Abstract
In order to examine an association between porcine circovirus type-2 (PCV2) infection and reproductive failure in pigs, sera (n = 171) from stillborn fetuses were collected from 3 different farms with prolonged histories of reproductive problems. These sera were tested for the presence of antibodies to PCV2 using an immunoperoxidase monolayer assay. Of the 171 sera tested, 28 had PCV2 antibody titers of ≥ 1:16. When these 28 samples were tested by a polymerase chain reaction assay, 13 were found to contain PCV2 viral DNA. Of these 13 samples containing both PCV2 antibodies and viral DNA, 9 yielded PCV2 on virus isolation. Amino acid sequences comprising open reading frame 2 of PCV2 from 2 of these isolates were compared to PCV2 isolates from cases of post-weaning multi-systemic wasting syndrome (PMWS). The amino acid sequences of the 2 isolates from stillborn pigs were shown to be nearly identical to each other, as well as to other PCV2 isolates associated with reproductive failure. When compared with PMWS isolates, the isolates from the stillborn fetuses showed differences of at least 2 amino acids. These results confirm previous findings that transplacental infection of PCV2 occurs in the field and that stillbirths in pigs may be associated with PCV2 infections. At present, the significance of minor differences in amino acid sequences is not known.
Introduction
Porcine circovirus type-2 (PCV2) has been associated with several disease syndromes in pigs. The virus was first identified in tissues of piglets suffering from post-weaning multi-systemic wasting syndrome (PMWS) (1). Although PMWS has now been reproduced using PCV2 alone (2), co-infection with another pathogen, activation of the immune system, or both appear necessary to reproduce severe clinical disease (3). Porcine circovirus type 2 has also been detected in cases of porcine dermatitis and nephropathy syndrome (PDNS) (4,5), and porcine respiratory disease complex (6). Recently, PCV2 has been found to infect swine fetuses and cause fetal deaths. Several reports have suggested that PCV2 may be associated with swine reproductive failure (7,8,9,10,11).
The primary objective of this study was to examine sera of stillborn fetuses collected in the field for the presence of PCV2. The sera were first tested for the presence of PCV2 specific antibodies. The antibody-positive samples were then examined for the presence of the virus, viral DNA, or both. In addition, 2 PCV2 isolates from stillborn fetuses were genetically characterized and compared with previously reported PCV2 strains.
Materials and methods
Sample collection
Fetal sera were collected from swine farms with a history of prolonged and recurring reproductive problems. Farms selected for inclusion in this study experienced ≥ 10% born-dead (stillborn and mummified) piglets for a period of more than 6 mo. Three different farms were included, and blood samples from stillborn fetuses were collected on 5 different visits between October 1999 and October 2000. Fetal sera (n = 171) were collected and tested for the presence of PCV2 antibody.
Serology
An immunoperoxidase monolayer assay (IPMA) was performed to detect antibodies to PCV2 in fetal sera, as previously described (12). Briefly, PCV2-infected PK-15 cell monolayers were prepared in 96-well mirotitration test plates. The plates were fixed and stored at −20°C until they were used. For the IPMA procedure, each test serum was serially diluted 4-fold in phosphate buffered saline (PBS, pH 7.2). Each diluted sample was transferred to the test plate and incubated for 45 min at 37°C. After incubation, the plates were washed 3 times with PBS. An anti-swine immunoglobulin G (IgG), conjugated with peroxidase (Cappel Organon Teknika Corporation, West Chester, Pennsylvania, USA) was added, and plates were incubated for 1 h at 37°C. Following removal of the conjugate and washing the plates 3 times with PBS, a substrate solution containing 3-amino-9-ethylcarbazole and hydrogen peroxide was added, and plates were incubated at room temperature for 10 to 15 min. Substrate was removed and the plates were washed. The highest serum dilution showing specific staining was considered to be the IPMA antibody titer, and titers of ≥1:16 were considered positive.
