Abstract
Forty-two Salmonella isolates obtained from diseased swine were genetically characterized for the presence of specific antimicrobial resistance mechanisms. Twenty of these isolates were characterized as S. Typhimurium DT104 strains. Pulsed-field gel electrophoresis was used to determine genetic relatedness and revealed 20 distinct genetic patterns among the 42 isolates. However, all DT104 isolates fell within 2 closely related genetic clusters. Other Salmonella isolates were genetically grouped together according to serotype. All DT104 isolates displayed the penta-resistance phenotype to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline. Resistance to sulfamethoxazole, tetracycline, streptomycin, kanamycin, and ampicillin was most common among the non-DT104 Salmonella isolates. All DT104 strains contained 2 chromosomal integrons of 1000 and 1200 base pairs. The DNA sequencing revealed that the 2 integrons contained genes encoding a resistance to streptomycin and ampicillin, respectively. None of the non-DT104 strains showed the same pattern, although several strains possessed integrons of 1000 base pairs or larger. However, the majority of non-DT104 Salmonella strains did not possess any integrons. Two Salmonella isolates displayed tolerance to the organic solvent cyclohexane, indicating the possibility that they are overexpressing chromosomal regulatory genes marA or soxS or the associated multidrug efflux pump, acrAB. This research suggests that integrons contribute to antimicrobial resistance among specific swine Salmonella serotypes; however, they are not as widely disseminated among non-Typhimurium swine Salmonella serotypes as previously thought.
Introduction
The ease with which bacteria become resistant to antimicrobial agents continues to concern clinicians, public health officials, and researchers. Antimicrobial resistance is a problem of both national and international importance, with resistance mechanisms having been described for all known antibiotics that are currently available (1). Although the spread of resistant microorganisms is disturbing, the association of resistance determinants with mobile DNA elements; such as, plasmids, transposons, and integrons, is also of concern. These mobile DNA elements assist in the rapid dispersion of resistance genes within bacterial species and between different species (2,3). Many antimicrobial resistance genes in Escherichia coli and Salmonella species are on large, transferable plasmids. Another type of mobile DNA element, termed the transposon, also often carries antimicrobial resistance genes. In turn, plasmids and transposons, coding multiple drug resistance, often possess another genetic element, the integron.
Integrons contain 1 or more resistance genes present as mobile gene cassettes and inserted into various arrangements between 2 conserved DNA regions, creating arrays of different antimicrobial resistance genes (3). Over 60 gene cassettes and 4 distinct classes of integrons have been identified to date (2,3). Cassette-associated genes conferring resistance to beta-lactams, aminoglycosides, trimethoprim, chloramphenicol, and quaternary ammonium compounds used as antiseptics and disinfectants have been found (2). Also, class I integrons include a sulfonamide resistance gene (sul1) in the backbone structure (2). Integrons were the focus of our study, since little is known about the potential contribution of integrons to maintaining multiple antimicrobial resistance among clinical swine Salmonella species. To date, there is limited information regarding emergence and characterization of integron-mediated, multiple antimicrobial resistance among non-Typhimurium DT104 swine Salmonella serotypes from North America. The majority of work published on resistance genes in Salmonella has been obtained from European isolates (4,5).
All isolates were further examined for phenotypic characteristics associated with mutation of the marRAB locus (6,7). The marRAB locus is 1 of 2 operons (the other being soxRS), that have been associated with chromosomal based resistance to multiple antibiotics in E. coli and S. Typhimurium (8,9). Mutations in these chromosomal loci in E. coli are associated with the acquisition of low-level resistance to certain antimicrobials and tolerance to certain chemicals, including cyclohexane (6,8,9,10,11). Antibiotic resistance mediated by the soxRS and marRAB regulons is directed via both the over-expression of the acrAB-encoded efflux pump and down-regulation of the outer membrane porin OmpF (6,7,12). There is increasing evidence that the expression of soxS, marA, and acrAB contributes to clinical antimicrobial resistance in E. coli and Salmonella (13,14,15,16).
