Abstract
Conventional kinesin and Eg5 are essential nanoscale motor proteins. Single-molecule and presteady-state kinetic experiments indicate that both motors use similar strategies to generate movement along microtubules, despite having distinctly different in vivo functions. Single molecules of kinesin, a long-distance cargo transporter, are highly processive, binding the microtubule and taking 100 or more sequential steps at velocities of up to 700 nm/s before dissociating, whereas Eg5, a motor active in mitotic spindle assembly, is also processive, but takes fewer steps at a slower rate. By dissecting the structural, biochemical and mechanical features of these proteins, we hope to learn how kinesin and Eg5 are optimized for their specific biological tasks, while gaining insight into how biochemical energy is converted into mechanical work.
Introduction
Microtubule-based kinesin superfamily motors are involved in diverse cellular processes including intracellular transport, mitosis and meiosis, regulation of microtubule dynamics, and signal transduction [1]. Here we consider conventional kinesin, (‘kinesin’, the Kinesin-1 subfamily) and Eg5 (the Kinesin-5 subfamily), both of which have conserved N-terminal catalytic motor domains and walk processively toward the plus-end of microtubules, hydrolyzing one ATP per 8-nm step ([2,3,4••], reviewed in [5•,6,7,8•,9]).
In both, the motor domain is followed by a 12–15 amino acid residue neck linker leading to the coiled-coil stalk. However, there are important structural differences (Figure 1). Kinesin is a homodimer, composed of two heavy chains, with two light chains associating with the C-terminal cargo-binding domain. By contrast, Eg5 is a homotetramer: two polypeptides first dimerize to form a parallel coiled-coil, and then two dimers form an anti-parallel coiled-coil tetramer containing four motor domains. As a tetramer, Eg5 can crosslink two adjacent microtubules such that each dimeric motor unit interacts with a single protofilament on each microtubule [10•,11••].
Figure 1.
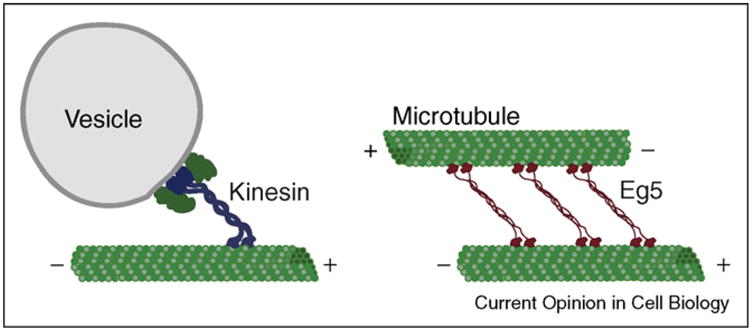
Schematic of Kinesin-1 towing its cargo along a microtubule (left) while Eg5 crosslinks two anti-parallel microtubules (right). By walking to the plus-ends of each microtubule, Eg5 can slide the two microtubules apart.
Kinesin was discovered in 1985 [12] and new single molecule assays [13,14] and presteady-state kinetic experiments [15] soon followed. Since then, thousands of experiments have been performed, allowing a consensus model to emerge (Figure 2). By contrast, mechanochemical data for Eg5 are just beginning to appear. Here, we review the current model for kinesin processivity, summarizing the evidence for a strain-gated hand-over-hand mechanism, and then examine what we currently know about the kinetics and mechanics of Eg5 (Figure 2, Table 1), high-lighting recent advances and unresolved questions for each.
Figure 2.