Polymerase chain reaction (PCR) assay
Antibody-positive samples were examined for the presence of PCV2 DNA by a PCR assay (13). Viral DNA was extracted from serum samples (TriZol extraction method; Invitrogen, Carlsbad, California, USA). Two primers were designed to amplify a PCV2 target sequence using published GenBank sequence data. Primer #1 (5'-TATTGTAGTCCTGGTCGTAT-3') is located in the genomic position 1099-1118 of PCV2 isolate IAF-4370 (AF-118097) (14). Primer #2 (5'-ACCCCCGCCACCGCTACC-3') is located between positions 1627–1644. These primers amplify a 545 base pair (bp) fragment, and were confirmed as PCV2 by partial sequencing. For the PCR reaction, 4 μL of extracted DNA was added to a PCR mixture with final concentrations of 1.25 mM MgCl2, 1 × PCR buffer, 0.2 mM dNTP, 1.0 mL of each primer and 2.5 mL Taq DNA polymerase per 50 mL. Amplification of DNA was achieved by 35 cycles with the following parameters; denaturing at 94°C for 30 s, annealing at 63°C for 30 s and elongating at 72°C for 60 s. The PCR was completed with a final extension step of 10 min at 72°C. PCR amplified products were visualized by staining with 0.5 μL/mL ethidium bromide in a 0.7% agarose gel.
Virus isolation
Serum samples positive for both PCV2 antibody and viral DNA, were tested for the presence of PCV2 by virus isolation. The PK-15 cells that were free of PCV-1 and porcine parvovirus (PPV) were used for virus isolation. One-day-old PK-15 cell monolayers in 24-well plates were treated with 300 mM D-glucosamine, as previously described (15). Then, 0.1 mL/well of each test serum was added in duplicate onto the cell monolayers. Following adsorption for 1 h at 37°C, Eagle's minimal essential medium supplemented with 5% fetal bovine serum was added to each well. Plates were incubated for 5 d, and then frozen and thawed 3 times. Each culture was blindly passaged 3 times on PK-15 cell monolayers, as described above. The last passage for each sample was tested for the presence of PCV2 using the PCR assay. Additionally, the last passage was inoculated onto PK-15 cell monolayers, and PCV2 infected cells were detected by IPMA using a reference PCV2 antiserum. The reference serum was obtained from swine fetuses following experimental inoculation in utero with PCV2 (16).
Nucleotide sequencing and phylogenetic analysis
To determine the extent of genetic similarity among PCV2 isolates, a target segment encompassing the open reading frame 2 (ORF2) of the PCV2 genome (17) was amplified and sequenced from 2 isolates from stillborn fetuses in this study (SB-1, SB-2). These isolates were demonstrated as free of PCV1, PPV, and porcine reproductive and respiratory syndrome virus (PRRSV) by their respective PCR assays (18,19). Two primers were designed to amplify the entire ORF2 of PCV2. Primer ORF2-F (5'-CGCTAT GACGTATCCAAGG-3') is located in the genomic position 1720–1738 of PCV2 isolate IAF-4370 (AF-118097) (14). Primer ORF2-R (5'-TTATTT TTCATT TAGGGGT-3') is located between positions 1023–1041. Together, these primers amplify a 705 bp fragment, which encompasses the putative capsid protein (ORF2) of PCV2 (17). Previously published PCV2 nucleotide sequences, used for comparison with isolates sequenced for this study, were obtained from the GenBank database of the National Center for Biotechnology Information (NCBI). All sequences came from PCV2 isolates, with 1 exception (AF071879 from PCV type-1) (20). Two sequences (AJ293867, AJ293868) came from PCV2 isolated from aborted fetuses (10). One sequence (AJ293869) came from a PDNS associated isolate (10), and another sequence (AF109397) originated in a bovine lung. The remaining sequences came from PMWS isolates. In addition to the Minnesota isolates sequenced for this study, 2 isolates originated from France (AF055393, AF055394), 1 from the United Kingdom (AJ293869), 1 from Taiwan (AF166528), 1 from China (AF381177), 1 from California (AF055391), and the rest from Canada (AF027217, AF055392, AF118097, AF408635, AJ293867, and AJ293868).