Despite much research into the characterization of resistance mechanisms among Salmonella, the prevalence and relative contribution of both mar- and integron-mediated, multiple antimicrobial resistance among various veterinary Salmonella serotypes is unclear. Thus, our underlying goal was to advance the knowledge concerning the development and dissemination of antimicrobial resistance among bacterial pathogens important in swine husbandry and emerging as foodborne pathogens contaminating pork.
Materials and methods
Bacterial isolates
Forty-two isolates of Salmonella isolated from diseased swine were included in the study. Salmonella serotypes assayed included 20 S. Typhimurium DT104, 2 S. Typhimurium non-DT104, and 2 S. Typhimurium Copenhagen non-DT104 strains. The remaining Salmonella isolates include representatives of the Anatum (n = 3), Choleraesuis-kunzendorf (n = 4), Derby (n = 3), Heidelburg (n = 3), Infantis (n = 3), and Mbandaka (n = 2) serotypes. Isolates were obtained from the National Veterinary Services Laboratory (NVSL), in Ames, Iowa, and were representative of the most prevalent Salmonella serotypes submitted to NVSL from diseased swine throughout the United States during the year 2000. Salmonella isolates assayed were recovered from swine from 18 states with the majority submitted from North Carolina (n = 7) and Illinois (n = 6). Bacteria were grown on MacConkey agar (Difco Laboratories, Detroit, Michigan, USA) and stored in trypticase soy broth (TSB) (Difco) containing 50% glycerol at −80°C until use.
Antimicrobial susceptibility determination
Antimicrobial minimum inhibitory concentrations (MIC) of Salmonella isolates were determined with an automated antimicrobial susceptibility system according to the manufacturer's instructions (Sensititre automated antimicrobial susceptibility system; Trek Diagnostic Systems, Westlake, Ohio, USA) and interpreted according to the National Committee Clinical Laboratory Standards (NCCLS) standards for broth microdilution methods (17,18). Escherichia coli ATCC 25922 and 35218, Staphylococcus aureus ATCC 29213, Pseudomonas aeruginosa ATCC 27853, and Enterococcus faecalis ATCC 29212 were used as quality control microorganisms. A comprehensive antibiogram was determined for the Salmonella isolates, using a customized antimicrobial panel of 17 antibiotics with the range of concentrations employed in the National Antimicrobial Resistance Monitoring System (NARMS) established by the Center for Disease Control and Prevention (CDC), United States Department of Agriculture (USDA), and the Food and Drug Administration (FDA) (19).
Bacterial DNA preparation and PCR and DNA sequencing
To determine the extent of integron-mediated multiple antimicrobial resistance, PCR primers homologous to conserved integron sequences were used (3) to assay the swine Salmonella isolates. Template DNA from the Salmonella isolates was prepared and purified using routine procedures (5). Integron PCR primers and amplification conditions employed have been previously described (3,4,5). Amplicons were separated by horizontal gel electrophoresis and visualized under ultraviolet (UV) light. Appropriate amplicons were identified by size, excised from the agarose, and purified (Wizard PCR Clean Up System; Promega, Madison, Wisconsin, USA). Sequencing of PCR products was performed according to manufacturer's protocol for cycle sequencing using the cycle sequencer (Model 377; Perkin Elmer Applied Biosystems, Foster City, California, USA) at the University of Maryland Center for Agricultural Biotechnology. The DNA sequence data were analyzed using the Genetics Computer Group (GCG) suite of software (Genetics Computer Group, Madison, Wisconsin, USA), and compared, using the NCBI-BLAST program (20), with published GenBank DNA sequences.
