Abstract
Multiplex nested polymerase chain reactions (PCRs) were developed for the simultaneous detection and differentiation of genomic material of porcine circovirus 1 (PCV1), porcine circovirus 2 (PCV2), and porcine parvovirus (PPV) in formalin-fixed, paraffin-embedded tissues. Multiplex conventional and nested PCR and in situ hybridization were compared for their ability to detect the 3 viruses in such tissues. Xylene deparaffinization followed by proteinase K digestion yielded DNA of sufficient quality for reliable and consistent PCR analyses. The DNA from PCV1, PCV2, and PPV was detected by both multiplex nested PCR and in situ hybridization in lymph-node tissue from 12 pigs experimentally co-infected with the 3 viruses, as well as in formalin-fixed, paraffin-embedded lymph-node tissue from 30 pigs with naturally occurring postweaning multisystemic wasting syndrome; the agreement rates for the 2 methods were 100% in both groups of pigs. Thus, multiplex nested PCR could be applied successfully to formalin-fixed, paraffin-embedded tissues for simultaneous detection of these 3 porcine viruses.
Porcine circovirus (PCV) was first isolated in 1974, as a contaminant of the continuous porcine kidney cell line PK-15 (ATCC CCL31) (1). This virus, now referred to as PCV1, is nonpathogenic (2). However, PCV1 has been isolated or amplified from pigs with a wasting disease, often shortly after weaning (3,4). Co-infection with both PCV1 and PCV2 can occur (5). PCV2, which differs markedly from PCV1, is commonly found in pigs with postweaning multisystemic wasting syndrome (PMWS) (6,7,8,9). This syndrome affects mainly nursery and early-growing pigs and is clinically characterized by poor body condition, dyspnea, pallor of the skin, and sometimes icterus (10). Recently, co-infection with PCV2 and PPV has been demonstrated in a significant proportion of field cases of PMWS in pigs in Korea (9,11,12) and in Canada (4). It has been shown experimentally that co-infection with PCV2 and PPV causes more severe lesions and clinical disease in PWMS than does infection with PCV2 alone (13,14,15). Therefore, the development of diagnostic tools that can simultaneously detect and differentiate among PCV1, PCV2, and PPV in the same samples would be of significant importance in epidemiologic surveillance programs to predict the severity of PMWS in swine herds.
Diagnosis of PMWS is by microscopic examination of tissues, followed by identification of PCV2 within tissue lesions by immunohistochemistry or in situ hybridization, the tissues being formalin-fixed and paraffin-embedded (10). Detection of PCV2 in tissues by the polymerase chain reaction (PCR) does not necessarily confirm a diagnosis of PMWS, as this virus is highly prevalent in healthy pigs (10,11). However, when PCR detects PCV2 in formalin-fixed, paraffin-embedded tissues from histopathologically identified lesions of PMWS, it is diagnostic for PMWS.
Although several multiplex PCRs can differentiate between PCV1 and PCV2 or between PCV2 and PPV in fresh and formalin-fixed tissues (11,16,17,18), no reports exist of studies that used multiplex PCR for the simultaneous detection and differentiation of 3 viruses in formalin-fixed, paraffin-embedded tissues. Therefore, the 1st objective of this study was to develop a multiplex nested PCR for the simultaneous detection and differentiation of PCV1, PCV2, and PPV DNA in such tissues. The 2nd objective was to compare multiplex nested PCR with conventional PCR and in situ hybridization for the detection of the 3 viruses, since PCR and in situ hybridization can detect viral DNA in formalin-fixed, paraffin-embedded tissues.
Tissue-culture-propagated PCV1 (strain SNUVR000462), PCV2 (strain SNUVR000463), and PPV (strain SNUVR000464) were used as the sources of virus. For inoculation, a PCV1 pool containing a 1.6 × 105 tissue culture infective dose50 (TCID50)/mL, a PCV2 pool containing a 1.2 × 105 TCID50/mL, and a PPV pool containing a 1.3 × 105 TCID50/mL was prepared as previously described (15). We randomly divided 24 colostrum-deprived conventional 28-d-old pigs into 2 groups. All the pigs were seronegative for PCV, PPV, and porcine reproductive and respiratory syndrome virus (PRRSV). One group was inoculated with a mixture (3 mL) of an equal volume of a 1:20 dilution of each virus pool. The other group served as a negative-control group; the pigs were inoculated with PCV-free PK-15 cell lysates. The groups were held in separate isolators and examined at regular intervals. Six pigs from each group were humanely killed at 27 and 35 d postinoculation (dpi) by electrocution. Superficial inguinal lymph nodes were tested for PCR after being fixed in formalin for 24 h, then embedded in paraffin. The experiment was approved by the Institutional Animal Care and Use Committee of Seoul National University.