The PCR products of expected size were purified from low melting point agarose gel with a QIAEXII kit (Qiagen, Valencia, California, USA) according to manufacturer's instructions. Nucleotide sequencing of the amplified products was carried out at the Advanced Genetic Analysis Center of the University of Minnesota with a DNA sequencer (Model 377; Applied Biosystems, Perkin-Elmer, Foster City, California, USA) and a Taq Dye Deoxy terminator cycle sequencing kit (Applied BioSystem). The sequences were resolved with the ABI PRISM collection program (Perkin-Elmer, Foster City, California, USA). Target segments were verified as PCV2 using the BLAST option found at the NCBI homepage. Sequences were compiled and aligned using the ClustalX program (21). Phylograms were prepared using the neighbor-joining algorithm, and then plotted using NJplot (22).
Results
Serology and virus detection
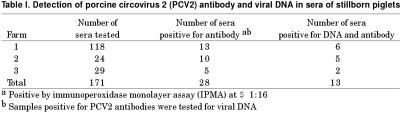
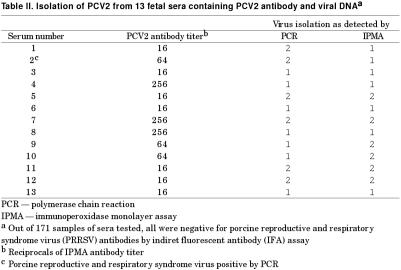
A summary of serology results for PCV2 antibody is shown in Table I. Of the 171 sera tested, 28 (16.4%) were antibody positive. The IPMA titers ranged from 1:16 to > 1:256. Of the 28 antibody-positive sera, PCV2 DNA was demonstrated in 13 samples, with PCR positive samples yielding bands of the expected size. The 28 PCV2 antibody positive samples were also examined for both PPV and PRRSV by respective PCR assays. All were negative except 1 sample that was positive for PRRSV. None of the 28 PCV2 antibody-positive samples were positive for PRRSV antibodies, as detected by indirect fluorescent antibody (IFA) assay. The results of virus isolation are summarized in Table II. Porcine circovirus 2 was isolated from 9 out of 13 sera that were positive for both viral DNA and antibody. The presence of PCV2 in infected cells was confirmed by PCR and IPMA. Five isolates were positive by both procedures and 2 samples each were positive by either PCR or by IPMA.
Table I.
Table II.
Genetic characterization
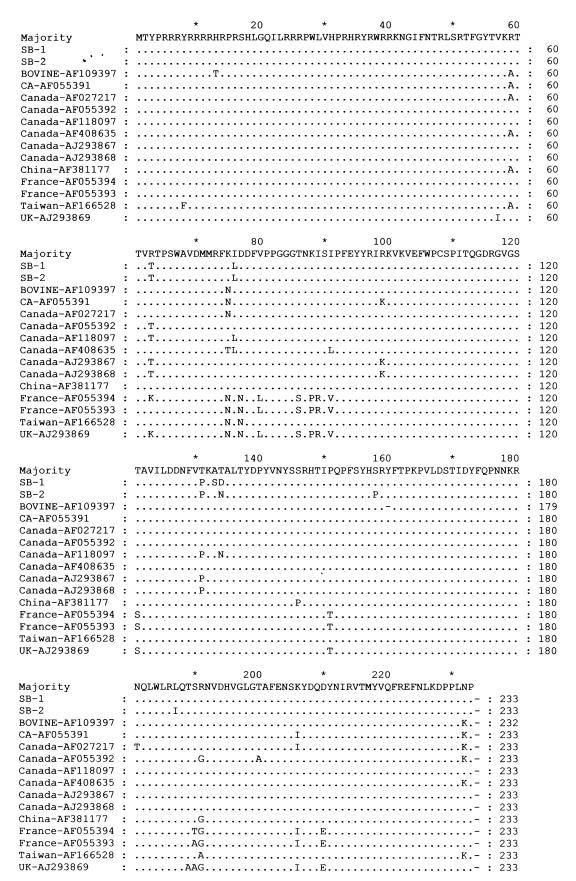