Pulsed-field gel electrophoresis (PFGE) of Salmonella isolates
The PFGE was used to compare DNA fingerprinting profiles of Salmonella isolates. The PFGE procedure was performed according to the protocol developed by the CDC. Briefly, bacteria were grown on trypticase soy agar (TSA) blood agar (Becton Dickinson Microbiology System, Cockeysville, Maryland, USA) at 37°C for 18 h. Bacterial colonies were suspended in cell suspension buffer (100 mM Tris HCl, 100 mM EDTA, pH 8.0) and adjusted to 0.48 to 0.52 optical density (OD) (Dade MicroScan Turbidity Meter; Dade Behring Inc., West Sacramento, California, USA). The cell suspension (200 μL) was mixed with 10 μL of proteinase K (10 mg/mL) and an equal volume of melted 1% SeaKem Gold agarose (FMC BioProducts, Rockville, Maine, USA) containing 1% sodium dodecyl sulfate (SDS). The mixture was carefully dispensed into a sample mold (Bio-Rad Laboratories, Hercules, California, USA). After solidification, the plugs were transferred to a tube containing 5 mL of lysis buffer (50 mM Tris HCl, 50 mM EDTA, pH 8.0, plus 1% Sarcosyl) and 0.1 mg/mL of proteinase K. Cells were lysed overnight in a water bath at 54°C with vigorous agitation. After lysis, the plugs were washed twice with deionized water and 4 times with tris-EDTA (TE) buffer (10 mM Tris HCl, 1 mM EDTA, pH 8.0) for 15 min per wash at 50°C with vigorous agitation. Agarose-embedded with DNA was digested with 50 U of XbaI (Boehringer Mannheim Corporation, Indianapolis, Indiana, USA) overnight in a water bath at 37°C. The plugs were then placed in a 1% SeaKem Gold agarose (FMC) gel and restriction fragments were separated by electrophoresis in 0.5 × tris-borate-EDTA (TBE) buffer at 14°C for 18 h using a Chef Mapper (Bio-Rad) with pulse times of 2.16 to 63.8 s. The gel was stained with ethidium bromide, and DNA bands were visualized with UV transillumination. The PFGE images were then analyzed (Molecular Analyst Fingerprinting Plus Software; Bio-Rad).
Organic solvent tolerance assays
Tolerance to certain organic solvents is associated with acquisition of low-level antimicrobial resistance via the overexpression of the marRAB and acrAB loci. Salmonella strains were grown to late logarithmic phase and diluted to a concentration of approximately 107 cfu/mL. A 5-μL aliquot of the bacterial suspension was spotted onto Luria-Bertani (LB) agar plates and allowed to dry. Cyclohexane, n-hexane, or n-pentane (Aldrich Chemicals, Milwaukee, Wisconsin, USA) were added to the plates at a depth of 2 to 3 mm. Plates were sealed with petroleum jelly and parafilm to prevent evaporation of solvents, and incubated at 30°C for up to 48 h (10). Plating was done in duplicate and solvent tolerance was measured as a function of bacterial growth. Growth was recorded as confluent growth (++), visible growth (<100 colonies; +), or no growth (−) after 24 h (10). Control strain E. coli AG100 demonstrated confluent growth under hexane and did not grow in the presence of cyclohexane whereas E. coli AG102 (Mar mutant) grew in the presence of both hexane and cyclohexane.
Northern blot analysis
Overnight cultures of Salmonella were diluted 100-fold in fresh LB broth and grown to the mid-logarithmic phase at 30°C with shaking. Total RNA was extracted from a 50-mL culture (Qiagen RNA Midiprep kit; Qiagen, Chatsworth, California, USA) and the concentration was determined spectrophotometrically at 260/280 OD. The RNA was transferred to nylon membranes (Ambion, Austin, Texas, USA) using a transfer system (Turboblotter Transfer System; Schleicher and Schuell, Keene, New Hampshire, USA) and cross-linked (Gene Linker UV chamber; Bio-Rad). Hybridization of the radiolabeled DNA probe to the membrane-bound RNA (10 μg/lane) was performed at 42°C overnight according to the specifications of the membrane manufacturer (Ambion, Austin, Texas, USA). The marA probe was a 387 base pairs (bp) PCR fragment containing the complete marA gene amplified from E. coli AG100 chromosomal DNA. The marA probe was purified with a gel extraction kit (QIAEXII gel extraction kit; Qiagen) and labeled with [α−32P]dCTP (High Prime DNA labeling kit; Boehringer Mannheim). The RNA blots were washed twice with 2 × saline-sodium citrate (SSC) buffer/0.1% SDS at room temperature and twice with 0.2 × SSC/0.1% SDS at 68°C. Membranes were air dried and exposed to X-ray film (Kodak BioMax MS film; Eastman Kodak, New Haven, Connecticut, USA) for 48 h and then manually visualized.