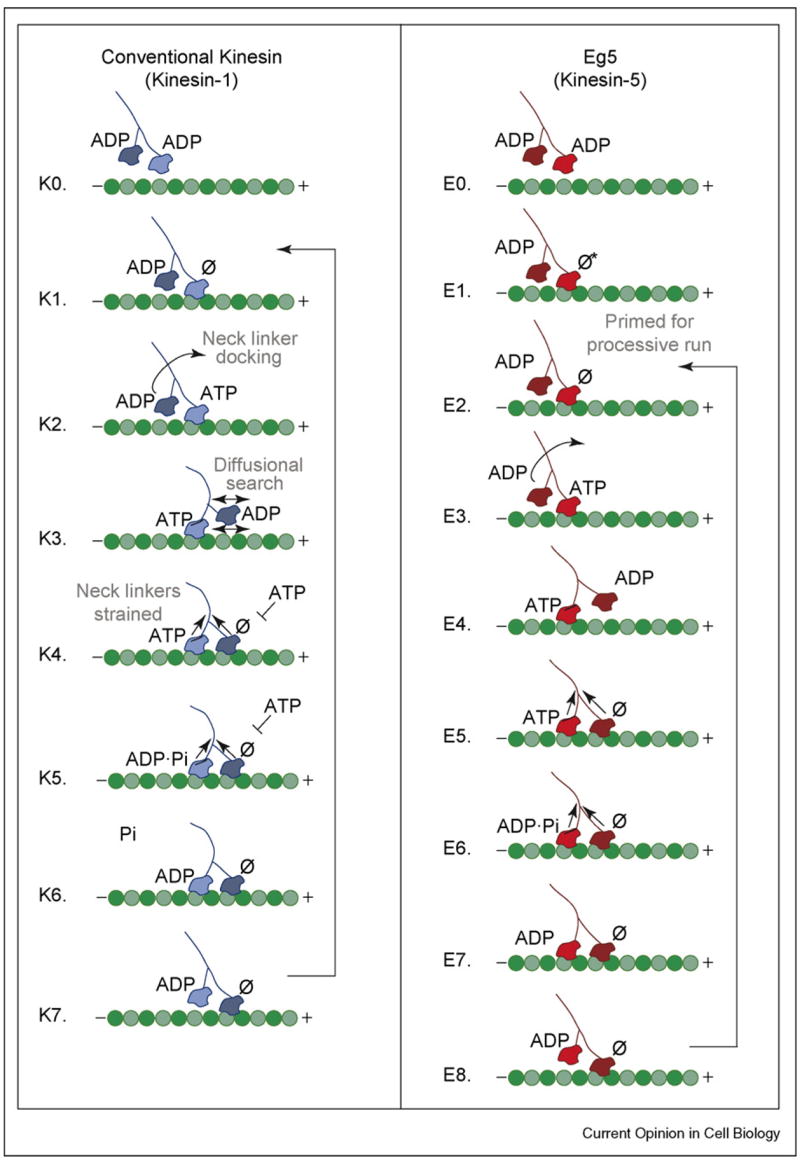
Stepping models for Kinesin and Eg5. For kinesin, a processive run begins when a motor head collides with the microtubule and releases its ADP. ATP binding at the forward head leads to neck linker docking and plus-end directed movement of the rearward head that promotes binding at the next site on the microtubule. Release of ADP at step K4 results in a state with both heads bound tightly to the microtubule and the development of intermolecular strain between the two motor domains. ATP hydrolysis occurs on the rearward head followed by phosphate release and rearward head detachment, which returns the motor to K1, having taken a single step. For kinesin, phosphate release is rate-limiting. In contrast to kinesin, Eg5 collides with the microtubule but cannot bind ATP until a conformational change occurs, which is believed to involve movement of the neck linker. This isomerization establishes a processive run (step E2), allowing ATP binding on the forward head. Successive steps in the cycle are similar to those for kinesin but with rate-limiting ATP hydrolysis.
Table 1.
