Abstract
Background
Rhodnius prolixus is the main vector of Chagas disease in Venezuela. Here, domestic infestations of poor quality rural housing have persisted despite four decades of vector control. This is in contrast to the Southern Cone region of South America, where the main vector, Triatoma infestans, has been eliminated over large areas. The repeated colonisation of houses by silvatic populations of R. prolixus potentially explains the control difficulties. However, controversy surrounds the existence of silvatic R. prolixus: it has been suggested that all silvatic populations are in fact Rhodnius robustus, a related species of minor epidemiological importance. Here we investigate, by direct sequencing (mtcytb, D2) and by microsatellite analysis, 1) the identity of silvatic Rhodnius and 2) whether silvatic populations of Rhodnius are isolated from domestic populations.
Methods and Findings
Direct sequencing confirmed the presence of R. prolixus in palms and that silvatic bugs can colonise houses, with house and palm specimens sharing seven cytb haplotypes. Additionally, mitochondrial introgression was detected between R. robustus and R. prolixus, indicating a previous hybridisation event. The use of ten polymorphic microsatellite loci revealed a lack of genetic structure between silvatic and domestic ecotopes (non-significant FST values), which is indicative of unrestricted gene flow.
Conclusions
Our analyses demonstrate that silvatic R. prolixus presents an unquestionable threat to the control of Chagas disease in Venezuela. The design of improved control strategies is essential for successful long term control and could include modified spraying and surveillance practices, together with housing improvements.
Author Summary
Chagas disease is spread by blood-feeding insects (triatomine bugs) that colonise poor-quality houses. Disease control relies primarily on killing domestic bugs by spraying dwellings with residual insecticide. In Venezuela, sustained control has proved difficult despite four decades of campaigns. Considered the main vector in Venezuela, the bug Rhodnius prolixus may also infest palm trees and might repeatedly recolonise houses from palms. A complication is that a morphologically similar species, R. robustus, also infests palms but is of minor medical importance. Therefore, confusion exists as to the true identity of palm bugs and their importance in disease transmission.
We applied two molecular methods (sequencing DNA of the cytochrome b gene, and analysing microsatellites) to triatomines collected in Venezuela so that we could identify unequivocally the species of palm-dwelling Rhodnius and establish their role in maintaining house infestations. We demonstrated that R. prolixus is indeed present in palms, and that such silvatic populations can colonise houses and are a threat to the successful control of Chagas disease in Venezuela. This finding resolves a longstanding controversy of fundamental epidemiological importance. It is also an example of the application of molecular epidemiology to correct vector identification and successful disease control.
Introduction
Chagas disease is a chronic parasitic disease transmitted by triatomine bugs (Reduviidae: Triatominae) and limited in distribution to the Americas. The causative agent is the protozoan Trypanosoma cruzi. Rhodnius prolixus is the primary vector in Venezuela and Colombia and is one of the main targets of the Andean Pact and Central American initiatives, together with the secondary vectors Triatoma dimidiata in Central America, Rhodnius pallescens in Panama and Rhodnius ecuadoriensis in northern Peru [1]. In Venezuela R. prolixus occurs in all States, where it colonises poor quality housing and exhibits high infection rates with T. cruzi.
Significant progress has been made in reducing the incidence of Chagas disease in Venezuela through four decades of triatomine control [2]. Nevertheless, domestic infestations of R. prolixus persist and recent data indicate that transmission of T. cruzi may be increasing [3]. In contrast, in the Southern Cone region of South America the main vector, Triatoma infestans, has been eliminated over large areas following control efforts [1]. Triatoma infestans is considered to be a primarily domestic species, with the exception of Bolivian Andes and Gran Chaco region (Bolivia and northern Argentina) where silvatic populations were found [4]. Further studies are needed to evaluate the risk these populations pose to effective control in these regions. In comparison R. prolixus is reported to have a widespread silvatic distribution in Venezuela, found most commonly in palm trees and birds nests and more rarely in other sites such as dry trees [5]–[7]. The reinvasion of sprayed houses by silvatic R. prolixus, together with localised control failures could be maintaining disease transmission in Venezuela [3]. However, the existence of silvatic R. prolixus populations has been questioned due to the identification of the closely related species Rhodnius robustus in palm trees in Venezuela [8]. Rhodnius robustus poses a problem as it is virtually indistinguishable morphologically from R. prolixus but this species it is of minor epidemiological importance as it does not colonise houses, although flying adults may enter domestic areas attracted by light [8],[9]. Confusion has been fuelled by conflicting results of studies investigating the taxonomic status of R. prolixus and R. robustus, with morphometric and isoenzyme studies failing to detect interspecific differences [10]–[15]. However, recent DNA sequencing analyses has not only supported the validity of R. robustus but also indicated the existence of more than one cryptic species [16]–[18]. Additionally in a preliminary finding for this present study four Rhodnius specimens collected in a palm in Guarico State Venezuela were identified as R. prolixus [17].
Here we investigated the genetic structure of 34 populations of R. prolixus, including five adjacent populations, from silvatic, domestic and peridomestic ecotopes in six Venezuelan States. Our aim was to contribute to the control of Chagas disease in Venezuela, through the provision of information that might allow the design of improved control strategies. We finally resolve this controversy over the existence of silvatic R. prolixus and the interaction between silvatic and domestic populations. Our analyses demonstrate that silvatic R. prolixus presents an unquestionable threat to the control of Chagas disease in Venezuela and that successful long term control could benefit from modified spraying and surveillance practices, together with housing improvement.
Materials and Methods
Bug collection
For the purpose of this study field work was carried out in 2001–2004 in the Venezuelan States of Lara, Portuguesa, Guarico, Cojedes, Barinas, and Trujillo (see Figure 1, Table 1, Table 2). Fieldwork involved the survey of palms, chicken huts and houses in localities in these States in collaboration with the Ministry of Health field inspectors.
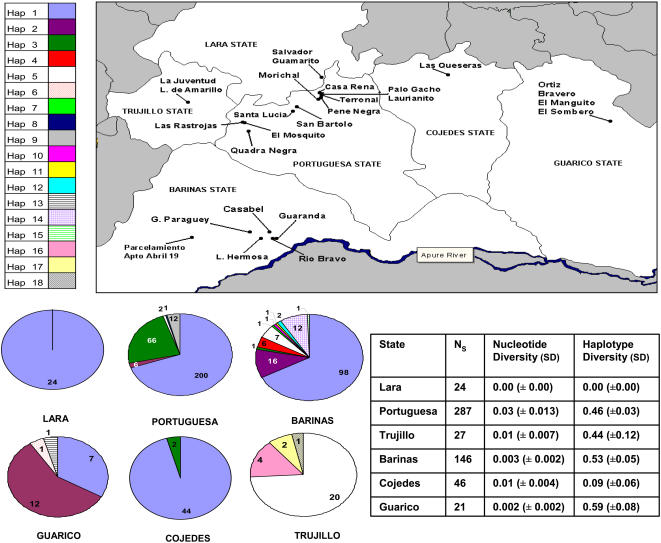
Figure 1. Genetic diversity (table) and haplotype distribution (pie charts) in the sampled States.
The map illustrates 27 of the sampled localities in this study.
Table 1. Details of the 34 populations analysed by direct sequencing.
| Pop ID | State | Locality | Ecotope | Collection Date | NS | Haplotypes Detected | Nucleotide Diversity (SD) | Gene Diversity (SD) |
| Pop 1 | Portuguesa | Terronal | House 1 | 2001 | 22 | 1,3 | 0.03 (±0.02) | 0.46 (±0.08) |
| Pop 2 | Portuguesa | Terronal | House 2 | 2001 | 27 | 1,3 | 0.03 (±0.02) | 0.46 (±0.07) |
| Pop 3 | Portuguesa | Terronal * | Palm 1 by pop 2 | 2001 | 30 | 1,2,3 | 0.02 (±0.01) | 0.32 (±0.09) |
| Palms by pop 2 | 2001 | 7 | ||||||
| Pop 4 | Portuguesa | Terronal * | House 1 | 2003 | 10 | 1,3 | 0.037 (±0.02) | 0.53 (±0.06) |
| House 2 | 2003 | 4 | ||||||
| Pop 5 | Portuguesa | Terronal | Palm 2 by pop 2 | 2003 | 35 | 1,3 | 0.02 (±0.01) | 0.33 (±0.08) |
| Pop 6 | Portuguesa | Los Rastrojos | House | 2004 | 22 | 1,2 | 0.0004 (±0.001) | 0.17 (±0.10) |
| Pop 7 | Portuguesa | Los Rastrojos | Palm | 2004 | 10 | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 8 | Portuguesa | Palo Gacho * | Palms | 2001 | 10 | 1,3 | 0.04 (±0.02) | 0.53 (±0.10) |
| Pop 9 | Portuguesa | San Bartolo * | House 1 (House 25) | 2002 | 16 (2) | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| House 2 (Chicken hut) | 2002 | 13 (2) | ||||||
| Pop 10 | Portuguesa | Santa Lucia * | House 89 | 2002 | 14 | 1,9 | 0.001 (±0.001) | 0.44 (±0.10) |
| Other houses | 2002 | 3 | ||||||
| Pop 11 | Portuguesa | Quebrada Negra * | Houses | 2001 | 12 | 1,2,5,8 | 0.003 (±0.002) | 0.56 (±0.15) |
| Pop 12 | Portuguesa | Peña Negra | House | 2001 | 10 | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 13 | Portuguesa | Casarena | House | 2003 | 10 | 3 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 14 | Barinas | Carreterón | Palm 1 (Palm 2) | 2003 | 7 (1) | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 15 | Barinas | Carreterón * | Houses | 2003 | 13 | 1,3,4,5,7 | 0.013 (±0.008) | 0.69 (±0.12) |
| Pop 16 | Barinas | Cascabel | Chicken hut | 2003 | 9 | 1,2,4 | 0.002 (±0.002) | 0.64 (±0.13) |
| Pop 17 | Barinas | Cascabel | House | 2003 | 8 | 1,2,5 | 0.001 (±0.001) | 0.46 (±0.20) |
| Pop 18 | Barinas | Cascabel | Palm | 2003 | 15 | 1,2,5,12 | 0.002 (±0.002) | 0.66 (±0.08) |
| Pop 19 | Barinas | El Guamito | House | 2003 | 5 | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 20 | Barinas | El Guamito | Palm | 2003 | 12 | 1,10,2 | 0.001 (±0.001) | 0.32 (±0.16) |
| Pop 21 | Barinas | Laguna Hermosa | House | 2003 | 11 | 1,5 | 0.0004 (±0.001) | 0.18 (±0.14) |
| Pop 22 | Barinas | Laguna Hermosa | Chicken hut | 2003 | 7 | 1,5,12 | 0.002 (±0.002) | 0.52 (±0.21) |
| Pop 23 | Barinas | Laguna Hermosa | Palm | 2003 | 9 | 1,2,5,11 | 0.002 (±0.002) | 0.58 (±0.18) |
| Pop 24 | Barinas | G. Paguey * | House 2 (House 1) | 2003 | 8 (3) | 1,2,5 | 0.001 (±0.001) | 0.46 (±0.20) |
| Pop 25 | Barinas | Parcelamiento * | Palm | 2003 | 10 | 1,14,15 | 0.001 (±0.001) | 0.51 (±0.16) |
| Pop 26 | Barinas | 19 Abril | Chicken hut | 2003 | 10 | 14 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 27 | Barinas | Rio Bravo II | Chicken hut | 2003 | 6 | 1,2 | 0.001 (±0.001) | 0.33 (±0.22) |
| Pop 28 | Barinas | Rio Bravo II | Palm | 2003 | 9 | 1,2 | 0.001 (±0.001) | 0.40 (±0.16) |
| Pop 29 | Cojedes | Las Queseras | House | 2004 | 22 | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Pop 30 | Cojedes | Las Queseras | Palm | 2004 | 24 | 1, 3 | 0.01 (±0.006) | 0.16 (±0.10) |
| Pop 31 | Lara | Guamito | House 1 | 2001 | 22 | 1 | 0.00 (±0.00) | 0.00 (±0.00) |
| Salvador | House 2 | 2001 | 2 | |||||
| Pop 32 | Guarico | Bravero, Ortiz | Various palms | 2001 | 21 | 1,2,6,13 | 0.002 (±0.002) | 0.59 (±0.08) |
| El Manguito, | ||||||||
| El Sombero | ||||||||
| Pop 33 | Trujillo | L. de Amarillo | House | 2003 | 21 | 5,16 | 0.003 (±0.002) | 0.095 (±0.08) |
| Pop 34 | Trujillo | La Juventud | Palm | 2003 | 3 | 16,17 | ||
| Insectary ˆ | Palm | 1995 | 3 | 16,18 | 0.002 (±0.002) | 0.73 (±0.16) | ||
| Other | Portuguesa | Terronal * | Chicken hut by pop2 | 2001 | 1 | 1 | - | - |
| Other | Portuguesa | Terronal | Palms by pop 1 | 2003 | 6 | 1 | - | - |
| Other | Portuguesa | Casarena * | Palm | 2001/2003 | 5 | |||
| Chicken hut | 2003 | 2 | 1,3 | - | - | |||
| Other | Portuguesa | El Mosquito * | Houses | 2001 | 8 | 1,2 | - | - |
| Other | Portuguesa | Palmarito * | House 1 (Palm) | 2001 | 5 (1) | 1,3 | - | - |
| Other | Portuguesa | Morichal * | House 10.1 (House 10) | 2001 | 5 (1) | 1,3 | - | - |
| Other | Barinas | Various ∞ | House, Palm, Chicken hut | 2001/3 | 7 | 1,4 | - | - |
Note NS = Total no of specimens sequenced * insects from more than one sample site combined and analysed as one population, in parenthesis the no. of specimens from other populations included in the total number sequenced and the ecotope in which they originated; ˆ insectary reared bugs, originally collected in palms (source University of Los Andes, Trujillo); Other = specimens sequenced but not included in population analysis due to small numbers or multiple sample sites. ∞ the localities Acequita, Santa Elena de la Caramuca, Obispos and San Isidro. See Table S1, S3, and S4 for all FST values.