Results and discussion
Antimicrobial resistance patterns in swine Salmonella
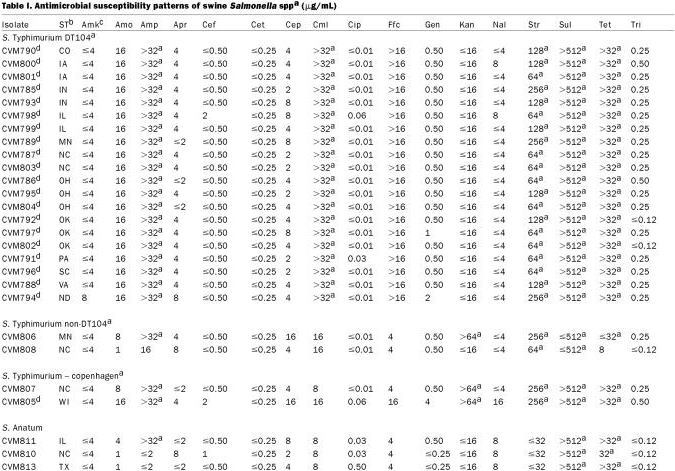
Forty-two Salmonella isolates recovered from diseased swine were tested for their resistance to antimicrobial agents of human and veterinary significance, using a broth microdilution method and interpreted using NCCLS standards (Table I). All S. Typhimurium DT104 isolates displayed the penta-resistance phenotype to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (ACSSuT). Three out of 4 non-DT104 S. Typhimurium strains displayed resistance to ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline. Ninety-eight percent (41/42) of Salmonella isolates were resistant to at least 2 different antimicrobial classes. One out of 2 S. Infantis isolates (CVM814) was resistant to amoxicillin, apramycin, cephalothin, chloramphenicol, gentamicin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline (Table I). Only 1 isolate, S. Mbandaka, was resistant to sulfamethoxazole and trimethoprim. Among the non-DT104 Salmonella isolates, resistance was most often observed to sulfamethoxazole, tetracycline, streptomycin, kanamycin, and ampicillin, as has been reported by other investigators (21). Interestingly, 95% (n = 40/42) and 88% (n = 37/42) of all Salmonella isolates exhibited resistance to sulfamethoxazole and tetracycline, respectively. None of the DT104 strains displayed resistance to kanamycin; however, 41% (n = 9/22) of non-DT104 Salmonella strains exhibited resistance to this antimicrobial. Sulfonamides, tetracyclines, and aminoglycosides are widely used in the swine production environment for treatment and prevention of disease, and for growth promotion (21,22,23). Therefore, it is not unexpected to see increased resistance to these antimicrobials among swine Salmonella serotypes.
Table I.
Table I continued.