Constants for the microtubule–kinesin and Eg5 ATPase pathways from solution kinetic studies
| Constants | Kinesina | Eg5-513b | |
|---|---|---|---|
| ATP binding | k+1k+1′ | 2–5 μM−1 −1 | 5.8 μM−1 s−1 |
| k+1′ | 240 s−1 | 52 s−1 | |
| Kd,ATP | 80 mM | 9 μM | |
| K−1 | 120 s−1 | 1–2 s−1 | |
| ATP hydrolysis | k2 | 100 s−1 | 5–10 s−1 |
| Pi release | k3 | 40 s−1 | ≫ 10 s−1 |
| ATP-promoted dissociation | k+5 | 40–60 s−1 | ≫ 10 s−1 |
| MT•motor association | k+4 | 11 mM−1s−1 | 2.8 μM−1 s−1 |
| k−4 | 10 s−1 | ||
| MT-activated ADP release | koff,ADP | Head 1: >200 s−1 | Head 1: 28 s−1 |
| Head 2: >100 s−1 | Head 2: ≫10s−1 | ||
| MT-activated ATPase | kcat | 25 s−1 per site | 0.5 s−1 |
| Km,ATP | 80 μM | 8 μM | |
| K1/2,Mt | 0.9 μM | 1.8 μM | |

| |||
A consensus mechanochemical cycle for dimeric kinesin
In solution, each of kinesin’s motor heads contains a tightly bound ADP, which is released upon collision with the microtubule. This rapid release is biphasic: the first ADP is released at >200 s−1, while the release of the second ADP is nucleotide-dependent, with ATP-stimulated release at >100 s−1 (Table 1, [16–19]). When interpreted in the context of the Rice et al. neck linker model [20–22], these data lead to the following pathway (Figure 2, steps K1–K4). The first motor head collides with the microtubule and releases its ADP. The second head remains tethered until ATP binding on the microtubule-bound head causes the neck linker to dock along its catalytic core toward the plus-end of the microtubule. Single-molecule measurements support the proposal that ATP-binding precedes the force-dependent translocation. The distance to the transition state for the mechanical step is ~2.7 nm, significantly smaller than the full 8-nm step, perhaps indicating that diffusion also plays a role [23].
Once the neck-linker-driven conformational change propels the tethered head toward its forward microtubule-binding site, the second ADP is released, forming an intermediate with both heads tightly bound to the microtubule and neck linkers strained. Exit from step K4 must be tightly controlled, since premature binding of ATP to the empty forward head would desynchronize the catalytic cycles of the two motor heads, leading to eventual microtubule dissociation. To maintain processivity, ATP hydrolysis and product release from the rear head must precede ATP binding on the forward head, leading to a state in which the empty head is tightly bound to the microtubule and the ADP-bound head is tethered, restarting the biochemical cycle. Experiments with mutants that uncouple phosphate (Pi) release and motor detachment from the microtubule were used to determine the order of steps K5–K7, in which Pi release is rate-limiting [24]. With each cycle, the two motor heads exchange position, progressing in an asymmetric hand-over-hand fashion [25–27].
Two broad classes of models have been proposed to explain how exit from step K4 may be regulated by strain between the two heads. In the first, strain weakens the microtubule-binding affinity of the rearward head, promoting Pi release and detachment (the ‘rear-gated head’ model) [28,29]. In the second, strain prevents ATP binding to the forward head until the rearward head detaches (the ‘front-gated head’ model) [24,30–32]. Although there is experimental evidence for both models — which are by no means mutually exclusive — new data strongly reinforce the front-gated head model. A single-molecule study by Guydosh and Block used beryllium fluoride (BeFx) to induce long pauses in normal kinesin stepping. ADP•BeFx BeFx occupies the rear head, inducing a neck linker-docked tight-binding ATP state on the microtubule analogous to the configuration shown in step K4, with no nucleotide bound at the forward head [33•]. Processive stepping only resumed after an obligatory ATP-dependent backstep that swapped the positions of the forward and rear heads. This result indicates preferential release of BeFx from the forward head, supporting the model in which ATP binding to the forward head is gated by strain. Strain may also slow the rate of ATP hydrolysis at step K5, because ATP hydrolysis is significantly faster for monomers (>300 s−1) than in dimeric kinesin (100 s−1) [34–36].