Table 2. Details of the 33 populations analysed by microsatellites.
| Pop ID | State | Locality | Ecotope | Collection Date | NM | NL | LP | AM | AR | NA | HO | HE | FIS |
| Pop 1 | Portuguesa | Terronal | House 1 | 2001 | 26 | 9 | 9 | 2.6 | 2.3 | 3 | 0.3 | 0.4 | 0.2 |
| Pop 2 | Portuguesa | Terronal | House 2 | 2001 | 18 | 9 | 9 | 2.7 | 2.4 | 1 | 0.4 | 0.5 | 0.1 |
| Pop 3 | Portuguesa | Terronal | Palm by pop2 | 2001 | 26 | 9 | 9 | 2.4 | 2 | 2 | 0.2 | 0.3 | 0.3 |
| Pop 4 | Portuguesa | Terronal | House 1 | 2003 | 10 | 9 | 9 | 2.5 | 2.5 | 2 | 0.4 | 0.5 | 0.1 |
| Pop 5 | Portuguesa | Terronal | Palm by pop2 | 2003 | 39 | 9 | 9 | 3.0 | 2.4 | 5 | 0.4 | 0.5 | 0.1 |
| Pop 6 | Portuguesa | Los Rastrojos | House | 2004 | 24 | 10 | 9 | 2.6 | 2.3 | 1 | 0.3 | 0.4 | 0.03 |
| Pop 7 | Portuguesa | Los Rastrojos | Palm | 2004 | 12 | 10 | 10 | 2.9 | 2.7 | 1 | 0.4 | 0.4 | 0 |
| Pop 8 | Portuguesa | Palo Gacho * | Palms | 2001 | 15 | 9 | 8 | 2.6 | 2.3 | - | 0.3 | 0.4 | 0.2 |
| Pop 9a | Portuguesa | San Bartolo | House 1 | 2002 | 14 | 9 | 7 | 2.4 | 2.4 | 1 | 0.4 | 0.4 | 0.1 |
| Pop 9b | Portuguesa | San Bartolo | House 2 | 2002 | 14 | 9 | 7 | 2.3 | 2.3 | 3 | 0.3 | 0.4 | 0.3 |
| Pop 10 | Portuguesa | Santa Lucia | House 89 | 2002 | 13 | 9 | 6 | 1.9 | 1.7 | - | 0.2 | 0.3 | 0.1 |
| Pop 11 | Portuguesa | Qda Negra | Houses | 2001 | NA | ||||||||
| Pop 12 | Portuguesa | Peña Negra | House | 2001 | NA | ||||||||
| Pop 13 | Portuguesa | Casarena | House | 2003 | 11 | 9 | 7 | 2.2 | 2.2 | - | 0.3 | 0.4 | 0.02 |
| Pop 14 | Barinas | Carreterón | Palms | 2003 | NA | ||||||||
| Pop 15 | Barinas | Carreterón | Houses | 2003 | NA | ||||||||
| Pop 16 | Barinas | Cascabel | Chicken hut | 2003 | 11 | 10 | 10 | 2.8 | 2.7 | 1 | 0.4 | 0.5 | 0.1 |
| Pop 17 | Barinas | Cascabel | House | 2003 | 10 | 10 | 10 | 3.1 | 3.1 | 2 | 0.5 | 0.6 | 0.1 |
| Pop 18 | Barinas | Cascabel | Palm | 2003 | 24 | 10 | 10 | 4.6 | 3.6 | 1 | 0.5 | 0.6 | 0.1 |
| Pop 19 | Barinas | El Guamito | House | 2003 | 11 | 10 | 10 | 3.3 | 3.2 | 1 | 0.4 | 0.5 | 0.1 |
| Pop 20 | Barinas | El Guamito | Palm | 2003 | 20 | 10 | 10 | 4.0 | 3.3 | 3 | 0.5 | 0.6 | 0.02 |
| Pop 21 | Barinas | Laguna Hermosa | House | 2003 | 16 | 10 | 10 | 3.6 | 3.2 | 1 | 0.6 | 0.6 | -0.1 |
| Pop 22 | Barinas | Laguna Hermosa | Chicken hut | 2003 | 13 | 10 | 10 | 2.7 | 2.6 | - | 0.4 | 0.5 | 0 |
| Pop 23 | Barinas | Laguna Hermosa | Palm | 2003 | 17 | 10 | 10 | 4.3 | 3.6 | 1 | 0.5 | 0.6 | 0.04 |
| Pop 24a | Barinas | G. Paguey | House 1 | 2003 | 11 | 10 | 10 | 3.1 | 3.0 | - | 0.4 | 0.6 | 0.2 |
| Pop 24b | Barinas | G. Paguey | House 2 | 2003 | 12 | 10 | 10 | 3.4 | 3.2 | - | 0.4 | 0.6 | 0.2 |
| Pop 24c | Barinas | G. Paguey | Palm 1 | 2003 | 12 | 10 | 10 | 3.4 | 3.2 | - | 0.5 | 0.6 | 0.1 |
| Pop 24d | Barinas | G. Paguey | Palm 2 | 2003 | 11 | 10 | 10 | 3.4 | 3.3 | - | 0.5 | 0.6 | 0.01 |
| Pop 25 | Barinas | Parcelamiento* | Palms | 2003 | 13 | 10 | 10 | 3.5 | 3.2 | 4 | 0.5 | 0.6 | 0.1 |
| Pop 26 | Barinas | 19 Abril | Chicken hut | 2003 | 13 | 10 | 10 | 2.7 | 2.5 | - | 0.4 | 0.5 | 0.1 |
| Pop 27 | Barinas | Rio Bravo II | Chicken hut | - | 17 | 10 | 10 | 3.0 | 2.7 | 3 | 0.5 | 0.5 | 0.01 |
| Pop 28 | Barinas | Rio Bravo II | Palm | - | 10 | 10 | 10 | 3.6 | 3.5 | 2 | 0.5 | 0.6 | 0.1 |
| Pop 29 | Cojedes | Las Queseras | Palm | 2004 | 24 | 10 | 10 | 3.0 | 2.6 | 3 | 0.4 | 0.5 | 0.1 |
| Pop 30 | Cojedes | Las Queseras | House | 2004 | 24 | 10 | 10 | 2.2 | 2.1 | 1 | 0.3 | 0.4 | 0.2 |
| Pop 31 | Lara | Guamito | House 1 | 2001 | 15 | 9 | 9 | 2.2 | 2.0 | 1 | 0.3 | 0.4 | 0.2 |
| Salvador | House 2 | 2 | |||||||||||
| Pop 32 | Guarico | Brav., Ortiz | Various palms | 2001 | NA | ||||||||
| El Man. El Som. | |||||||||||||
| Pop 33 | Trujillo | Loma de Amarillo | House | 2003 | 26 | 9 | 9 | 3.0 | 2.2 | 3 | 0.2 | 0.3 | 0.3 |
| Pop 34 | Trujillo | La Juv./Insectary | Palm | 2003/1995 | NA | ||||||||
| Pop 35 | Portuguesa | Laurianito | Chicken hut | 2003 | 21 | 9 | 9 | 3.1 | 2.7 | - | 0.4 | 0.5 | 0.1 |
Note * insects from more than one sample site combined and analysed as one population; NM = Total no. of specimens analysed by microsatellites; NL = no of loci analysed; Lp = no. of polymorphic loci; AM = mean no of alleles detected averaged over all loci, AR = allele richness averaged over all loci; NA = Null alleles; HO, HE = Observed and Expected Heterozygosity averaged over all loci; FIS = Inbreeding Coefficient averaged over all loci. NA = population not analysed by microsatellites. See Tables S1, S3, and S4 for all FST values.
Sampling methods
Silvatic collections were made with Noireau live bait traps [19]. Palm dissection was also used with the consent of landowners. The palm was cut at the base and cleared from the base up to the crown using a machete, removing and inspecting each layer. Domestic and peridomestic collections were made by the traditional search and capture method, with prior consent of householders. All bugs collected were placed in collection tubes, noting date and place of collection. Specimens were identified using the keys of Lent and Wygodzinsky (1979) [20].
In Portuguesa State
Bugs were collected in 12 localities from houses, chicken huts and palms. Positive houses were primarily of the traditional ‘rancho’ type, constructed of wattle and daub with palm and corrugated iron roofs. A total of 287 specimens were analysed by direct sequencing and 243 by microsatellite analysis (pop 1 through pop 13 and pop 35; see Table 1, Table 2 for population details).
In Barinas State
Bugs were collected in 13 localities from houses, chicken huts and palms. In these localities houses had walls of wood or cement blocks, with metal or palm roofs. A total of 146 specimens from domestic, silvatic and peridomestic ecotopes in this State were analysed by direct sequencing and 221 by microsatellite analysis (pop 14 through pop 28; see Table 1, Table 2).