Integron mediated antimicrobial resistance in swine Salmonella
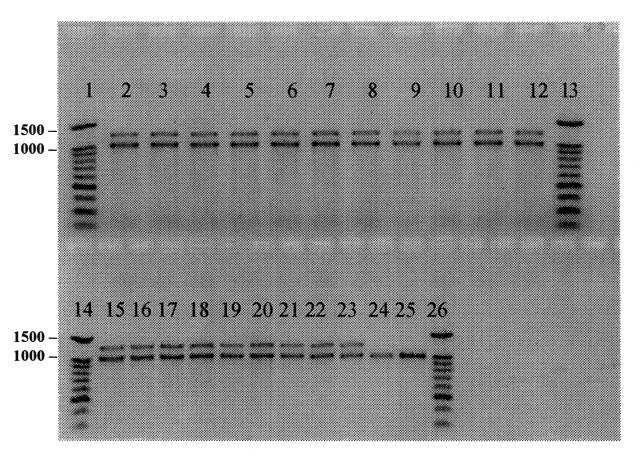
As was expected, all 20 swine DT104 strains exhibiting the penta-resistance phenotype, possessed 2 integron PCR amplicons, of 1000 bp and 1200 bp in size (Figure 1). The DNA sequencing revealed that the 1000 bp integron contained a gene encoding resistance to streptomycin (aadA2 aminoglycoside 3′ adenyltransferase), whereas the 1200 bp integron contained a gene encoding resistance to ampicillin (bla PSE-1 β-lactamase). These have been previously described by other investigators and have been shown to be located on the chromosome rather than on transmissible plasmids (4,5,24,25). None of the non-DT104 strains showed this same pattern although 2 strains (CVM805; S. Typhimurium var Copenhagen and CVM817; S. Derby), produced a 1000 bp integron upon PCR amplification (Figure 1). The DNA sequencing revealed that the 1000 bp integron that recovered from CVM805-S. Typhimurium var Copenhagen contained the aadA gene, encoding resistance to streptomycin and spectinomycin. Whereas the 1000 bp integron amplified from CVM817-S. Derby contained the aadA2 gene as described previously, also encoding resistance to streptomycin and spectinomycin. Integrons containing streptomycin resistant determinants have been previously described in a wide range of Salmonella serotypes including S. Enteritidis, S. Derby, S. Anatum, S. Typhimurium non-DT104, and numerous others (24,26,27,28). It is interesting to note that many of the Salmonella isolates were resistant to streptomycin but did not possess integrons suggesting the presence of alternative resistance mechanisms for this antimicrobial. The majority of non-S. Typhimurium strains did not possess integrons as determined by PCR (Table I), suggesting that integrons are not as widely disseminated among non-S. Typhimurium swine Salmonella serotypes, as had been previously thought. Therefore, it is of interest to continually screen for these DNA elements as part of any future epidemiological investigation aimed at characterizing the dissemination of antimicrobial resistance among Salmonella serotypes.
Figure 1. Polymerase chain reaction (PCR) amplification of integrons among swine Salmonella isolates. The PCR products generated using integron specific primers (5) were run on a 1% agarose gel. A 100 base pairs (bp) ladder (Promega) was used as the molecular size standard in lanes 1,13,14, and 26. Lanes 2 to 12 and 15 to 23 are S. Typhimurium DT104 isolates (CVM785 and 804, respectively). Lanes 24 to 25 are S. Typhimurium var Copenhagen (CVM805) and S. Derby (CVM817).
Characterization of Salmonella Mar mutants
Salmonella isolates were assayed for growth in the presence of the organic solvent cyclohexane. Cyclohexane tolerance in E. coli and Salmonella has been linked to increased antibiotic resistance due to upregulation of efflux pump mechanisms via overexpression of the marA and soxS regulatory genes (10,11). Cyclohexane tolerance was observed in 1 DT104 strain and 1 S. Typhimurium Copenhagen non-DT104 isolate (CVM798 and 805, respectively). Interestingly, these 2 strains differed from other Salmonella strains tested in that they displayed decreased susceptibility to ceftiofur and ciprofloxacin (minimal inhibitory concentration (MIC) of 2 μg/mL and 0.06 μg/mL to ceftiofur and ciprofloxacin, respectively) (Table I). This decrease in susceptibility to ciprofloxacin is especially troublesome since some researchers have noted that DT104 strains may acquire resistance to this drug (29,30). Emergence of resistance to ciprofloxacin in Salmonella is potentially disturbing since it is often the drug of choice for treating human salmonellosis (29).