Determining the orientation and mobility of the neck linkers for different nucleotide states is essential to confirming models of strain gating and neck linker docking [20,22,37,38,39••,40,41]. In a key experiment, Skiniotis et al. [37] engineered an active dimeric kinesin with an SH3 domain (a ~7 kDa globular β barrel) at the transition between the neck linker and the neck coiled-coil, thus enhancing the visibility of the neck region in cryoelectron micrographs (cryoEM). They showed binding of both heads along the same microtubule protofilament with neck linkers extended in opposite orientations, verifying the ability of the neck linkers to mediate strain in a two-heads-bound state. More recently, Asenjo et al. [39••] used fluorescence polarization microscopy to measure neck-linker configuration in dimers. They demonstrated that the neck linker was mobile during stepping and also in the absence of nucleotide. However, the mobility decreased in the presence of AMPPNP (a non-hydrolyzable nucleotide analog that mimics an ATP-like state) as well as in the presence of ADP•AlF4− (which mimics the ADP•Pi state). Moreover, the neck linker region was roughly parallel to the microtubule axis. These results provide additional evidence for the neck linker orientations displayed in steps K4 and K5.
Kinesin processivity: unresolved questions
Although the model presented in Figure 2 is representative of published results, there remain points of disagreement and steps that require stronger experimental support. Between the conformational changes that drive processive stepping, the motor pauses in a ‘waiting’ state, the nature of which is largely unknown but which is likely to depend on the ATP concentration and applied load. One point of controversy is the position of the tethered head with respect to the bound head and microtubule lattice. Recent biochemical experiments have indicated that the rear ADP-containing head of kinesin (steps K1 and K6–K7) can infrequently synthesize ATP, suggesting that tight microtubule binding occurs, at least transiently [42]. However, because ATP resynthesis is energetically costly and inhibits processivity, it is possible that entry into the synthesis-competent state is also strain-regulated, limiting the probability of ATP resynthesis, which is measured to be <3% [42]. Single-molecule imaging of the fluorescently labeled kinesin motor domains at low ATP levels indicates a predominantly two-heads-bound state [27]. However, unbinding force measurements in the presence of ADP indicate a one-head-bound state [43], and recent mechanical data [44••] and kinetic data [45] suggest that, during the waiting state, one head grips the microtubule while the other diffuses. Reconciling these data demands the differentiation of strongly bound force-bearing states and weakly bound electrostatically tethered states of each motor head, and new experimental methods may be required.
A second, related controversy surrounds the ability of kinesin to step backwards along the microtubule toward its minus-end. Infrequent backsteps appear to be a normal part of kinesin’s kinetic cycle, and have been observed in single-molecule optical trapping studies at a variety of ATP and force conditions [46]. A single kinesin motor will step forward against hindering loads of up to ~6–7 pN, with the velocity slowing until the motor finally stalls [23]. Near this maximal force, the frequency of forward and backward stepping is roughly equal. Carter and Cross recently showed that at much higher superstall forces, kinesin will step processively backwards at an ATP-dependent rate [44••]. They propose a model in which the diffusional search of the tethered head in step K3 can be sufficiently biased by rearward load to reverse motor direction. Yet important questions remain. Does processive backward stepping synthesize ATP — a true cycle reversal as detected by Hackney et al. [42]? Is ATP hydrolysis required? Is ATP binding driving a neck-linker-dependent conformational change that promotes mobility of the tethered head? Finally, the observation by Guydosh and Block that analog-induced pauses were relieved only through back-stepping is intriguing [33•]. Is it possible that the baseline-levels of backstepping observed in physiological buffers and substall forces also provide a means of error correction? For example, forcing the heads to swap positions could promote the dissociation of an incorrect nucleotide or reorient an incorrectly positioned neck linker to reset the coordination between the two motor heads. In this way, backstepping might actually provide a novel means of promoting forward processivity.