To examine the effect of fixation time on the ability of multiplex nested PCR to detect PCV1, PCV2, and PPV DNA, we allowed tissues from pigs experimentally co-infected with PCV1, PCV2, and PPV to remain in formalin before being embedded in paraffin. At intervals of 1, 15, 30, 60, 90, 120, 150, and 180 d, we removed a representative sample from each tissue and, with standard histologic procedures, processed the tissue and embedded it in paraffin.
A total of 30 archived formalin-fixed, paraffin-embedded lymph-node specimens from pigs with naturally occurring PMWS were also used in this investigation. Specimens were selected on the basis of clinical signs and microscopic lesions, as described previously (9,11,12,19). The age of the tissues was 5, 4, 3, 2, and 1 y for 6 specimens each. In addition, 3 archived formalin-fixed, paraffin-embedded lymph-node specimens from colostrum-deprived pigs aged 5 y were used as negative controls; 2 specimens were 5 y old and 1 was 3 y old.
For each pig, a 10 μm-wide section of lymph node was prepared from the paraffin-embedded block, and excess paraffin was trimmed. Sections were placed in 1.5-mL sterile tubes. The microtome blade, tweezers, and other equipment that could come into contact with the samples were carefully cleaned before processing of the next tissue block. Extraction of DNA from the tissue was conducted as previously described, with xylene deparaffinization followed by proteinase K digestion (20).
For detecting all 3 types of viral DNA by nested PCR, reverse primers were designed with the use of computer software (Oligo 4.0 program; National Biosciences, Plymouth, Minnesota, USA); in addition, the reverse primers of conventional PCR were used.
For PCV1, the forward primer for conventional PCR was 5'-CTCG GCAGCGTCAGTGAAAA-3' (nucleotide positions 800 to 819), and the reverse primer was 5'-AAATTACGGGCCCACTGGCT-3' (nucleotide positions 1350 to 1369). For nested PCR the forward primer was 5'-CCTTCCGAGGAGGAGAAAAAC-3' (nucleotide positions 879 to 900). The nested primers amplified a 491-base-pair (bp) fragment of the 570-bp region in open reading frame (ORF) 1 (GenBank access no. Y09921) amplified in the first reaction.
For PCV2, conventional PCR was performed with primers previously described (21). The forward primer was 5'-CGGATATTGTAGTC CTGGTCG-3' (nucleotide positions 1095 to 1115), and the reverse primer was 5'-ACTGTCAAGGCTACCACAGTCA-3' (nucleotide positions 1549 to 1570). The nested forward primer was 5'-GGTTTGGGT GTGAAGTAACGGG-3' (nucleotide positions 1242 to 1263). The nested primers amplified a 329-bp fragment of the 481-bp region of ORF 2 (GenBank access no. AF027217) amplified in the first reaction.
For PPV, conventional PCR was performed with primers previously described (22). The forward and reverse primers were 5'-CCAGCAGC TAACACAAGAAAAGGTTATCAC-3' (nucleotide positions 3708 to 3730) and 5'-GTCCATGTTGGTAATCCATTGTAAATC-3' (nucleotide positions 3906 to 3928), respectively. The nested forward primer was 5'-TACACAGAAGCAACAGCAAT-3' (nucleotide positions 3756 to 3775) (GenBank access no. D00623). The nested primers amplified a 178-bp fragment of the 226-bp region amplified in the first reaction. As PCR templates, 10 μL of the supernatant containing extracted DNA was used for the 1st reaction, and 10 μL of the product was used for the 2nd reaction. The amplification was performed in a 50-μL reaction mixture containing 1.25 mM MgCl2, 1 × PCR buffer, 0.2 mM of each dNTP, 1 μM of each primer, and 2.5 U of Taq DNA polymerase (PerkinElmer Inc., Orlando, Florida, USA/Applied Biosystems, Foster City, California, USA). The 2 reactions were run in a thermocycler (GeneAmp PCR System 9600; PerkinElmer/Cetus, Norwalk, Connecticut, USA) under the same conditions: 35 cycles of denaturation at 95°C for 1 min, primer annealing at 65°C for 1 min, and extension at 72°C for 1 min. The PCR was ended with a final extension step at 72°C for 10 min. The reactions were performed in triplicate. Control DNA from the reference strain for PCV1, PPV2, and PPV was included in each reaction. PCR products were purified with a 30-kD cutoff membrane ultrafiltration filter. The nucleotide sequences of the purified PCR products were determined by the use of BigDye chemistry with the ABI Prism Sequencer (PerkinElmer/Applied Biosystems).