Expression of marA was evaluated by Northern blot analysis in isolates that demonstrated increased organic solvent tolerance. The RNA from the 2 isolates (CVM798 and 805), displaying increased organic solvent tolerance and wild type E. coli AG100 and Mar mutant E. coli AG102, were probed with an E. coli-derived marA gene probe. The marA gene encodes a transcriptional activator of the marRAB operon (14,31). The marA gene was not overexpressed in these 2 isolates (data not shown). Failure to detect marA expression in organisms exhibiting a mar-like phenotype suggests that these 2 Salmonella isolates may be overexpressing other genes; such as, soxS, the efflux pump acrAB, or both (13,15). Further studies are needed to determine the molecular basis of the observed Mar phenotype in these 2 isolates.
Pulsed-field gel electrophoresis (PFGE) profiles
To assess genetic relatedness among the swine Salmonella isolates, pulsed-field gel electrophoresis (PFGE) was used. The PFGE revealed 20 distinct genetic patterns among the 42 isolates (Figure 2). All S. Typhimurium DT104 isolates fell within 2 closely related genetic clusters, confirming other studies which have demonstrated the highly clonal nature of S. Typhimurium DT104 (24,32,33). No correlation was observed between the state of origin of the DT104 isolate and genetic clustering. Two out of the 3 S. Heidelburg isolates (CVM822 and 815) possessed both identical PFGE and antimicrobial resistance patterns (kanamycin, streptomycin, sulfamethoxazole, and tetracycline), but were isolated from diseased swine in Illinois and North Carolina, respectively (Figure 2). Identical PFGE patterns with different antimicrobial susceptibility patterns were observed between 2 out of 4 S. Choleraesuis isolates and 2 out of 3 S. Infantis isolates. In both cases, however, isolates originated from diseased swine in different states; Kansas and Illinois for S. Choleraesuis, and Idaho and North Dakota for S. Infantis (Figure 2). One out of the 2 S. Infantis isolates (CVM814) that shared identical PFGE patterns also displayed multiple resistance to amoxicillin-clavulanic acid, streptomycin, apramycin, cephalothin, chloramphenicol, gentamicin, kanamycin, sulfamethoxazole, and tetracycline. The other S. Infantis isolate (CVM812) that shared the same PFGE pattern as this multidrug resistant strain was only resistant to sulfamethoxazole. Since both S. Infantis isolates that shared the same PFGE pattern were negative for integrons, this suggests that CVM814 has acquired the multidrug resistance phenotype from some type of mobile DNA element, most likely a plasmid. Overall, PFGE typing grouped the majority of isolates according to serotype, with 1 S. Mbandaka strain being untypeable by PFGE and 1 S. Derby (CVM819) appearing to be more related to the S. Infantis cluster than the other Derby cluster (Figure 2). This data supports the findings of other investigators that PFGE, using XbaI restriction endonuclease, is a sensitive method for fingerprinting diverse Salmonella serotypes (32,34,35). As has been reported with isolates from other animals, our results show that S. Typhimurium DT104 isolates from diseased swine in the United States are clonal in origin and geographically widely distributed. Although only a small number of isolates were analyzed in this study, our preliminary data suggests that there are specialized swine pathogenic clones of other Salmonella serotypes (for example, Heidelburg, Choleraesuis, and Infantis) circulating among diseased swine in the United States.
Figure 2. Genetic relatedness among swine Salmonella isolates. Dendrogram of pulsed-field gel electrophoresis (PFGE) patterns of Salmonella isolates recovered from diseased swine, and their association with serotype, state of origin, antimicrobial resistance profile, and integron content.
The PFGE patterns of Salmonella species were cleaved with restriction enzyme Xba1. The percent similarity among the tested Salmonella isolates is represented by a dendrogram generated using Molecular Analyst Fingerprinting Plus Software (Bio-Rad). The percent similarity between PFGE types is shown at the top of the figure; scale at 100 means identical.
a A, ampicillin; Ap, apramycin; Ce, cephalothin; C, chloramphenicol; G, gentamicin; K, kanamycin; S, streptomycin; Su, sulfamethoxazole; T, tetracycline; T/S, trimethoprim/sulfamethoxazole.
b Size of integron/resistance gene: aadA, aminoglycoside adenyltransferase; aadA2, aminoglycoside adenyltransferase; blaPSE-1, beta-lactamase.