In Cojedes State
A single house infestation was detected in the locality Las Queseras. A dissected palm adjacent to the infested house was also positive. A total of 46 specimens were analysed by direct sequencing and 48 by microsatellites (pop 29, pop 30; see Table 1, Table 2).
In Lara State
Two houses were found infested in the localities Guamarito and Salvador, while palm searches proved negative. A total of 24 specimens from this State were examined by direct sequencing, 17 by microsatellite analysis (pop 31; see Table 1, Table 2).
In Guarico State
Specimens were collected in 4 localities (El Sombero, El Manguito, Bravero, Ortiz). All houses inspected were negative and samples were isolated from palms only. In these areas the traditional rancho was replaced by cement block structures as part of the National Programme for housing improvement in the 1960s. A total of 21 specimens were analysed by direct sequencing only (pop 32; see Table 1).
In Trujillo State
A single house was found infested in the locality Loma de Amarillo. A single palm was dissected in the locality La Juventud and was found positive. A total of 27 specimens were analysed by direct sequencing, including 3 insectary specimens derived from palms. Twenty-six domestic specimens were analysed with microsatellites (pop 33, 34; see Table 1, Table 2).
Genomic DNA was isolated from specimens using Qiagen Dneasy extraction kit following the manufacturer's protocol for isolation of DNA from animal tissues.
Species identity and genetic relatedness
Cytb sequencing
In order to confirm which species of Rhodnius were present and to examine the genetic relatedness of R. prolixus and R. robustus populations in Venezuela, a total of 551 specimens were analysed from 6 States by direct sequencing of a fragment of the mitochondrial gene cytochrome b (cytb) (Table 1). Eight published cytb nucleotide sequences from the study of Monteiro et al. 2003 were included as reference specimens (see Figure 2) [17]. In Monteiro et al. 2003 specimens of R. robustus and R. prolixus were distinguished using a combination of the following criteria (1) the morphology of late nymphal stages, as described by Lent and Wygodsinsky (1979, [20]) (2) by the inclusion in cytb typing of R. robustus specimens originally collected from areas close to the suggested ‘type localities’ of the species and specimens from the Brazilian Amazon where silvatic R. prolixus is not believed to occur (3) and by the inclusion of R. prolixus specimens collected from houses in Central America (Honduras/Guatamala), beyond the geographical distribution of silvatic R. prolixus or R. robustus [17]. To test for mtDNA introgression between these closely related species, a fragment of the D2 variable region of 28S RNA was sequenced for nine specimens, characterised by the mtcytb analysis as R. robustus or R. prolixus. Five D2 sequences were also available in GenBank (see Figure 3).
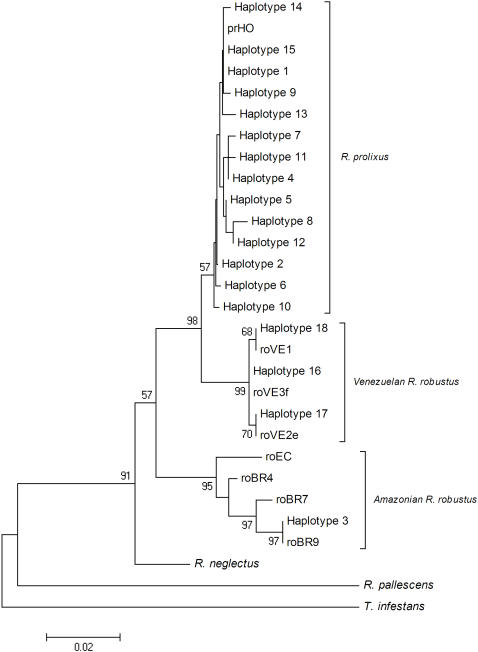
Figure 2. Phylogenetic tree of the 18 cytb haplotypes detected in the study and sequences from GenBank.
Figure 3. The polymorphic sites of the 18 cytb haplotypes and 9 specimens sequenced for D2.
We amplified a 682 bp fragment of the cytb gene and a 633 bp fragment of the D2 region with the following primers: Forward cytb7432F 5′-GGACG(AT)GG(AT)ATTTATTATGGATC; Reverse cytb7433R 5′-GC(AT)CCAATTCA(AG)GTTA(AG)TAA; Forward D2F 5′-GCGAGTCGTGTTGCTTGATAGTGCAG; Reverse D2R 5′-TTGGTCCGTGTTTCAAGACGGG [17],[21]. Reaction conditions were: cytb: 95°C 5 min; 35 cycles of 95°C 30 s, 50°C 45 s, and 72°C 45 s; final extension of 72°C for 5 min. D2: 25 cycles 94°C 1 min, 50°C 2 min, 72°C 2 min. Amplicons were purified using Qiaquick kit (Qiagen) or Quick-clean (Bioline), as specified by the manufacturers. Purified PCR products were sequenced by fluorescent dye terminator chemistry using ABI Prism Bigdye (Applied Biosystematics), on an ABI Prism 377 automated DNA sequencer (PE Applied Biosystematics) or on a 48 capillary ABI 3730 DNA analyser. Forward and reverse sequences were aligned using Sequence Navigator V1.01 (Perkin-Elmer) or BioEdit V7.0.4.1 [22] and a consensus sequence produced. Sequence identity was confirmed by comparison with data in GenBank.
The number of variable sites was determined using Mega v 2.1 software [23]. A neighbour-joining tree was created in Mega v 2.1 using the Kimura-2 parameter model of sequence evolution [23]. Statistical support for clades was assessed by the bootstrap method (1000 replications; [24]). Outgroup sequences were taken from GenBank: R. pallescens AF045720, R. neglectus AF045716 and T. infestans AF045721. All sites were equally weighted.
Data deposition footnote
Cytb haplotype genBank accession numbers. EF043576, EF043577, EF043578, EF043579, EF043580, EF043581, EF043582, EF043583, EF043584, EF043585, EF043586, EF043587, EF043588.
Genetic variation and population structure
Cytb analysis
For population analysis using cytb haplotypes specimens were placed into 34 population groups as listed in Table 1. Groups were determined by the collection site (ecotope). Ideally population groups consisted of specimens isolated from a single ecotope, however when only a few specimens were collected, populations from different houses or palms from the same locality and State were combined (Table 1). Intrapopulation population comparisons, was investigated using the index of population heterogeneity FST (Weir & Cockerhams 1984 unbiased estimator) generated in Arlequin v3.1 [25],[26]. The FST null distribution is obtained by permuting the haplotypes between the compared populations (10,000 times), given a null hypothesis of no difference between the populations (FST = 0). The p-value generated is the fraction of these permutations with an FST larger than or equal to the original estimate, if the given p-value is smaller than the nominated significance level, then the compared populations are considered to be significantly different. The nominal significance level was adjusted for multiple comparisons using the sequential Bonferroni procedure [27]. This consists of setting a lower threshold for the nominal significance level, i.e., for cytb analysis k = 561, p1 = 0.05/561 and p≤0.0001. Population geneflow was evaluated at different geographic levels 1) comparison of adjacent ecotopes, 2) comparison of populations within localities, 3) comparison of populations within and between States.
The genetic divergence of the populations was also estimated by an analysis of molecular variance (AMOVA see Table 3) using Arlequin v3.1 [25],[28]. Genetic divergence was based on pairwise differences between haplotypes and structure was evaluated at different geographic levels as above. Total genetic variance was partitioned into variation due to the differences between individuals within populations (within population polymorphism) and that caused by the differences among populations (among population polymorphism). Pairwise differences between haplotypes were used to calculate related F statistic analogues, while significance levels for these indices (p = 0.05) were calculated by non-parametric permutation (10,000).
Table 3. Results of hierarchical analysis (AMOVA) of population.
| Structure | Populations within Group | FST | Among Populations% | Within Populations% | p-Value * |
| Adjacent populations | |||||
| cytb | |||||
| 1 | pop 2, pop 3 | 0.05 | 4.7 | 95.3 | 0.14 |
| 1 | pop 2, pop 5 | 0.01 | 1.3 | 98.7 | 0.25 |
| 1 | pop 3, pop 5 | −0.02 | −2.4 | 102.4 | 0.76 |
| 1 | pop 29, pop 30 | 0.04 | 3.83 | 96.1 | 0.49 |
| 1 | pop 6, pop 7 | −0.01 | −0.46 | 100.46 | 0.56 |
| 1 | pop 27, pop 28 | −0.15 | −15.01 | 115.01 | 1.0 |
| microsatellite | |||||
| 1 | pop 2, pop 3 | 0.2 | 20.0 | 80.0 | 0.00 |
| 1 | pop 2, pop 5 | 0.003 | 0.3 | 99.7 | 0.39 |
| 1 | pop 3, pop 5 | 0.17 | 17.1 | 82.9 | 0.00 |
| 1 | pop 29, pop 30 | 0.15 | 14.7 | 85.3 | 0.00 |
| 1 | pop 6, pop 7 | 0.04 | 4.5 | 95.5 | 0.01 |
| 1 | pop 27, pop 28 | 0.03 | 3.4 | 96.6 | 0.07 |
| Within localities | |||||
| cytb | |||||
| 1 | pop 14, pop 15 | 0.03 | 2.7 | 97.3 | 0.39 |
| 1 | pop 16, pop 17, pop 18 | −0.03 | −2.7 | 102.7 | 0.55 |
| 1 | pop 19, pop 20 | −0.04 | −4.26 | 104.3 | 0.08 |
| 1 | pop 21, pop 22, pop 23 | 0.01 | 1.07 | 98.9 | 0.35 |
| microsatellite | |||||
| 1 | pop 9a, pop 9b | −0.02 | −2.4 | 102.4 | 0.99 |
| 1 | pop 16, pop 17, pop 18 | 0.02 | 2.4 | 97.6 | 0.05 |
| 1 | pop 16, pop 17 | 0.04 | 4.4 | 95.6 | 0.08 |
| 1 | pop 16, pop 18 | 0.04 | 4.2 | 95.8 | 0.01 |
| 1 | pop 17, pop 18 | −0.004 | −0.4 | 100.4 | 0.64 |
| 1 | pop 19, pop 20 | −0.01 | −0.8 | 100.8 | 0.77 |
| 1 | pop 21, pop 22, pop 23 | 0.03 | 3.1 | 96.9 | 0.003 |
| 1 | pop 21, pop 22 | 0.06 | 5.8 | 94.2 | 0.001 |
| 1 | pop 21, pop 23 | 0.004 | 0.4 | 99.6 | 0.3 |
| 1 | pop 22, pop 23 | 0.02 | 2.5 | 97.5 | 0.05 |
| 1 | pop 24a, 24b, 24c, 24d | 0.02 | 2.0 | 98.0 | 0.07 |
| Within States | |||||
| cytb | |||||
| 1 | pop 1–pop 13 | 0.38 | 38.9 | 61.1 | 0.00 |
| 1 | pop 14–pop 28 | 0.15 | 14.9 | 85.1 | 0.00 |
| microsatellite | |||||
| 1 | pop 1–10, pop 13, pop 35 | 0.11 | 11.4 | 88.6 | 0.00 |
| 1 | pop 16– pop 28 | 0.03 | 3.3 | 96.7 | 0.00 |
| Among States | |||||
| cytb | |||||
| 1 | Portuguesa, Barinas, Lara Cojedes, Trujillo, Guarico | 0.15 | 15.5 | 84.5 | 0.00 |
| microsatellite | |||||
| 1 | Portuguesa, Barinas, Cojedes, Trujillo, Lara | 0.07 | 7.3 | 92.7 | 0.00 |
| All populations | |||||
| cytb | |||||
| 1 | All 34 | 0.44 | 43.61 | 56.39 | 0.00 |
| microsatellites | |||||
| 1 | All 33 | 0.11 | 11.3 | 88.7 | 0.00 |
Note: p-value corresponds to the probability of obtaining random values larger or equal than the observed value.
