37:362–366. [DOI] [PMC free article] [PubMed]
- 10.Hampson DJ, Trott DJ, Clarke IL, Mwaniki CG, Robertson ID. Population Structure of Australian isolates of Streptococcus suis. J Clin Microbiol 1993;31:2895–2900. [DOI] [PMC free article] [PubMed]
- 11.Mwaniki CG, Robertson ID, Trott DJ, Atyeo RF, Lee BJ, Hampson DJ. Clonal analysis and virulence of Australian isolates of Streptococcus suis type 2. Epidemiol Infect 1994; 113:321–334. [DOI] [PMC free article] [PubMed]
- 12.Beaudoin M, Harel J, Higgins R, Gottschalk M, Frenette M, MacInnes JI. Molecular analysis of isolates of Streptococcus suis capsular type 2 by restriction-endonuclease-digested DNA separated on SDS-PAGE and by hybridization with an rDNA probe. J Gen Microbiol 1992;138:2639–2645. [DOI] [PubMed]
- 13.Mogollon JD, Pijoan C, Murtaugh MP, Kaplan EL, Collins JE, Cleary PP. Characterization of prototype and clinically defined strains of Streptococcus suis by genomic fingerprinting. J Clin Microbiol 1990;28:2462–2466. [DOI] [PMC free article] [PubMed]
- 14.Rasmussen SR, Aarestrup FM, Jensen NE, Jorsal SE. Associations of Streptococcus suis serotype 2 ribotype profiles with clinical disease and antimicrobial resistance. J Clin Microbiol 1999;37:404–408. [DOI] [PMC free article] [PubMed]
- 15.Okwumabua O, Staats J, Chengappa MM. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping). J Clin Microbiol 1995;33:968–972. [DOI] [PMC free article] [PubMed]
- 16.Staats JJ, Brandon L, Plattner BL, Nietfeld J, Dritz S, Chengappa MM. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J Clin Microbiol 1998;36:15–19. [DOI] [PMC free article] [PubMed]
- 17.Smith HE, Rijnsburger M, Stockhofe-Zurwieden N, Wisselink HJ, Vetch U, Smith MA. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J Clin Microbiol 1997;35:1049–1053. [DOI] [PMC free article] [PubMed]
- 18.Allgaier A, Goethe R, Wisselink HJ, Smith HE, Valentin-Weigand P. Relatedness of Streptococcus suis isolates of various serotypes and clinical background as evaluated by macrorestriction analysis and expression of potential virulence traits. J Clin Microbiol 2001;39:445–453. [DOI] [PMC free article] [PubMed]
- 19.Berthelot-Herault F, Marois C, Gottschalk M, Kobich M. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J Clin Microbiol 2002;40:615–619. [DOI] [PMC free article] [PubMed]
- 20.Torremorell M, Calsamiglia M, Pijoan C. Colonization of suckling pigs by Streptococcus suis with particular reference to pathogenic serotype 2 strains. Can J Vet Res 1998;62:21–26. [PMC free article] [PubMed]
- 21.Torremorell M, Pijoan C. Prolonged persistence of an epidemic Streptococcus suis strain in a closed pig population. Vet Rec 1998;143:394–395. [DOI] [PubMed]
- 22.Maslow JN, Mulligan ME, Arbeit RD. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis 1993;17:153–164. [DOI] [PubMed]
- 23.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 1990;18:7213–7218. [DOI] [PMC free article] [PubMed]
- 24.Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 1990;18:6531–6535. [DOI] [PMC free article] [PubMed]
- 25.Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol 2000;76:259–272. [DOI] [PubMed]
- 26.Gottschalk M, Higgins R, Boudreau M. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol 1993;31:2192–2194. [DOI] [PMC free article] [PubMed]
- 27.Higgins R, Gottschalk M. An update on Streptococcus suis identification. J Vet Diagn Investig 1990;2:249–252. [DOI] [PubMed]
- 28.Gottschalk M, Lebrun A, Wisselink HJ, Dubreuil JD, Smith HE, Vetch U. Production of virulence-related proteins by Canadian strains of Streptococcus suis capsular type 2. Can J Vet Res 1998;62:75–79. [PMC free article] [PubMed]
- 29.Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 1989;8:151–156.
- 30.Rivas AL, Gonzalez RN, Wiedmann M, et al. Diversity of Streptococcus agalactiae and Staphylococcus aureus ribotypes recovered from New York dairy herds. Am J Vet Res 1997; 58:482–487. [PubMed]
- 31.Reams RY, Harrington DD, Glickman LT, Thacker HL, Bowersock TL. Multiple serotype and strains of Streptococcus suis in naturally infected swine herds. J Vet Diagn Investig 1996; 8:119–121. [DOI] [PubMed]
- 32.Müller-Graf CDM, Whatmore AM, King SJ, Trzcinski K, et al. Population biology of Streptococcus pneumoniae isolated from oropharyngeal carriage and invasive disease. Microbiol 1999;145: 3283–3293. [DOI] [PubMed]
- 33.Berthelot-Hérault F, Gottschalk M, Labbé A, Cariolet R, Kobich M. Experimental airborne transmission of Streptococcus suis capsular type 2 in pigs. Vet Microbiol 2001;82:69–80. [DOI] [PubMed]