In summary, this research adds to the growing body of knowledge concerning antimicrobial resistance among swine Salmonella and demonstrates that multiple mechanisms, including integrons, contribute to resistance. Although we did not detect overexpression of marA, a known regulator of multi-drug efflux pumps, our data imply the presence of an active efflux in 2 multi-resistant strains (CVM 798 and 805), as evidenced by their tolerance to organic solvents. Large differences in susceptibility profiles between strains with identical PFGE patterns suggests the involvement of multi-drug resistance plasmids. Unlike plasmid-mediated resistance, which may disappear in the absence of continued selective pressure, chromosomally mediated resistance is often maintained; thus, it is necessary to implement measures to eliminate the resistant strain to prevent transfer among animals and between animals and humans. In an effort to reduce the prevalence of multi-drug resistant Salmonella in swine, additional preventive animal health management factors, besides antimicrobial use, should be considered; such as, increased biosecurity, administration of probiotics, prebiotics, and vaccination (36).
Because our findings are based on a limited sample size, further research with a larger number of non-DT104 strains is needed to better assess both the clonality of geographically dispersed isolates and the array of resistance mechanisms operative in swine Salmonella.
Footnotes
Acknowledgments
The authors thank Kathleen Ferris, Bacterial Identification Group, National Veterinary Services Laboratory, Ames, Iowa, USA, for the strains used in this research. We also thank the National Pork Board (#1998-218) for their generous support.
Address all correspondence and reprint requests to Dr. D.G. White; telephone: (301) 827-8037; fax: (301) 827-8127; e-mail: dwhite@cvm.fda.gov
Received January 29, 2002. Accepted June 25, 2002.
References
- 1.Gold HS, Moellering RC Jr. Antimicrobial-drug resistance. N Engl J Med 1996;335:1445–1453. [DOI] [PubMed]
- 2.Hall RM. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found Symp 1997;207:192–202. [DOI] [PubMed]
- 3.Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 1995;39:185–191. [DOI] [PMC free article] [PubMed]
- 4.Ridley A, Threlfall EJ. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella Typhimurium DT 104. Microb Drug Resist 1998;4:113–118. [DOI] [PubMed]
- 5.Sandvang D, Aarestrup FM, Jensen LB. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett 1998;160:37–41. [DOI] [PubMed]
- 6.Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 1997;41:2067–2075. [DOI] [PMC free article] [PubMed]
- 7.Cohen SP, Hachler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol 1993;175:1484–1492. [DOI] [PMC free article] [PubMed]
- 8.Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother 1994;38:1773–1779. [DOI] [PMC free article] [PubMed]
- 9.Sulavik M, Dazer CM, Miller PF. The Salmonella Typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol 1997;179:1857–1866. [DOI] [PMC free article] [PubMed]
- 10.Oethinger M, Kern WV, Goldman JD, Levy SB. Association of organic solvent tolerance and fluoroquinolone resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother 1998;41:111–114. [DOI] [PubMed]
- 11.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol 1997; 179:6122–6126. [DOI] [PMC free article] [PubMed]
- 12.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 1996;178:306–308. [DOI] [PMC free article] [PubMed]
- 13.Koutsolioutsou A, Martins EA, White DG, et al. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (Serovar typhimurium). Antimicrob Agents Chemother 2001;45:38–43. [DOI] [PMC free article] [PubMed]
- 14.Oethinger M, Podglajen I, Kern WV, Levy SB. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother 1998;42:2089–2094. [DOI] [PMC free article] [PubMed]
- 15.Piddock LJ, White DG, Gensberg K, Pumbwe L, Griggs DJ. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 2000;44:3118–3121. [DOI] [PMC free article] [PubMed]
- 16.Webber MA, Piddock LJ. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother 2001;45:1550–1552. [DOI] [PMC free article] [PubMed]
- 17.National Committee for Clinical Laboratory Standards (M7-A5). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—5th ed; Approved Standard. National Committee for Clinical Laboratory Standards, Villanova, Pennsylvania. 2001.