Microsatellite analysis
A total of 555 R. prolixus specimens, from silvatic, domestic and peridomestic ecotopes in five States were used for microsatellite amplification. Specimens were grouped into 33 populations determined by the collection site (ecotope) as listed in Table 2. Specimens were analysed at a total of 9 microsatellite loci, and at a 10th locus for a subset of 20 populations (Table 2). The 10 primers, flanking dinucleotide repeats, were isolated and amplified as described elsewhere [29]. Linkage disequilibrium was tested between all pairs of loci in each population using the program GENEPOP version 3.4 [30]. These results will be reported elsewhere (Fitzpatrick et al in preparation) but in brief significant linkage disequilibrium was detected between three loci pairs after Bonferroni correction in three populations; LIST14-017 and LIST14-042 in pop 9a, LIST14-010 and LIST14-013 in pop 20, LIST14-010 and LIST14-025 in pop 29 (Table 2). As these microsatellite loci did not exhibit significant linkage in each of the 33 population analysed, they were determined to be in linkage equilibrium. Observed (HO) and expected heterozygosity (HE) were calculated for each locus using the program Arelquin V2.000 [25]. Allele richness was calculated using FSTAT version 2.932 [31]. Deviation from Hardy-Weinberg equilibrium (HWE) was tested at each locus within each individual populations using a modified Markov chain randomisation method of Guo and Thompson (1992) (Arlequin V3.1, 10,000 steps; [25],[32]). Wright's inbreeding coefficient FIS was also calculated at each locus following Weir and Cockerham (1984) (GENEPOP version 3.4; [26],[30]). Genetic diversity in each population was measured in four ways: (i) Expected heterozygosity (He); (ii) mean number of alleles (iii) allele richness; and (iv) polymorphic loci.
Intrapopulation comparisons were based on the indices of population homogeneity FST (Weir & Cockerham's 1984), as previously detailed [25],[26]. Nominal significance level was adjusted, as previously with k = 528, p1 = 0.05/528, p≤0.0001 for 9 loci and k = 190, p1 = 0.05/190, p≤0.0003 for 10 loci [27]. Population geneflow was evaluated at different geographic levels 1) comparison of adjacent ecotopes, 2) comparison of populations within localities, 3) comparison of populations within and between States.
The genetic divergence of the populations was also estimated by an analysis of molecular variance (AMOVA) using Arlequin v3.1 [25],[28]. Genetic divergence was based on the number of different alleles detected (FST-like) and populations evaluated at different geographic levels as above (Table 3). The total genetic variance was partitioned into variation due to the differences between individuals within populations (within population polymorphism) and that caused by the differences among populations (among population polymorphism). Significance levels (p = 0.05) for the F statistic analogues were calculated by non-parametric permutation (10,000).
The relationship between geographical and genetic distance over the study area was assessed by testing the correlation between FST/(1−FST) and log transformed (ln) geographic distances. Rousset (1997) showed that a linear relationship occurs between natural log of geographical distance and FST/(1−FST) in two dimensional habitats [33]. The significance of the correlation was examined by a Mantel test using a permutation procedure (9,999 permutations) in GenAlex [34].
Results
Species identity and genetic relatedness
Cytb haplotypes
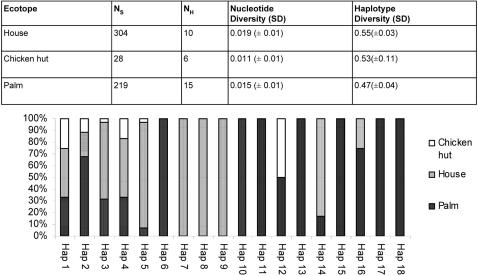
A total of 551 specimens were analysed from six States by direct sequencing of a fragment of the mitochondrial gene cytochrome b (cytb). This included 304 specimens from houses, 219 from palms and 28 from chicken huts (Figure 4). A 415 bp consensus sequence was produced for 541 specimens (Figure 3); and a slightly shorter consensus (392–408 bp) for 10 specimens. There were 18 cytb haplotypes; 14 of which were unique to single States and eight occurred once (Figure 1). The haplotypes varied at 46 sites (11.1% polymorphism). All variable sites were point mutations; 16 sites were parsimony informative (3.9%). Haplotype frequencies varied by State (Figure 1). A single haplotype was detected in Lara (haplotype 1), whereas 11 haplotypes were found in Barinas, including seven unique to that State; in Portuguesa State haplotypes 1 (67%) and 3 (33%) were dominant. Overall, haplotype 1 was the most common haplotype in the study (68% of specimens) and was present in all States, apart from Trujillo. Nucleotide diversity was highest in Portuguesa State, and lowest in Lara State, while haplotype diversity was highest in Guarico State, and lowest in Lara State.
Figure 4. Genetic diversity and haplotype distribution in the sampled ecotopes.
Species identity
In comparison with published sequences in GenBank our 18 haplotypes shared greatest similarity with R. prolixus (14 haplotypes) and R. robustus cytb sequences (4 haplotypes). Identity scores for haplotypes 1, 2, 4–15 were highest for a R. prolixus specimen from Honduras (99–100%; prHo AF421339) whilst haplotype 3 and haplotypes 16–18 were most similar to R. robustus from the Amazon (98–100%; roBR7 AF421343) and Venezuela (roVE1 AF421340). The most common haplotype in the study (haplotype 1) was identical to R. prolixus from Honduras (prHO AF421339).
The identity of silvatic Rhodnius
Silvatic and domestic specimens were collected in each State, with the exception of Lara, where palms were negative and Guarico, where houses were negative. Haplotype distribution varied by ecotope with seven haplotypes found exclusively in palms and three exclusively in houses (Figure 4). Nucleotide and haplotype diversity was similar in both houses and palms (Figure 4). Significantly 11 of the R. prolixus haplotypes were identified in palms, thus confirming the existence of silvatic R. prolixus. In addition five R. prolixus haplotypes were common to both palms and houses (Figure 4). Importantly, both nymphs and adults were detected in houses for the shared haplotypes 1, 2 and 5 (109 nymphs in total) indicating that silvatic R. prolixus is capable of domestic colonisation.
However, silvatic R. robustus was also identified in this study in the Andean state of Trujillo. Specimens from a palm dissected in the locality La Juventud were determined as Venezuelan R. robustus (haplotypes 16–18) (pop 34). In a previous study in this area adult Rhodnius specimens, thought to be R. robustus, were found to enter houses at night to feed, attracted by light but not to colonise [35]. In our study a single R. robustus adult (haplotype 16) was found in an infested house in the Locality Loma de Amarillo (pop 33), however all of the other domestic specimens were R. prolixus (haplotype 5), including all nymphs, thus indicating that this R. robustus adult may also have arrived to feed but had not colonised the house.
Mitochondrial introgression
The R. robustus haplotype 3 was the second most frequent haplotype (13%), although this was limited in distribution to Portuguesa State, with the exception of three specimens. As mentioned the species R. robustus is not known to colonise houses, we were therefore surprised to find 14 nymphs of R. robustus haplotype 3 in four houses in the localities Terronal, Casarena and Palmarito thus suggesting that this silvatic species is capable of domestic colonisation in this Venezuelan State (see Table 1; pop 1, pop 2, pop 13, other). Accordingly, we investigated mitochondrial introgression between R. robustus haplotype 3 and R. prolixus by sequencing a fragment of the nuclear target D2. The D2 sequence alignments (519 bp) revealed three haplotypes, varying at four sites (Figure 3). Strikingly, R. robustus (haplotype 3) had a D2 haplotype that was identical to R. prolixus (haplotype 1, 5) (Figure 3), while Amazonian R. robustus from GenBank, roBR4 and roBR8, presented two different D2 haplotypes. Thus indicating an introgression event, and that the 14 nymphs above were R. prolixus with introgressed R. robustus mitochondrial haplotype 3.
Genetic relatedness and phylogenetics
From the alignment of the polymorphic sections of the 18 haplotypes it is clear that R. robustus haplotype 3 and haplotypes 16–18 are the most divergent (genetic distance 0.07–0.09 and 0.03–0.09 respectively, Kimura-2 parameter Figure 3). While R. prolixus haplotypes 1, 2 and haplotypes 4–15 were very similar, separated by only 1–4 base pair changes (genetic distance 0.002–0.015; Kimura-2 parameter, Figure 3).
In our phylogenetic tree the 18 haplotypes divided into two major clades with high bootstrap values (Figure 2, clades I and II). Within clade I two main groups were visible, 1) R. prolixus haplotypes 1, 2, 4–15 and prHO and 2) Venezuelan R. robustus haplotypes 16–18, roVE1, roVE2e and roVE3f (99% bootstrap support). While clade II is composed of Amazonian R. robustus haplotypes. Within this group, haplotype 3 was identical to roBR9 (R. robustus from the Brazilian Amazon). These results indicate a closer genetic relationship between Venezuelan R. robustus and R. prolixus than R. robustus from the Amazon region giving further support to the existence of cryptic species within R. robustus [17].
Genetic variation and population structure
Our specific interest, in the context of detecting movement between silvatic and domestic Rhodnius populations, was to genotype adjacent silvatic and domestic populations, before examining the relationship between more geographically distant populations.
Mitochondrial DNA and population structure
For population analysis using cytb haplotypes specimens were placed into 34 population groups as listed in Table 1.
Comparisons between adjacent ecotopes
To test for possible geneflow between silvatic, domestic and peridomestic areas, comparisons were made initially between five population pairs in adjacent ecotopes. (Table 4, see Table S1 for all FST values). Pairwise FST values indicated a lack of population division between four adjacent house and palm populations; 1) pop 29 and pop 30 ( FST = 0.04, p = 0.49); 2) pop 6 and pop 7 (FST = −0.005, p = 0.55); 3) pop 2 and pop 5 (FST = 0.01, p = 0.26) and 4) pop 2 and pop 3 (FST = 0.05, p = 0.14) and between adjacent palm and chicken hut ecotopes, pop 27 and pop 28 (FST = −0.15, p = 1.0). Additionally the two palms adjacent to pop 2, but sampled in different years, were not genetically different (pop 3 and pop 5; FST = −0.02, p = 0.77). These cytb results indicated that bugs can move between silvatic, peridomestic and domestic habitats, thus indicating geneflow. The divergence of adjacent populations was also estimated by AMOVA (see Table 3). The amount of variation due to within population polymorphism was greater than between populations indicating that no heterogeneity is present and suggesting a lack of population structure between ecotopes (FST range = −0.02 to 0.05).