The sensitivity of PCV1, PCV2, and PPV detection was also estimated through serial dilutions of DNA extracts prepared from cell culture. In specificity studies, PRRSV, porcine epidemic diarrhea virus, transmissible gastroenteritis virus, rotavirus, and classic swine fever virus were tested with primers for PCV1, PCV2, and PPV. In situ hybridization for PCV1, PCV2, and PPV was used as previously described (9,19).
The nucleotides for the PCR products of PCV1, PCV2, and PPV were determined. A comparison of the sequences with those previously published (GenBank access nos. Y09921, AF027217, and D00623, respectively) revealed 100% homology for all 3 viruses.
Multiplex conventional PCR detected PCV1, PCV2, and PPV DNA up to a dilution of 10−3 (corresponding to 1.7 × 10−3, 1.8 ×10−3, and 2.2 × 10−3 TCID50/mL, respectively). Multiplex nested PCR, on the other hand, detected PCV1, PCV2, and PPV DNA up to a dilution of 10−5 (corresponding to 2.4 × 10−4, 2.1 × 10−4, and 2.3 × 10−4 TCID50/mL, respectively). Neither the outer nor the inner primers cross-reacted with any of the viruses tested.
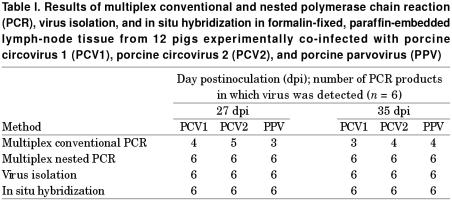
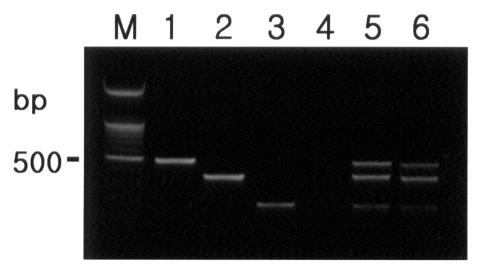
Six multiplex conventional and nested PCR products from each virus were sequenced and their identity confirmed as PCV1, PCV2, or PPV. As Table I shows, multiplex nested PCR detected all 3 viruses (Figure 1) at both 27 and 35 dpi in 6 out of 6 lymph nodes. In contrast, multiplex conventional PCR detected PCV1 DNA in 4/6 at 27 dpi and 3/6 at 35 dpi, PCV2 DNA in 5/6 at 27 dpi and 4/6 at 35 dpi, and PPV DNA in 3/6 at 27 dpi and 4/6 at 35 dpi.
Table I.
Figure 1. Detection of porcine circovirus 1 (PCV1), porcine circovirus 2 (PCV2), and porcine parvovirus (PPV) in formalin-fixed, paraffin-embedded lymph-node tissue by multiplex nested polymerase chain reaction (PCR). Lane M: 100 base-pair (bp) DNA ladder; lane 1: positive control for PCV1; lane 2: positive control for PCV2; lane 3: positive control for PPV; lane 4: negative control; lane 5: positive results for the 3 viruses in tissues from pigs experimentally co-infected with the 3 viruses; lane 6: positive results for the 3 viruses in tissues from pigs naturally co-infected with the 3 viruses.
Positive hybridization signals for PCV1, PCV2, and PPV were detected in the cytoplasm of macrophages and multinucleated giant cells in the lymph nodes from all 12 pigs experimentally co-infected with the 3 viruses. The host-cell structure was preserved in spite of the incubation procedure. Positive cells typically exhibited a dark brown to black reaction product, mainly in the cytoplasm but occasionally in the nucleus, without any background staining. The agreement rates for multiplex nested PCR and in situ hybridization were 100% for the detection of PCV1, PCV2, and PPV in the pigs experimentally co-infected (Table I).
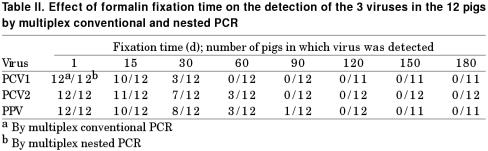
To determine whether prolonged fixation in formalin affected the detection ability of multiplex conventional and nested PCR, 96 tissue samples from the 12 co-infected pigs were examined at intervals up to 180 d after formalin fixation. PCV1 DNA was detected in 25 (26%) and 93 (96.9%) of the 96 samples, PCV2 DNA in 33 (34.4%) and 96 (100%), and PPV DNA in 34 (35.4%) and 94 (97.9%) by conventional and nested PCR, respectively. Acceptable signals were detected in tissues fixed for up to 15 and 180 d by conventional and nested PCR, respectively (Table II).