To examine the extent of homology among PCV2 isolates, amino acid sequences of ORF2 were aligned (Figure 1). A pair-wise comparison of aligned amino acid sequences was developed (MegAlign program; DNASTAR, Madison, Wisconsin, USA) (not shown). Amino acid identities of PCV2 isolates SB-1 and SB-2 (GenBank accession numbers AY129154 and AY129155) showed 98% similarity. The SB-1 showed 91.0 to 98.6% homology with other PCV2 isolates and 62.7% homology with PCV1. The SB-2 showed 90.6 to 98.7% homology with the other PCV2 isolates and 61.4% homology with PCV1. Compared with the amino acid sequences of other North American PCV2 isolates, SB-1 and SB-2 isolates were closely related showing from 94.9 to 98.7% homology. The 2 French isolates and 1 United Kingdom isolate (AF055393, AF055394, and AJ293869) share 91% amino acid identity with SB-1 and SB-2. The PCV1 isolate had the largest divergence from all PCV2 isolates showing only 61.4% to 63.9% homology.
Figure 1. Amino acid sequence alignment of the capsid protein open reading frame 2 (ORF2) of porcine circovirus 2 PCV2 isolates. Amino acid deviations from the consensus sequence are indicated by one letter abbreviation. Isolates SB-1 and SB-2 (GenBank accession numbers AY129154 and AY129155) were sequenced for this study, other isolates are cited in the text.
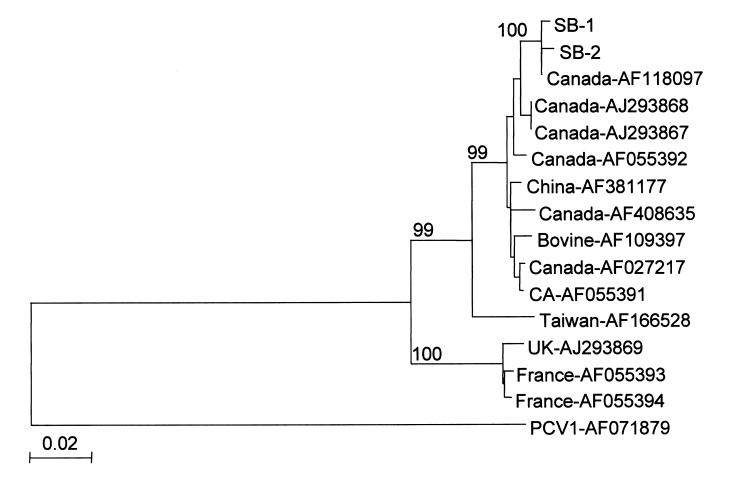
Alignment of nucleotide sequences from ORF2 of PCV2 resulted in a phylogenetic tree (Figure 2). Visual inspection of the tree shows the PCV2 isolates are grouped together in 1 large cluster with 3 minor branches. The largest branch contains all of the North American isolates with the exception of an isolate from China (AF381177), and an isolate from a bovine lung, which is from an unknown location. The 2nd largest branch contains the European isolates, 2 from France (AF055393, AF055394), and 1 from the United Kingdom (AJ293869). The 3rd branch contains only 1 Taiwanese isolate (AF166528). The 2 isolates sequenced for this study appear nearly identical and are grouped with the other North American PCV2 isolates.
Figure 2. Phylogenetic tree based on the nucleotide sequences of open reading frame 2 (ORF2) from 16 porcine circovirus 2 (PCV2) isolates. Nucleotide sequences were aligned using ClustalX (21) and phylograms were generated by the neighbor-joining method using NJplot (22). The percent bootstrap values for each node are shown in each tree. The scale represents the number of substitutions per nucleotide. Isolates SB-1 and SB-2 (GenBank accession numbers AY129154 and AY129155) were sequenced for this study, other isolates are cited in the text.