Table 4. Summary of geneflow.
| Population | State | Locality | Compared Ecotope | NL | HS | FST Values ** | |
| Cytb | Microsatellites | ||||||
| Pop 29,Pop 30 * | Cojedes | Las Queseras | Palm, House | 9 | 1 | Yes, FST = 0.04 p = 0.49 | No, FST = 0.15 p = <0.0001 |
| Pop 27,Pop 28 * | Barinas | Rio Bravo II | Chicken hut, Palm | 10 | 1, 2 | Yes, FST = −0.15 p = 1.0 | Yes, FST = 0.04 p = 0.045 |
| Pop 6,Pop 7* | Portuguesa | Los Rastrojos | House, Palm | 10 | 1 | Yes, FST = −0.005 p = 0.55 | Yes, FST = 0.04 p = 0.007 |
| Pop 2,Pop 3 * | Portuguesa | Terronal | House, Palm | 9 | 1, 3 | Yes, FST = 0.05 p = 0.19 | No, FST = 0.2 p = <0.0001 |
| Pop 2,Pop 5 * | Portuguesa | Terronal | House, Palm | 9 | 1, 3 | Yes, FST = 0.01 p = 0.26 | Yes, FST = 0.002 p = 0.43 |
| Pop 3,Pop 5* | Portuguesa | Terronal | Palm, Palm | 9 | 1, 3 | Yes, FST = −0.02 p = 0.77 | No, FST = 0.17 p = <0.0001 |
| Pop 1,Pop 4 | Portuguesa | Terronal | House, House | 9 | 1, 3 | Yes, FST = −0.03 p = 0.71 | Yes, FST = 0.046 p = 0.023 |
| Pop 1,Pop 2 | Portuguesa | Terronal | House, House | 9 | 1, 3 | Yes, FST = 0.18, p = 0.02 | Yes, FST = 0.032 p = 0.02 |
| Pop 4,Pop 3 | Portuguesa | Terronal | House. Palm | 9 | 1, 3 | Yes, FST = 0.30, p = 0.006 | Yes, FST = 0.061 p = 0.023 |
| Pop 4,Pop 5 | Portuguesa | Terronal | House, Palm | 9 | 1, 3 | Yes, FST = 0.24 p = 0.015 | Yes, FST = 0.071 p = 0.0015 |
| Pop 9a,Pop 9b | Portuguesa | San Bartolo | House | 9 | 1 | - | Yes, FST = −0.023 p = 0.99 |
| Pop 17,Pop 18 | Barinas | Cascabel | House, Palm | 10 | 1,2,5 | Yes, FST = 0.009 p = 0.37 | Yes, FST = 0.004 p = 0.40 |
| Pop 16,Pop 17 | Barinas | Cascabel | House, Chicken hut | 10 | 1,2 | Yes, FST = 0.038 p = 0.29 | Yes, FST = 0.034 p = 0.105 |
| Pop 16,Pop 18 | Barinas | Cascabel | Chicken hut, Palm | 10 | 1,2 | Yes, FST = −0.08 p = 0.99 | Yes, FST = 0.047 p = 0.006 |
| Pop 19,Pop 20 | Barinas | El Guamito | House, Palm | 10 | 1 | Yes, FST = −0.04 p = 0.77 | Yes, FST = −0.008 p = 0.74 |
| Pop 21,Pop 23 | Barinas | L. Hermosa | House, Palm | 10 | 1,5 | Yes, FST = 0.04 p = 0.32 | Yes, FST = 0.001 p = 0.38 |
| Pop 21,Pop 22 | Barinas | L. Hermosa | House, Chicken hut | 10 | 1,5 | Yes, FST = 0.03 p = 0.43 | Yes, FST = 0.06 p = 0.0006 |
| Pop 23,Pop 22 | Barinas | L. Hermosa | Palm, Chicken hut | 10 | 1,5 | Yes, FST = −0.03 p = 0.59 | Yes, FST = 0.034 p = 0.013 |
| Pop 24a,Pop 24c | Barinas | G. Paguey | House, Palm | 10 | - | - | Yes, FST = 0.026 p = 0.145 |
| Pop 24a,Pop 24d | Barinas | G. Paguey | House, Palm | 10 | - | - | Yes, FST = 0.032 p = 0.078 |
| Pop 24b,Pop 24c | Barinas | G. Paguey | House, Palm | 10 | - | - | Yes, FST = 0.005 p = 0.483 |
| Pop 24b,Pop 24d | Barinas | G. Paguey | House, Palm | 10 | - | - | Yes, FST = 0.017 p = 0.19 |
| Pop 24a,Pop 24b | Barinas | G. Paguey | House, House | 10 | 1 | - | Yes, FST = 0.018 p = 0.279 |
| Pop 24c,Pop 24d | Barinas | G. Paguey | Palm, Palm | 10 | - | - | Yes, FST = 0.021 p = 0.132 |
*: Adjacent populations, NL = no of loci amplified, HS = shared haplotypes.
**: Sequential Bonferroni correction applied to FST p values (For cytb k = 561, p1 = 0.05/561, p≤0.0001; Microsatellite: for 9 loci k = 528, p1 = 0.05/528 p≤0.0001, for 10 loci k = 190, p1 = 0.05/190 p≤0.0003).; yes = geneflow, no = no geneflow, -population not sequenced, see Table 1, Table 2 and S1, S3, S4 for all population comparisons FST values.
Comparisons within localities
To detect possible geneflow between more geographically distant populations, non adjacent populations within individual localities were compared (Table 3, Table 4, see Table S1 for all FST values). Pairwise FST values indicated a lack of population division between a palm and house sampled within the locality El Guamito (pop 19 and pop 20; FST = −0.04, p = 0.78), between a house, chicken hut and palm in the locality Cascabel (pop 16, 17 and 18; FST range = −0.08 to 0.04, p = 0.29 to 0.99), also within the locality Laguna Hermosa (pop 21, 22 and 23; FST range = −0.03 to 0.04, p = 0.32 to 0.6) and between houses in the locality Terronal (pop 1 and pop 4; FST = −0.03, p = 0.72). The population divergence within localities was also estimated by AMOVA (see Table 3). Again the variation due to within population polymorphism was greater than between population polymorphism in individual localities indicating a lack of structure (FST range = −0.04 to 0.03) (Table 3).
Comparison within States
Portuguesa State
A total of 287 specimens from domestic, silvatic and peridomestic ecotopes in this State were analysed by direct sequencing. For population analysis specimens were divided into 13 populations (Table 1). A hierarchical analysis of all populations within Portuguesa detected a greater within population diversity (61%) than between population diversity (39%), however FST indicated structure does exist between populations in this State (FST = 0.38, p = 0, Table 3). Detected heterogeneity in this State was primarily related to domestic populations from Santa Lucia (pop 10), Casarena (pop 13), and palm population (pop 1) (FST range = 0.12 to 1.0, pairwise FST). When populations in Portuguesa were further analysed in ecotope groupings variation was greatest within populations (64%) in comparison to among groups (−9.2) or among populations within groups (45%) (FST = 0.41 p = 0, FSC = 0.36 p = 0, FCT = −0.1 p = 0.7; AMOVA 2 groups, house <pop 1,2,4,6, 9,10,11,12,13> palm <3,5,7,8>).
Barinas State
A total of 146 specimens from domestic, silvatic and peridomestic ecotopes in this State were analysed by direct sequencing. For population analysis specimens were divided into 15 groups (Table 1). A hierarchical analysis of all populations within Barinas detected a greater within population diversity (85%) than between populations (15%), however FST indicated that structure does exist between populations in this State (FST = 0.15, p = 0, (Table 3). In population comparisons (pairwise FST) detected heterogeneity was due to a peridomestic population (pop 26), which was different from the majority of populations in Barinas (FST range = 0.28 to 1.0). When populations in Barinas were further analysed in ecotope groupings variation was greatest within populations (85%) in comparison to among groups (1.2%) or among populations within groups (14%) (FST = 0.15 p = 0, FSC = 0.14 p = 0, FCT = 0.001 p = 0; AMOVA 3 groups, house <15,17,19,21,24> palm <14,18,20,23,25,28> chicken hut <16,22,26,27>).
Trujillo State
A total of 27 specimens of domestic (pop 33) and silvatic origin (pop 34) were analysed by direct sequencing. Gene flow not evident between these two ecotopes (FST = 0.91, p<0.0001).
Comparison between States
A hierarchical analysis of all 34 populations analysed by cytb revealed population structure, with similar level of polymorphism detected within (56%) and between populations (44%) (FST = 0.44, p = 0, Table 3). When specimens were analysed further by their State of collection (1 group: Portuguesa, Barinas, Guarico, Cojedes, Trujillo, Lara) genetic isolation was detected (FST = 0.15 p = 0), however variation was greater within individual States than between States (15.5%) (Table 3). Additional hierarchal analysis between State populations was carried out (5 groups; Portuguesa <pop 1–13>, Barinas <pop 14–28> Cojedes <pop 29, 30>, Trujillo <pop 33, 34> Other <pop 31, 32>). Again variation was greatest within populations (55%) in comparison to among groups (8%) or among populations within groups (37%) (FST = 0.45 p = 0, FSC = 0.4 p = 0, FCT = 0.08 p = 0.1).
Microsatellite analysis and population structure
In parallel with mitochondrial analyses, population structures, in particular for adjacent domestic and silvatic populations, were re-examined using high resolution microsatellites. A total of 33 populations were analysed (Table 2). The number of polymorphic loci in populations ranged from 6–10, with 85% of all populations polymorphic at all loci (Table 2). Monomorphic loci were detected in a number of populations, ranging from three loci in pop 10 (Santa Lucia) to one locus in pop 8 (Palo Gacho) (Table 2). The allele richness per population varied from 1.7 (pop 10) to 3.6 (pop 18) (Table 2). The number of private alleles detected in the study was low, nine in total, four of which occurred in a single domestic population in Loma de Amarillo, Trujillo State (pop 33). Mean observed heterozygosity ranged from 0.2 to 0.6 and expected heterozygosity between 0.3 to 0.6 (Table 2). Loci in each population were tested for significant departure from Hardy-Weinberg equilibrium (HWE); six loci in 17 populations were significant after sequential Bonferroni correction (see Tables S2a, S2b). Departures were primarily related to excess homozygosity at locus LIST14-017 (12 populations). FST values generated including and excluding List14-017 were significantly correlated [Mantel test R2 = 0.9, p<0.001, 34] and this locus was therefore included in the analysis. Departures from HWE were also related to excess heterozygosity at locus LIST14-013 (1 population) and LIST14-056 (2 populations). Null alleles can be problematical in microsatellite analysis and can result in departures from HWE. Here 54 specimens consistently failed to amplify at single locus and 2 specimens at two loci in 23 populations (Table 2).
Comparisons between adjacent ecotopes
Geneflow between our five adjacent ecotope pairs was re-examined using microsatellite analysis (Table 2, Table 3, see Table S3 and Table S4 for all FST values). Pairwise FST comparisons indicated a lack of population structure between three of the adjacent ecotopes; 1) between a house and palm (pop 2 and pop 5; FST = 0.002 p = 0.43), 2) pop 6 and pop 7 (FST = 0.04 p = 0.007) and 3) between a palm and chicken hut (pop 27 and pop 28; FST = 0.04 p = 0.045). These results reaffirm that bugs move between silvatic, peridomestic and domestic ecotopes. However, further heterogeneity was uncovered by microsatellite analysis between the remaining adjacent populations, in contrast to cytb analysis; between a palm and house (pop 29 and pop 30; FST = 0.15 p<0.0001) and pop 2 and pop 3 (FST = 0.2 p<0.0001), also between two palm populations (pop 3 and pop 5; FST = 0.17 p<0.0001).