Table II.
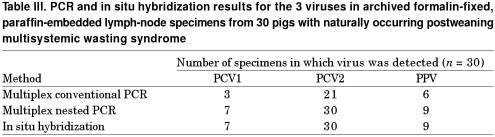
Table III summarizes the results for the 30 archived specimens of formalin-fixed lung tissue from pigs with naturally occurring PWMS. The specimens found to be positive by multiplex conventional PCR were always positive by multiplex nested PCR: 3 and 7 of the 30 samples were positive for PCV1, 21 and 30 were positive for PCV2, and 6 and 9 were positive for PPV by conventional and nested PCR, respectively. Among the 30 specimens, PCV2 DNA was detected in all 6 of those 1, 2, 3, 4, or 5 y old. Positive hybridization signals for PCV1, PCV2, and PPV were detected in 7 (age 1 or 2 y for 2 specimens each and age 3, 4, or 5 y for 1 specimen each), 30, and 9 (age 1, 2, 4, or 5 y for 2 specimens each and age 3 y for 1 specimen) specimens, respectively. Positive cells typically exhibited a dark brown to black reaction product, mainly in the cytoplasm but occasionally in the nucleus, without any background staining. The agreement rates for multiplex nested PCR and in situ hybridization were 100% for the detection of PCV1, PCV2, and PPV in the pigs with naturally occurring PMWS.
Table III.
Diagnosis of PMWS is based on 3 criteria: the presence of compatible clinical signs, characteristic microscopic lesions, and typical virus within the lesions (10). Characteristically, the microscopic lesions show depletion of lymphoid tissue and lymphohistiocytic to granulomatous inflammation in the lung, liver, kidney, and lymphoid tissues (10,12,23). In order to establish an etiologic diagnosis for PMWS, techniques that correlate the presence of virus and specific tissue lesions are required; these include immunohistochemistry (23) and in situ hybridization (8,19), but not PCR (11), virus isolation, and serologic tests (24), because PCV2 is highly prevalent in healthy pigs (10,11). However, PCR can be applied to the detection of PCV2 in formalin-fixed, paraffin-embedded tissues containing known histopathologic lesions of PMWS, as described here. When PCR results are interpreted in conjunction with the characteristic histopathologic changes, PCR should be a valuable confirmatory diagnostic tool.
The main obstacle in preparing DNA suitable for PCR amplification is removal of paraffin (20). A previous study demonstrated that xylene produces better results than any other solvent (20). Another obstacle to the reproducibility of PCR results is the strong degradation of DNA extracted from formalin-fixed, paraffin-embedded tissues, which is affected by the time between surgical removal of the tissue and fixation, the fixative used, and the duration of fixation (25). At the molecular level, formalin fixation leads to cross-linking of DNA to tissue constituents, which makes it difficult to extract genomic DNA templates of good quality (for example, with a low level of strand breaks, DNA breakdown, and PCR inhibitors). Moreover, degradation of DNA may occur before adequate fixation, and the processing conditions may vary extensively for various formalin-fixed, paraffin-embedded tissues (25). Therefore, the protocol for the detection of viral DNA in routinely processed samples must be rigorously optimized.
Pathological specimens for microscopic evaluation are routinely formalin-fixed and paraffin-embedded by standard methods prior to evaluation. These samples are an invaluable resource for molecular studies, and the ability to amplify specific regions of DNA from formalin-fixed, paraffin-embedded tissues by PCR has a profound impact on diagnostic pathology. In comparison with existing identification methods, such as virus isolation and in situ hybridization, PCR amplification of viral sequences from formalin-fixed, paraffin-embedded tissue is a simple and sensitive method and should permit convenient and reliable detection of viral sequences in diagnostic pathology. Detection of PCV1, PCV2, and PPV by multiplex nested PCR will be useful both as a molecular diagnostic method and as a research tool for the study of the pathogenesis and epidemiology of these 3 porcine viruses in PMWS.
Footnotes
Acknowledgments
This research was supported by the Ministry of Agriculture, Forestry and Fisheries-Special Grants Research Program (MAFF-SGRP) and by the Brain Korea 21 Project, Republic of Korea.
Address all correspondence and reprint requests to Dr. Chanhee Chae; telephone: +82 2 880-1277; fax: +82 2 871-5821; e-mail: swine@plaza.snu.ac.kr
Received July 16, 2002. Accepted October 29, 2002.