Discussion
The results reported here indicate that PCV2 antibody and viral DNA can be detected in the sera of stillborn pigs. These results indicate that vertical transmission of PCV2 does occur, and PCV2 is capable of inducing fetal death under field conditions. These results are in agreement with previous studies in which PCV2 was shown to replicate in the porcine fetus (11) and to cause reproductive failures (7,8,9,10).
In a previous study, the pathogenicity of PCV2 in late-term swine fetuses was demonstrated in this laboratory (16). An attempt to reproduce PMWS by fetal inoculation with PCV2 resulted in reproductive failure manifested as stillborns, partial mummies, and weak-born along with clinically normal piglets. Other researchers have examined the effects of PCV2 in swine fetuses at 57, 75, and 92 d of gestation (11). They were able to observe gross lesions in fetuses inoculated at 57 d of age without any detectable PCV2 antibody. The PCV2 antibody was detected in the absence of gross lesions in fetuses inoculated at 75 and 92 d of gestation. These results indicate that PCV2 infection of fetuses may not always cause fetal death, and that the virus, antibody, or both could be isolated from clinically normal born piglets.
In the virus isolation procedure, the presence of PCV2 in the cell cultures can be demonstrated by either PCR assay of infected supernatant or staining of the cell monolayers. Results from the current study appear to suggest that the PCR used to detect PCV2 viral DNA may not always be as effective as detection of PCV2 viral antigen (IPMA) or vice-versa. The discrepancy between these 2 methods may indicate that a combination of both tests should be used to ensure accuracy.
Detection of the single PRRSV-positive sample among the PCV2 antibody and viral DNA positive samples (Table II) was an interesting result. While PRRSV is known to be transmitted both vertically and horizontally in utero (23), co-infection with PRRSV and PCV2 in utero has not yet been examined. At this time, it is not known if the PRRSV PCR positive sample was a truly positive one.
It appears that the ability of PCV2 to infect swine fetuses is obvious, but the ability of the virus to cause fetal pathogenicity is still not clear. The pathogenicity in the fetus could be due to PCV strain involved or fetal age at the time of infection. A recent study examined the presence of PCV2 in stillborn and nonviable neonatal piglets on a newly established sow farm which had experienced a significant increase in the proportion of stillborn and mummified fetuses along with increased preweaning mortality (7). The authors concluded that PCV2-like viruses isolated from cases of reproductive failure may be phenotypically or genetically different from PCV2 associated with PMWS and that such differences could well account for differing disease presentations from one virus. The hypothesis that different types of PCV2 may be responsible for different disease presentations was the subject of another recent study (10). In this study, PCV2 isolates from 2 cases of sow abortion and 1 case of PDNS were genetically characterized. Comparing their results with previously characterized PMWS associated PCV2 isolates, the authors did find some differences in the respective PCV2 genomes. Our study also found some small differences in PCV2 genomes originating from PMWS isolates and those of stillborn fetuses. At this time it remains unclear what significance these differences may have.
Results from the current study indicate that there are a small number of amino acid differences within the capsid protein of PCV2 isolates associated with reproductive failure when compared with PMWS isolates. Exactly what effects these differences may have remains under investigation. More studies are needed to determine different factors associated with PCV2 pathogenicity in swine fetuses. However, it is generally agreed that fetal infection with PCV2 could occur in the field and PCV2 can be considered a fetal pathogen. From the present study, detection of PCV2 antibody and virus in serum of stillborn piglets provides strong evidence that fetal infection with PCV2 and virus induced fetal death may occur in the field.
Footnotes
Address all correspondence and reprint requests to Dr. H.S. Joo; telephone: (612) 625-0235; fax: (612) 625-1210; e-mail: jooxx001@umn.edu
Received July 23, 2002. Accepted November 26, 2002.