The divergence of adjacent populations was also estimated by AMOVA (see Table 3). The amount of variation due to within population polymorphism was greater than between populations and geneflow was also confirmed, with the exception of pop 6 and pop 7 (house and palm). FST comparisons were significant in the absence of bonferroni correction (FST = 0.04, p = 0.01).
Comparisons within localities
Geographically distant populations from non adjacent ecotopes within individual localities were also re-examined. For example using microsatellite data panmixia was detected between a house, palm and chicken hut population within the locality Laguna Hermosa (pop 21, 22, 23; FST range = 0.002 to 0.06, p = 0.0006 to 0.38), also within the locality Cascabel (pop 16, 17, 18; FST range = 0.004 to 0.05, p = 0.006 to 0.40). In the locality El Guamito a single house and palm were homogenous (pop 19, pop 20; FST = −0.008, p = 0.74). FST comparisons detected population homogeneity within the locality G. Paguey between two house and two palm populations (pop 24a,24b,24c,24d; FST range = 0.005 to 0.03, p = 0.14 to 0.48). These results agreed with cytb analysis. Also panmixia was also evident within the locality Terronal; between houses (pop 1, pop 2; FST = 0.03, p = 0.02), between a house and palm (pop 3, pop 4; FST = 0.06 p = 0.02) and between populations collected from the same house in different years (pop 1, pop 4; FST = 0.05, p = 0.02). In the locality San Bartolo, no genetic structure was detected between two houses (pop 9a, pop 9b; FST = −0.03 p = 1.0).
The population divergence within localities was also estimated by AMOVA (see Table 3). Again the variation due to within population polymorphism was greater than between population polymorphism in individual localities indicating a lack of structure. However AMOVA analysis indicated a greater degree of population structure in the locality Cascabel between a palm and chicken hut (pop 16 pop 18; FST = 0.04, p = 0.01) and Laguna Hermosa (pop 22, pop 23; FST = 0.02, p = 0.05 and pop 21, pop 22 FST = 0.06, p = 0.001). FST comparisons significant in the absence of bonferroni correction.
Comparisons within States
Portuguesa State
A total of 243 specimens from Portuguesa State were divided into 13 populations and analysed at 9 or 10 microsatellite loci. These included 130 domestic, 92 silvatic and 21 peridomestic specimens. A hierarchical analysis of all populations within Portuguesa detected a greater within population diversity (89%) than between populations (11%), however the associated FST value indicated structure does exist within the State (FST = 0.11, p = 0), (Table 3). Pairwise comparisons (FST) indicate that a number of populations contributed to the detected heterogeneity in this State. A domestic population in Santa Lucia (pop 10) was different from the many of populations in Portuguesa possibly due to genetic drift (FST range = 0.13 to 0.42). Three microsatellite loci were monomorphic in this population and the mean number of alleles and allele richness was the lowest in the study (1.9 and 1.7). Domestic populations in the locality San Bartolo (pop 9a, 9b) were also different from the majority of other populations in Portuguesa (FST range = 0.04 to 0.26). Both populations were monomorphic at the two loci. Pairwise population comparisons (FST) indicated that geneflow also occurred between localities for example between a house in the locality Terronal and a palm in Palo Gacho (pop 2 and pop 8; FST = 0.002 p = 0.50). These results were also supported by cytb analysis.
When populations in Portuguesa were further analysed in ecotope groupings variation was greatest within populations (88%) in comparison to among groups (0.9%) or among populations within groups (11%) (FST = 0.12 p = 0, FSC = 0.11 p = 0, FCT = 0.01 p = 2; AMOVA; 2 groups; house <pop 1,2,4,6,9a,9b,10,13>, palm/ chicken hut <3,5,7,8, 35>).
Barinas State
A total of 221 specimens from Barinas State were divided into 16 populations and analysed at 10 microsatellite loci. These specimens included 60 domestic, 54 peridomestic and 107 silvatic specimens. Average allele richness was greater in Barinas State (3.1) than Portuguesa (2.3). Expected heterozygosity was higher and ranged from 0.5 to 0.6 (Table 2). A hierarchical analysis of all populations within Barinas detected a greater within population diversity (97%) than between populations diversity (3.3%), however structure does exist within the State (FST = 0.03, p = 0), (Table 3). When geneflow was examined by pairwise FST comparisons detected structure was primarily related to a peridomestic population in the locality 19 Abril (pop 26; FST range = 0.06 to 0.18; see Table S4) in agreement with cytb analysis.
When populations in Barinas were further analysed in ecotope groupings variation was greatest within populations (97%) in comparison to among groups (−0.03%) or among populations within groups (3.3%) (FST = 0.03 p = 0.5, FSC = 0.03 p = 0, FCT = −0.003 p = 5; AMOVA house <17,19,21,24a,24b> palm <18,20,23,24c,24d,25,28> chicken hut <16,22,26,27>).
Comparisons between States
Lara State
A single domestic population was analysed in this State (pop 31). Mean allele number and richness were low (2.2, 2.0). This population was different by pairwise comparisons from the majority of populations analysed (FST range = 0.07 to 0.33).
Cojedes State
The single domestic and silvatic population from the locality Las Quebralitas also differed from the majority other populations in the study (FST range = 0.08 to 0.35).
Trujillo State
The domestic population analysed from Trujillo (pop 33) wa distinct from the majority of populations (FST range = 0.09 to 0.42). Four private alleles were detected in this population, all in the single female adult identified as R. robustus by cytb analysis.
A hierarchical analysis of all 33 populations analysed by microsatellites revealed a greater level of polymorphism within (89%) than between populations (11%), however population structure was detected (FST = 0.11, p = 0, Table 3). When specimens were grouped by their State of collection (1 group; Portuguesa, Barinas, Cojedes, Trujillo, Lara) detected variation was greater within individual States than between States (7.3%), however State groups were distinct (FST = 0.07 p = 0) (Table 3).
Additional hierarchal analysis between States was carried out (4 groups; Portuguesa <pop 1–10,13,35>, Barinas <pop 16–28> Cojedes <pop 29,30>, Other <pop 31, 33>). Again variation was greatest within populations (87%) in comparison to among populations within groups (8.3%) or among groups (4.3%) (FST = 0.13 p = 0, FSC = 0.09 p = 0, FCT = 0.04 p = 0).
Isolation by distance (IBD)
Tests for IBD (FST /(1−FST) against log transformed (ln) distances were conducted at various hierarchical levels (1) between populations (2) between localities (3) within Portuguesa and within Barinas State and (4) between States. Patterns were weakly correlated but significant at population level (33 groups; R2 = 0.06 p-value = 0.0001), locality level (17 groups; R2 = 0.06 p-value = 0.0001) and non-significant at State level (5 groups; R2 = 0.01 p value = 0.64). Patterns were weakly correlated but significant within Portuguesa State (13 groups; R2 = 0.07 p-value = 0.01), within Barinas (16 groups; R2 = 0.02 p-value = 0.01). R2 values range from 0 to 1, with values close to 1 indicating a greater correlation between the compared variables.
Discussion
National surveys of Chagas disease endemic areas in Venezuela in the 1970s suggested that there were widespread silvatic foci of R. prolixus, particularly in palm trees [5]–[8],[36],[37]. It was suggested that such abundant silvatic populations could maintain Chagas transmission by reinvading domestic habitats after vector control campaigns. However, following the identification of the essentially silvatic R. robustus in palms in Venezuela, questions were raised as to the epidemiological importance of silvatic Rhodnius populations and additionally the taxonomic status of R. robustus.
We aimed to resolve the controversy regarding the identity of silvatic populations of Rhodnius and the interaction between silvatic and domestic populations, through mitochondrial and microsatellite analyses. Thus our interest and priority here is not in a global analysis of congruence between mitochondrial and microsatellite phylogenetic trees but in applying both methods, with differing resolution to search for continuity between Rhodnius populations, particularly between geographically adjacent silvatic and domestic populations. Both methods gave valuable and complementary insight, with different degrees of resolution. A similar picture of shared cytb haplotypes and microsatellite homogeneity indicated that silvatic and domestic populations are not isolated, and that gene flow does indeed occur.
Species identity and genetic relatedness
Mitochondrial DNA has been used previously in triatomine studies, including the tribe Rhodniini [16]–[18]. Here eighteen haplotypes were detected among the 551 Venezuelan specimens analysed and these were confirmed as both R. prolixus and R. robustus species.
Our data detected silvatic R. prolixus in palms in all States, except for Trujillo and Lara. We can therefore unequivocally reaffirm that R. prolixus is present in silvatic habitats in Venezuela. Silvatic R. robustus does also exist and was the only species detected in this study in palms in Trujillo State (pop 34). In this region the post-spray reinvasion of houses is therefore unlikely, and vector control may be more straightforward. Nevertheless, adult silvatic R. robustus have been implicated in the sporadic transmission of T. cruzi in western Venezuela [9] and the use of insecticide treated curtains may contribute to reducing sporadic cases of Chagas disease in this State [35].
From sequence analysis it is clear that common haplotypes occur across all ecotopes, with palm and house populations sharing five R. prolixus haplotypes. Three of these shared haplotypes were found in domestic nymphs, in addition to domestic adults, thus indicating these silvatic R. prolixus are capable of invading and importantly colonising houses.
The incongruence detected between nuclear (D2) and mitochondrial (mtcytb) analysis of haplotype 3 confirmed the introgression suspected after the discovery of domestic nymphs of “R. robustus”. Introgression has been recorded previously in triatomine species [38] and other haematophagus insects [39],[40]. In accord with colonisation behaviour, these “Amazonian R. robustus” are R. prolixus with introgressed R. robustus mitochondrial DNA. Additional support for introgression is the absence of unique microsatellite alleles in these haplotype 3 specimens, in contrast to our single domestic Venezuelan R. robustus adult (haplotype 16), which revealed four unique alleles.
Genetic variation and population structure
Mitochondrial DNA and population structure
In addition to shared haplotypes, population homogeneity was also evident by pairwise comparisons between house, palm and peridomestic sites (pairwise FST and AMOVA). This includes examples of geneflow between five adjacent ecotopes, also within localities in both Barinas and Portuguesa State. These results indicate that bugs are moving between houses and between palms, in addition to between palms and houses. Importantly, this is supported by recent data analysis from Sanchez-Martin et al., (2006) where infested palms (>10 palms) within 100m of a house were identified as risk factors for house and peridomestic infestation, in addition to palm roofs less than one year old [41]. Additionally a recent morphometric study in Barinas State comparing silvatic populations of R. prolixus with pre- and postspray peridomestic and domestic populations was unable to differentiate the silvatic specimens as a separate subpopulation [42]. These results also suggest that silvatic populations of R. prolixus are capable of invasion and colonisation and a threat to effective vector control.
When all 34 populations were compared structure was detected (AMOVA, FST = 0.44). Both pairwise FST and AMOVA analysis suggest that population heterogeneity was more pronounced within Portuguesa State (39% between population variation) than Barinas (15% between population variation) (Table 3). Interestingly hierarchical analysis indicated that a populations' ecotope is not a factor in determining population differentiation within both Portuguesa and Barinas States (FCT = −0.1, FCT = 0.001). Additionally detected within and among populations variance did not differ greatly between the comparisons all 33 population or populations in an ecotope group hierarchy. This suggests gene flow occurs between populations from different ecotopes. AMOVA analysis of cytb data also suggested that detected heterogeneity is not related to the State of origin of a population. Again detected within and among populations variance did not differ greatly between the comparison of all populations or populations in a State group hierarchy.