References
- 1.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Mikrobiol Hyg Ser A 1974;226:153–167. [PubMed]
- 2.Allan GM, McNeilly F, Cassidy JP, et al. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig fetal material. Vet Microbiol 1995;44:49–64. [DOI] [PubMed]
- 3.LeCann P, Albina E, Madec F, et al. Piglet wasting disease. Vet Rec 1997;141:660. [PubMed]
- 4.Ellis JA, Bratanich A, Clark EG, et al. Co-infection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J Vet Diagn Invest 2000;12:21–27. [DOI] [PubMed]
- 5.Allan G, Meehan B, Todd D, et al. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec 1998;142:467–468. [PubMed]
- 6.Meehan BM, McNeilly F, Todd D, et al. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol 1998;79:2171–2179. [DOI] [PubMed]
- 7.Morozov I, Sirinarumitr T, Sorden SD, et al. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol 1998;36:2535–2541. [DOI] [PMC free article] [PubMed]
- 8.Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Comp Pathol 1999;121:265–270. [DOI] [PubMed]
- 9.Choi C, Chae C. Distribution of porcine parvovirus in porcine circovirus 2 — infected pigs with postweaning multisystemic wasting syndrome as shown by in-situ hybridization. J Comp Pathol 2000;123:302–305. [DOI] [PubMed]
- 10.Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest 2000;12:3–14. [DOI] [PubMed]
- 11.Kim J, Choi C, Han DU, Chae C. Simultaneous detection of porcine circovirus 2 and porcine parvovirus in pigs with postweaning multisystemic wasting syndrome by multiplex PCR. Vet Rec 2001;149:304–305. [DOI] [PubMed]
- 12.Kim J, Chung H-K, Jung T, et al. Postweaning multisystemic wasting syndrome of pigs in Korea: prevalence, microscopic lesions and coexisting microorganisms. J Vet Med Sci 2002;64:57–62. [DOI] [PubMed]
- 13.Allan GM, Kennedy S, McNeilly F, et al. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol 1999;121:1–11. [DOI] [PubMed]
- 14.Kennedy S, Moffett D, McNeilly F, et al. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J Comp Pathol 2000;122:9–24. [DOI] [PubMed]
- 15.Krakowka S, Ellis JA, Meehan B, et al. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by co-infection with porcine circovirus 2 and porcine parvovirus. Vet Pathol 2000;37:254–263. [DOI] [PubMed]
- 16.Kim J, Han DU, Choi C, Chae C. Differentiation of porcine circovirus (PCV)-1 and PCV-2 in boar semen using multiplex nested polymerase chain reaction. J Virol Methods 2001;98:25–31. [DOI] [PubMed]
- 17.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol 1999;37:3917–3924. [DOI] [PMC free article] [PubMed]
- 18.Larochelle R, Antaya M, Morin M, Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. J Virol Methods 1999;80:69–75. [DOI] [PubMed]
- 19.Kim J, Chae C. Differentiation of porcine circovirus 1 and 2 in formalin-fixed, paraffin-embedded tissues from pigs with postweaning multisystemic wasting syndrome by in-situ hybridisation. Res Vet Sci 2001;70:265–269. [DOI] [PubMed]
- 20.Kim J, Chae C. Optimized protocols for the detection of porcine circovirus 2 DNA from formalin-fixed paraffin-embedded tissues using nested polymerase chain reaction and comparison of nested PCR with in situ hybridization. J Virol Methods 2001;92:105–111. [DOI] [PubMed]
- 21.Ellis J, Krakowka S, Lairmore M, et al. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest 1999;11:3–14. [DOI] [PubMed]
- 22.Arnauld C, Legeay O, Laurian Y, et al. Development of a PCR-based method coupled with a microplate colorimetric assay for the detection of porcine parvovirus and application to diagnosis in piglet tissues and human plasma. Mol Cell Probes 1998;12:407–416. [DOI] [PubMed]
- 23.Choi C, Chae C, Clark EG. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J Vet Diagn Invest 2000;12:151–153. [DOI] [PubMed]
- 24.Magar R, Muller P, Larochelle R. Retrospective serological survey of antibodies to porcine circovirus type 1 and type 2. Can J Vet Res 2000;64:184–186. [PMC free article] [PubMed]
- 25.Merkelbach S, Gehlen J, Handt S, Fuzesi L. Novel enzyme immunoassay and optimized DNA extraction for the detection of polymerase-chain-reaction-amplified viral DNA from paraffin-embedded tissue. Am J Pathol 1997;150:1537–1546. [PMC free article] [PubMed]