- 18.National Committee for Clinical Laboratory Standards (M31-A2). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. National Committee for Clinical Laboratory Standards, Villanova, Pennsylvania. 2002.
- 19.Tollefson L, Angulo FJ, Fedorka-Cray PJ. National surveillance for antibiotic resistance in zoonotic enteric pathogens. Vet Clin North Am Food Anim Pract 1998;14:141–150. [DOI] [PubMed]
- 20.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402. [DOI] [PMC free article] [PubMed]
- 21.Poppe C, Ayroud M, Ollis G, et al. Trends in antimicrobial resistance of Salmonella isolated from animals, foods of animal origin, and the environment of animal production in Canada, 1994–1997. Microb Drug Resist 2001;7:197–212. [DOI] [PubMed]
- 22.Friendship RM. Antimicrobial drug use in swine. In: Prescott JF, Baggot JD, Walker RD, eds. Antimicrobial Therapy in Veterinary Medicine. Ames, Iowa: Iowa State University Press, 2000:602–611.
- 23.Schwarz S, Kehrenberg C, Walsh TR. Use of antimicrobial agents in veterinary medicine and food animal production. Int J Antimicrob Agents 2001;17:431–437. [DOI] [PubMed]
- 24.Daly M, Buckley J, Power E, et al. Molecular characterization of Irish Salmonella enterica serotype typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl Environ Microbiol 2000;66:614–619. [DOI] [PMC free article] [PubMed]
- 25.Ng LK, Mulvey MR, Martin I, et al. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother 1999;43:3018–3021. [DOI] [PMC free article] [PubMed]
- 26.Brown AW, Rankin SC, Platt DJ. Detection and characterisation of integrons in Salmonella enterica serotype enteritidis. FEMS Microbiol Lett 2000;191:145–149. [DOI] [PubMed]
- 27.Daly M, Fanning S. Characterization and chromosomal mapping of antimicrobial resistance genes in Salmonella enterica serotype typhimurium. Appl Environ Microbiol 2000;66:4842–4848. [DOI] [PMC free article] [PubMed]
- 28.Guerra B, Soto S, Cal S, Mendoza MC. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob Agents Chemother 2000;44:2166–2169. [DOI] [PMC free article] [PubMed]
- 29.Glynn MK, Bopp C, Dewitt W, et al. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med 1998;338:1333–1338. [DOI] [PubMed]
- 30.Herikstad H, Hayes P, Mokhtar M, et al. Emerging quinolone-resistant Salmonella in the United States. Emerg Infect Dis 1997;3:371–372. [DOI] [PMC free article] [PubMed]
- 31.Martin RG, Jair KW, Wolf RE Jr, Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol 1996;178:2216–2223. [DOI] [PMC free article] [PubMed]
- 32.Malorny B, Schroeter A, Bunge C, et al. Evaluation of molecular typing methods for Salmonella enterica serovar Typhimurium DT104 isolated in Germany from healthy pigs. Vet Res 2001;32:119–129. [DOI] [PubMed]
- 33.Murphy TM, McNamara E, Hill M, et al. Epidemiological studies of human and animal Salmonella typhimurium DT104 and DT104b isolates in Ireland. Epidemiol Infect 2001;126:3–9. [DOI] [PMC free article] [PubMed]
- 34.Liebana E, Guns D, Garcia-Migura L, et al. Molecular typing of Salmonella serotypes prevalent in animals in England: assessment of methodology. J Clin Microbiol 2001;39:3609–3616. [DOI] [PMC free article] [PubMed]
- 35.Valdezate S, Echeita A, Diez R, Usera MA. Evaluation of phenotypic and genotypic markers for characterisation of the emerging gastroenteritis pathogen Salmonella hadar. Eur J Clin Microbiol Infect Dis 2000;19:275–281. [DOI] [PubMed]
- 36.Letellier A, Messier S, Lessard L, et al. Host response to various treatments to reduce Salmonella infections in swine. Can J Vet Res 2001;65:168–172. [PMC free article] [PubMed]