Microsatellite analysis and population structure
For higher resolution of relationships between silvatic and domestic populations of R. prolixus in Venezuela a panel of microsatellite markers was developed [29]. Microsatellites are suitable for population genetics and have proven to be highly polymorphic in species with low isoenzyme polymorphism [43],[44], as noted for R. prolixus [15]. Nine or ten loci were used; additional loci would be advantageous. Polymorphism was low to moderate for the majority of loci; and excess homozygosity at loci such as LIST14-017 may indicate null alleles, which might hide some diversity at that locus or a Walhund effect with restricted genetic exchange between grouped subpopulations.
As for cytb analysis population homogeneity was evident with non-significant pairwise comparisons detected between house, palm and peridomestic sites including adjacent ecotopes. However, some additional genetic diversity was revealed by microsatellites analysis. In the locality Las Queseras populations from an adjacent house and palm (pop 29, pop 30) were significantly different by microsatellites but not by cytb analysis. Additionally in the locality Terronal, a house (pop 2) and an adjacent palm (pop 3), and adjacent palms (pop 3, pop 5) were different by microsatellite analysis but indistinguishable by cytb analysis. AMOVA analysis also detected further structure between an adjacent palm and house (pop 6, pop 7) not evident in pairwise FST comparisons which are corrected for multiple comparisons.
Population homogeneity was detected between populations within localities in Portuguesa and Barinas. Both pairwise FST analysis and AMOVA analysis detected population homogeneity between palm and houses e.g. in the locality Cascabel (pop 17 and pop 18) and in Laguna Hermosa (pop 21, 23), between houses e.g. pop 9a, 9b in the locality San Bartolo, and between palms in the locality G. Paguey (pop 24c, 24d). These results can be explained by the movement of bugs not only between palms and houses but also between houses and between palms.
Comparisons over wider geographic areas revealed population structure (AMOVA, FST). Population structure was detected between all 33 populations (AMOVA, FST = 0.11). Distinct populations (pairwise FST) exhibited monomorphic loci and low allele richness, suggesting isolation and possible genetic drift. Hierarchical analysis also indicated that population heterogeneity was more pronounced within Portuguesa State (11% between population variation) than Barinas State (3% between population variation) (Table 3). In Portuguesa State populations were collected in mountainous terrain, possibly allowing for greater population isolation, this is in contrast to Barinas, where all localities were situated in flat lands, the Llanos, which could allow for easier mixing of populations. Heterogeneity within Barinas State was primarily related to a single peridomestic population (pop 26; pairwise FST). This population was situated at the extreme distribution of sampled sites in Barinas and in an area where T. maculata infestations were more common, factors which may have contributed to detected genetic isolation. The separation of the domestic R. prolixus population from Trujillo State (pop 34) from all other populations indicates that the Andes mountain range and the predominance of silvatic R. robustus may also act as barriers to gene flow.
Hierarchical analysis of microsatellite data also indicated that population ecotope is not a factor in determining population differentiation within both Portuguesa and Barinas State (FCT = 0.01, FCT = −0.003), thus suggesting geneflow occurs between populations from different ecotopes. Interestingly microsatellite analysis detected greater heterogeneity between populations from different State (FCT = 0.04) as compared to cytb analysis.
We investigate the relationship between genetic isolation and increasing geographic distances (IBD). However, while the relationship was significant between populations, between localities and within States, distance was not a critical factor influencing genetic differentiation as the detected correlations were very weak.
As expected a higher degree of population heterogeneity was detected with microsatellites than with the analysis of cytb sequences. Microsatellites are fast-evolving, neutral, noncoding loci, whereas the cytb is a protein-coding gene with important metabolic functions and thus may be subject to selective constraints [45]. Importantly, populations analysed from different ecotopes and localities, including Terronal, San Bartolo were homogeneous by both methods and distinct populations were also detected by both methods (Trujillo, Santa Lucia and 19 Abril). Occasionally microsatellites uncovered diversity not apparent by cytb typing e.g. pop 29, pop 30. Both or pairwise FST and AMOVA data for both methods are consistent with movement between silvatic and domestic habitats with ecotope not determining population structure and with greater population heterogeneity in Portuguesa than Barinas State.
Our results contrast a recent microsatellite study of 19 populations of T. infestans from domestic and peridomestic ecotopes in Argentina. The analysis indicated a strong population structure, with limited gene flow and genetic drift leading to genetic differentiation and suggested an important role for recrudescence in post control infestations rather than reinvasion from untreated areas [46].
Conclusions
Movement of bugs between silvatic, peridomestic and domestic ecotopes probably occurs both actively and passively. Risk factor analysis detected an association between new thatched palm roofs and infestation [41]. Female R. prolixus glue their eggs to palm fronds suggesting passive transport of bugs into houses on these fronds [6]. Restriction or elimination of palm roofs on dwellings must therefore be a key element of control strategies, although it is important that an appropriate substitute roofing material is readily available to the inhabitants. Active transport can also occur, flying adult triatomine bugs may enter a house attracted to light [9]. Rhodnius prolixus in Venezuela is known to be light attracted [47].
From our data it is clear that silvatic populations of R. prolixus in Venezuela represent a definite threat to successful control of Chagas disease, as suspected but controversially debated since populations of R. prolixus were reported in palm trees [5]. Results indicate that the current control programme in Venezuela is unlikely to achieve the level of success seen in the Southern cone, where T. infestans has been eliminated over large areas [1]. The control programme will have to deal with this continual threat, for example by more frequent spraying of houses, combined with community vigilance for reinfestations as an integral part of the control programme. The additional use of alternative control methods such as insecticide treated curtains [35] or bednets [48] would be beneficial. Increased housing improvements, although expensive, seem vital for long term control, by creating a domestic environment unsuitable for colonisation by silvatic bugs.
This study has made a fundamental contribution to the understanding of Rhodnius populations in the context of disease epidemiology and vector control in Venezuela. An important follow-up to this project would be to define population interaction more extensively, particularly in regions of Colombia, where silvatic and domestic Rhodnius populations also occur and reinvasion may be maintaining large domestic colonies of R. prolixus [49]. This would allow prioritisation of control interventions and tailoring of control strategies to regional circumstances. Additionally, modified control strategies to counteract the threat of reinvasion could be assessed, such as widespread provision of ideal low cost roofing, the treatment or removal of palms close to houses, and, improved spraying and surveillance, all with the aim of reducing the burden of Chagas disease in rural areas.
Supporting Information
The pairwise comparison of 34 populations from six Venezuelan States by cytb analysis; FST values below diagonal (p-values above) (Arlequin v3.1). Values in bold remain significant following sequential Bonferroni correction (k = 561, p1 = 0.05/561, p≤0.0001). See Table 1 for population details.
(0.05 MB PDF)
Summary of population microsatellite data. (A) Summary of population microsatellite data per locus (LIST14-056, LIST14-017, LIST14-042, LIST14-010, LIST14-064). N = number of specimens amplified, NA = number of alleles, HO, HE = Observed and Expected heterozygosity, P = exact probability for expected Hardy Weinberg equilibrium conditions for each locus/population combination (Arlequin v2.1), M = monomorphic. FIS = Weir & Cockerham (1984) (GENEPOP V3.4). Values in bold departures from HWE significant after Bonferroni correction (populations analysed at 9 loci k = 9, p1 = 0.05/9, at 10 loci k = 10, p1 = 0.05/10). See Table 2 for population details. (B) Summary of population microsatellite data per locus (LIST14-013, LIST14-021, LIST14-025, LIST14-037, LIST14-079). N = number of specimens amplified, NA = number of alleles, HO, HE = Observed and Expected heterozygosity, P = exact probability for expected Hardy Weinberg equilibrium conditions for each locus/population combination (Arlequin v2.1). FIS = Weir & Cockerham (1984) (GENEPOP V3.4). Values in bold departures from HWE significant after Bonferroni correction, populations analysed (9 loci k = 9, p1 = 0.05/9, at 10 loci k = 10, p1 = 0.05/10). ∧LIST14-079 amplified in subset of populations. M = monomorphic, NA = not amplified. See Table 2 for population details.
(0.02 MB PDF)
The pairwise comparison of 33 populations from six Venezuelan States at 9 microsatellite loci, FST values below diagonal (p-values above) (Arlequin v2.1). Values in bold significant after sequential Bonferroni correction k = 528, p1 = 0.05/528, p≤0.0001. See Table 2 for population details.
(0.05 MB PDF)
The pairwise comparison of a subset of 20 populations at 10 microsatellite loci FST values below diagonal (p-values above) (Arlequin v2.1). Values in bold significant after sequential Bonferroni correction k = 190, p1 = 0.05/190, p≤0.0003.
(0.04 MB PDF)
Acknowledgments
We thank Clive Davies for his support, Dr Michael Gaunt for his advice on computing, everyone in BIOMED Venezuela for their warm welcome. We would like to thank the staff of the Ministry of Health in each Venezuelan State for their cooperation and expertise, without which field work would have been impossible.
Footnotes
The authors have declared that no competing interests exist.