References
- 1.Ellis JA, Hassard L, Clark E, et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J 1998;39:44–51. [PMC free article] [PubMed]
- 2.Kennedy S, Moffett D, McNeilly F, et al. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol 2000;122:9–24. [DOI] [PubMed]
- 3.Krakowka S, Ellis JA, McNeilly F, Ringler S, Rings DM, Allan G. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet Pathol 2001;38:31–42. [DOI] [PubMed]
- 4.Gresham A, Thomson J. PMWS and PDNS in Great Britain. Vet Rec 2001;148:387. [PubMed]
- 5.Rosell C, Segales J, Ramos-Vara JA, et al. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec 2000;146:40–43. [DOI] [PubMed]
- 6.Harms PA, Halbur PG, Sorden SD. Three cases of porcine respiratory disease complex associates with porcine circovirus type 2 infection. J Amer Assoc Sw Vet 2002;10:27–30.
- 7.O'Connor B, Gauvreau H, West K, et al. Multiple porcine circovirus 2-associated abortions and reproductive failure in a multisite swine production unit. Can Vet J 2001;42:551–553. [PMC free article] [PubMed]
- 8.Bogdan J, West K, Clark E, et al. Association of porcine circovirus 2 with reproductive failure in pigs: a retrospective study, 1995–1998. Can Vet J 2001;42:548–550. [PMC free article] [PubMed]
- 9.West KH, Bystrom JM, Wojnarowicz C, et al. Myocarditis and abortion associated with intrauterine infection of sows with porcine circovirus 2. J Vet Diagn Invest 1999;11:530–532. [DOI] [PubMed]
- 10.Meehan BM, McNeilly F, McNair I, et al. Isolation and characterization of porcine circovirus 2 from cases of sow abortion and porcine dermatitis and nephropathy syndrome. Arch Virol 2001;146:835–842. [DOI] [PubMed]
- 11.Sanchez RE, Nauwynck HJ, McNeilly F, Allan GM, Pensaert MB. Porcine circovirus 2 infection in swine foetuses inoculated at different stages of gestation. Vet Microbiol 2001;83:169–176. [DOI] [PubMed]
- 12.Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest 2000;12:3–14. [DOI] [PubMed]
- 13.Allan GM, McNeilly F, Meehan BM, et al. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol 1999;66:115–123. [DOI] [PubMed]
- 14.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol 1999;37:3917–3924. [DOI] [PMC free article] [PubMed]
- 15.Tischer I, Peters D, Rasch R, Pociuli S. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch Virol 1987;96:39–57. [DOI] [PubMed]
- 16.Johnson CS, Direksin K, Choi YK, Yoon KJ, Joo HS. Experimental in utero inoculation of late-term swine fetuses with porcine circovirus type 2. J Vet Diagn Invest 2002;14:507–512. [DOI] [PubMed]
- 17.Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol 2000;81:2281–2287. [DOI] [PubMed]
- 18.Ellis JA, Bratanich A, Clark EG, et al. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J Vet Diagn Invest 2000;12:21–27. [DOI] [PubMed]
- 19.Cheon DS, Chae C. Comparison of virus isolation, reverse transcription-polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine reproductive and respiratory syndrome virus from naturally aborted fetuses and stillborn piglets. J Vet Diagn Invest 2000;12:582–587. [DOI] [PubMed]
- 20.Niagro FD, Forsthoefel AN, Lawther RP, et al. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol 1998;143:1723–1744. [DOI] [PubMed]
- 21.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876–4882. [DOI] [PMC free article] [PubMed]
- 22.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 1996;78:364–369. [DOI] [PubMed]
- 23.Mengeling WL, Lager KM, Vorwald AC. Temporal characterization of transplacental infection of porcine fetuses with porcine reproductive and respiratory syndrome virus. Am J Vet Res 1994;55:1391–1398. [PubMed]