This project received financial support from the Wellcome Trust Research Training Studentship in Biodiversity. Additional financial support was provided by the Ministry of Science and Technology, Venezuela (FONACIT- Agenda Salud-20000088), the Wellcome Trust (Project No. 062984/Z/00Z), and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dias JC, Silveira AC, Schofield CJ. The impact of Chagas disease control in Latin America–A Review. Mem Inst Oswaldo Cruz. 2002;97:603–612. doi: 10.1590/s0074-02762002000500002. [DOI] [PubMed] [Google Scholar]
- 2.Ache A, Matos AJ. Interrupting Chagas disease transmission in Venezuela. Rev Inst Med Trop Sao Paulo. 2001;43:37–43. doi: 10.1590/s0036-46652001000100008. [DOI] [PubMed] [Google Scholar]
- 3.Feliciangeli MD, Campbell-Lendrum D, Martinez C, Gonzalez D, Coleman P, et al. Chagas disease control in Venezuela: lessons for the Andean region and beyond. Trends Parasitol. 2003;19:44–49. doi: 10.1016/s1471-4922(02)00013-2. [DOI] [PubMed] [Google Scholar]
- 4.Noireau F, Cortez MG, Monteiro FA, Jansen AM, Torrico F. Can wild Triatoma infestans foci in Bolivia jeopardize Chagas disease control efforts? Trends Parasitol. 2005;21:7–10. doi: 10.1016/j.pt.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Gamboa CJ. Comprobacion de Rhonius prolixus extradomiciliario en Venezuela. Bol Oficina Sanit Panam. 1963;54:18–25. [PubMed] [Google Scholar]
- 6.Gamboa CJ. La población silvestre de Rhodnius prolixus en Venezuela. Arch Venez Med Trop Parasitol Med. 1973;5:321–352. [Google Scholar]
- 7.Zeledon R, Rabinovich JE. Chagas disease: an ecological appraisal with special emphasis on its insect vectors. Annu Rev Entomol. 1981;26:101–133. doi: 10.1146/annurev.en.26.010181.000533. [DOI] [PubMed] [Google Scholar]
- 8.Tonn R, Carcavallo RU, Ortega R. Notas sobre la biología, ecología y distribución geográfica de Rhodnius robustus Larrousse, 1927 (Hemiptera, Reduviidae). Bol Dir Malariol y San Amb. 1976;16:158–162. [Google Scholar]
- 9.Feliciangeli MD, Dujardin JP, Bastrenta B, Mazzarri M, Villegas J, et al. Is Rhodnius robustus (Hemiptera: Reduviidae) responsible for Chagas disease transmission in Western Venezuela? Trop Med Int Health. 2002;7:280–287. doi: 10.1046/j.1365-3156.2002.00853.x. [DOI] [PubMed] [Google Scholar]
- 10.Solano P, Dujardin JP, Schofield CJ, Romana C, Tibayrenc M. Isoenzymes as a tool for identification of Rhodnius species. Res Rev Parasitol. 1996;56:41–47. [Google Scholar]
- 11.Harry M, Galindez I, Cariou ML. Isozyme variability and differentiation between Rhodnius prolixus, R. robustus and R. pictipes, vectors of Chagas disease in Venezuela. Med Vet Entomol. 1992;6:37–43. doi: 10.1111/j.1365-2915.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 12.Harry M. Use of the median process of the pygophore in the identification of Rhodnius nasutus, R. neglectus, R. prolixus and R. robustus (Hemiptera: Reduviidae). Ann Trop Med Parasitol. 1993a;87:277–282. doi: 10.1080/00034983.1993.11812767. [DOI] [PubMed] [Google Scholar]
- 13.Harry M. Isozymic data question the specific status of some blood-sucking bugs of the genus Rhodnius, vectors of Chagas disease. Trans R Soc Trop Med Hyg. 1993b;87:492–493. doi: 10.1016/0035-9203(93)90054-t. [DOI] [PubMed] [Google Scholar]
- 14.Harry M. Morphometric variability in the Chagas disease vector Rhodnius prolixus. . Jpn J Genet. 1994;69:233–250. doi: 10.1266/jjg.69.233. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro FA, Lazoski C, Noireau F, Sole-Cava AM. Allozyme relationships among ten species of Rhodniini, showing paraphyly of Rhodnius including Psammolestes. Med Vet Entomol. 2002;16:83–90. doi: 10.1046/j.0269-283x.2002.00343.x. [DOI] [PubMed] [Google Scholar]
- 16.Monteiro FA, Wesson DM, Dotson EM, Schofield CJ, Beard CB. Phylogeny and molecular taxonomy of the Rhodniini derived from mitochondrial and nuclear DNA sequences. Am J Trop Med Hyg. 2000;62:460–465. doi: 10.4269/ajtmh.2000.62.460. [DOI] [PubMed] [Google Scholar]
- 17.Monteiro FA, Barrett TV, Fitzpatrick S, Cordon-Rosales C, Feliciangeli MD, et al. Molecular phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus. Mol Ecol. 2003;12:997–1006. doi: 10.1046/j.1365-294x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- 18.Lyman DF, Monteiro FA, Escalante AA, Cordon-Rosales C, Wesson DM, et al. Mitochondrial DNA sequence variation among Triatomine vectors of Chagas' disease. Am J Trop Med Hyg. 1999;60:377–386. doi: 10.4269/ajtmh.1999.60.377. [DOI] [PubMed] [Google Scholar]
- 19.Abad-Franch F, Noireau F, Paucar A, Aguilar HM, Carpio C, et al. The use of live-bait traps for the study of sylvatic Rhodnius populations (Hemiptera: Reduviidae) in palm trees. Trans R Soc Trop Med Hyg. 2000;94:629–630. doi: 10.1016/s0035-9203(00)90213-x. [DOI] [PubMed] [Google Scholar]
- 20.Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull. Am. Mus. Nat. Hist. 1979;163:125–520. [Google Scholar]
- 21.Porter CH, Collins FH. Phylogeny of nearctic members of the Anopheles maculipennis species group derived from the D2 variable region of 28S ribosomal RNA. Mol Phylogenet Evol. 1996;6:178–188. doi: 10.1006/mpev.1996.0070. [DOI] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment, editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 23.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Laval ExcoffierLG, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 26.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 27.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 28.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fitzpatrick SO. 2005. The analysis of the relationship between silvatic and domestic populations of Rhodnius prolixus/robustus (Hemiptera: Reduviidae) in Venezuela by morphometric and molecular methods. PhD Thesis. London School of Hygiene and Tropical Medicine, University of London, U.K. [Google Scholar]
- 30.Raymond M, Rousset F. GENEPOP, Version 1.2. Population genetics software for exact tests and ecumenicisms. J Hered. 1995;86:249–249. [Google Scholar]
- 31.Goudet J. FSTAT (VERSION 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 32.Guo SW, Thompson EA. A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet. 1992;51:1111–1126. [PMC free article] [PubMed] [Google Scholar]
- 33.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peakall R, Smouse PE. GenAlEx 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2005;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herber O, Kroeger A. Pyrethroid-impregnated curtains for Chagas' disease control in Venezuela. Acta Trop. 2003;88:33–38. doi: 10.1016/s0001-706x(03)00193-1. [DOI] [PubMed] [Google Scholar]
- 36.Feliciangeli de Pinero DF, Torrealba JW. Observaciones sobre Rhodnius prolixus (Hemiptera, Reduviidae) en su biotopo silvestre Copernicia tectorum. Bol Dir Malariol y San Amb. 1977;17:198–205. [Google Scholar]
- 37.Carcavallo RU, Tonn RJ, Ortega R, Betancourt P, Carrasquero B. Notas sobre la biología, ecología y distribución geográfica del Rhodnius prolixus Stal, 1859 (Hemiptera, Reduviidae). Bol Dir Malariol y San Amb. 1978;18:175–198. [Google Scholar]
- 38.Garcia BA, Powell JR. Phylogeny of species of Triatoma (Hemiptera: Reduviidae) based on mitochondrial DNA sequences. J Med Entomol. 1998;35:232–238. doi: 10.1093/jmedent/35.3.232. [DOI] [PubMed] [Google Scholar]
- 39.Testa JM, Montoya-Lerma J, Cadena H, Oviedo M, Ready PD. Molecular identification of vectors of Leishmania in Colombia: mitochondrial introgression in the Lutzomyia townsendi series. Acta Trop. 2002;84:205–218. doi: 10.1016/s0001-706x(02)00187-0. [DOI] [PubMed] [Google Scholar]
- 40.Thelwell NJ, Huisman RA, Harbach RE, Butlin RK. Evidence for mitochondrial introgression between Anopheles bwambae and Anopheles gambiae. Insect Mol Biol. 2000;9:203–210. doi: 10.1046/j.1365-2583.2000.00178.x. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Martin MJ, Feliciangeli MD, Campbell-Lendrum D, Davies CR. Could the Chagas disease elimination programme in Venezuela be compromised by reinvasion of houses by sylvatic Rhodnius prolixus bug populations? Trop Med Int Health. 2006;11:1585–1593. doi: 10.1111/j.1365-3156.2006.01717.x. [DOI] [PubMed] [Google Scholar]
- 42.Feliciangeli MD, Sanchez-Martin M, Marrero R, Davies C, Dujardin JP. Morphometric evidence for a possible role of Rhodnius prolixus from palm trees in house re-infestation in the State of Barinas (Venezuela). Acta Trop. 2007;101:169–77. doi: 10.1016/j.actatropica.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Hughes CR, Queller DC. Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol Ecol. 1993;2:131–137. doi: 10.1111/j.1365-294x.1993.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 44.Estoup A, Rousset F, Michalakis Y, Cornuet JM, Adriamanga M, et al. Comparative analysis of microsatellite and allozyme markers: a case study investigating microgeographic differentiation in brown trout (Salmo trutta). Mol Ecol. 1998;7:339–353. doi: 10.1046/j.1365-294x.1998.00362.x. [DOI] [PubMed] [Google Scholar]
- 45.Maingon RD, Ward RD, Hamilton JG, Noyes HA, Souza N, et al. Genetic identification of two sibling species of Lutzomyia longipalpis (Diptera: Psychodidae) that produce distinct male sex pheromones in Sobral, Ceara State, Brazil. Mol Ecol. 2003;12:1879–1894. doi: 10.1046/j.1365-294x.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 46.Perez de Rosas AR, Segura EL, Garcia BA. Microsatellite analysis of genetic structure in natural Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina: its implication in assessing the effectiveness of Chagas' disease vector control programmes. Mol Ecol. 2007;16:1401–1. doi: 10.1111/j.1365-294X.2007.03251.x. [DOI] [PubMed] [Google Scholar]
- 47.Tonn RJ, Espinola H, Mora E, Jimenez JE. Trampa de luz negra como metodo de captura nocturna de triatominos en Venezuela. Bol Dir Malariol y San Amb. 1978;18:25–30. [Google Scholar]
- 48.Kroeger A, Villegas E, Ordonez-Gonzalez J, Pabon E, Scorza JV. Prevention of the transmission of Chagas disease with pyrethroid-impregnated materials. Am J Trop Med Hyg. 2003;68:307–311. [PubMed] [Google Scholar]
- 49.Guhl F, Restrepo M, Angulo VM, Antunes CM, Campbell-Lendrum D, et al. Lessons from a national survey of Chagas disease transmission risk in Colombia. Trends Parasitol. 2005;21:259–262. doi: 10.1016/j.pt.2005.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The pairwise comparison of 34 populations from six Venezuelan States by cytb analysis; FST values below diagonal (p-values above) (Arlequin v3.1). Values in bold remain significant following sequential Bonferroni correction (k = 561, p1 = 0.05/561, p≤0.0001). See Table 1 for population details.
(0.05 MB PDF)
Summary of population microsatellite data. (A) Summary of population microsatellite data per locus (LIST14-056, LIST14-017, LIST14-042, LIST14-010, LIST14-064). N = number of specimens amplified, NA = number of alleles, HO, HE = Observed and Expected heterozygosity, P = exact probability for expected Hardy Weinberg equilibrium conditions for each locus/population combination (Arlequin v2.1), M = monomorphic. FIS = Weir & Cockerham (1984) (GENEPOP V3.4). Values in bold departures from HWE significant after Bonferroni correction (populations analysed at 9 loci k = 9, p1 = 0.05/9, at 10 loci k = 10, p1 = 0.05/10). See Table 2 for population details. (B) Summary of population microsatellite data per locus (LIST14-013, LIST14-021, LIST14-025, LIST14-037, LIST14-079). N = number of specimens amplified, NA = number of alleles, HO, HE = Observed and Expected heterozygosity, P = exact probability for expected Hardy Weinberg equilibrium conditions for each locus/population combination (Arlequin v2.1). FIS = Weir & Cockerham (1984) (GENEPOP V3.4). Values in bold departures from HWE significant after Bonferroni correction, populations analysed (9 loci k = 9, p1 = 0.05/9, at 10 loci k = 10, p1 = 0.05/10). ∧LIST14-079 amplified in subset of populations. M = monomorphic, NA = not amplified. See Table 2 for population details.
(0.02 MB PDF)
The pairwise comparison of 33 populations from six Venezuelan States at 9 microsatellite loci, FST values below diagonal (p-values above) (Arlequin v2.1). Values in bold significant after sequential Bonferroni correction k = 528, p1 = 0.05/528, p≤0.0001. See Table 2 for population details.
(0.05 MB PDF)
The pairwise comparison of a subset of 20 populations at 10 microsatellite loci FST values below diagonal (p-values above) (Arlequin v2.1). Values in bold significant after sequential Bonferroni correction k = 190, p1 = 0.05/190, p≤0.0003.
(0.04 MB PDF)