An emerging model of Eg5 mechanochemistry
To determine the mechanical and kinetic requirements for Eg5-promoted spindle assembly [47–49], a stable Eg5 dimer was developed [11••]. Eg5-513 promoted robust plus-end-directed microtubule gliding at a rate similar to that of native tetramers [11••,50]. Single Eg5 dimers were found to step processively in optical trapping experiments, taking approximately eight steps on average at speeds of up to 100 nm/s at saturating ATP levels and zero applied load [4••]. Moreover, Eg5 dimers continued to move processively against hindering loads of ~6–7 pN, and, in sharp contrast to kinesin, motors did not slow substantially and stall at higher loads. Instead, Eg5 dissociated from the filament at high loads with only a small decrease in velocity. A minimal kinetic model indicates that ATP-binding precedes the force-dependent translocation and predicts a distance to the transition state of 1.9 nm, smaller than that measured for kinesin and consistent with Eg5 velocity being less force-sensitive.
The single molecule results showed that a rate-limiting transition occurs at 11.9 s−1 [4••], significantly faster than steady-state turnover determined from solution studies, kcat = 0.5 s−1 ([11••], Table 1). A recent transient-state kinetic analysis has resolved this discrepancy [51••]. Upon collision with the microtubule, Eg5 rapidly releases ADP (Step E1, Figure 2). However, unlike kinesin, Eg5 must then undergo a slow conformational change at 0.5–1 s−1 before binding ATP in order to initiate a processive run (steps E1—E2). Because the single-molecule experiments measured velocity within a processive run, they are insensitive to this slow step. Rosenfeld et al. detected a similar isomerization in FRET studies of Eg5 monomers, although the rate was faster in the absence of the partner head [52•]. The X-ray crystal structure of the Eg5 ADP-bound monomer revealed a unique neck linker configuration perpendicular to the long edge of the protein, which was stabilized by a series of hydrogen bonds that are conserved among Kinesin-5-subfamily members [53]. We hypothesize that the observed slow isomerization required for ATP binding [51••,52•] may involve reorienting the neck linker from the perpendicular position, a transition that may be Eg5-specific.
Once primed for a processive run (step E2), we propose that Eg5 proceeds through a kinetic cycle similar to that of kinesin. Both ADP release and ATP binding steps are biphasic, indicating alternating-site catalysis [51••]. Structural and FRET studies suggest that ATP binding docks the neck linker toward the plus-end of the microtubule [11••,52•,54], eventually leading to a two-heads-bound intermediate as observed by cryoEM [11••]. By analogy to kinesin, we suspect that the two-heads-bound state is under tension and involved in strain-induced head–head communication. ATP hydrolysis on the rearward head is followed by rapid Pi release and rearward head detachment (steps E6–E8), restarting the biochemical cycle. This model assumes that the neck linker does not return to the perpendicular position during a processive run and that Eg5 steps hand-over-hand. The presteady-state kinetics revealed another surprise: ATP hydrolysis is rate-limiting at 5–10 s−1, in good agreement with the rate-limiting transition measured by single molecule methods [4••]. This result indicates that, unlike kinesin, which is limited by Pi release, Eg5’s stepping rate is controlled by ATP hydrolysis.
Eg5 processivity: the challenge awaits
Eg5 dimers are clearly processive, but take far fewer steps than kinesin under similar conditions. A key area of future research will be determining what limits the run length of Eg5. Two possibilities seem likely: that the microtubule-binding affinity of each Eg5 motor head is weak enough in all nucleotide states to promote frequent detachment from the microtubule, or, alternatively, that strong binding states exist, but that poor head–head communication prevents the tight alternation of the ATPase cycles required for long processive runs. Given the ability of single motors to sustain high loads without significant decrease in run length or velocity [4••], we favor the latter. Future experiments are required to explore this important issue.
In cells, the differential processivity of kinesin and Eg5 may be advantageous. As an isolated cargo-transporter, kinesin must maintain a tight grip on the microtubule even under significant load. By contrast, ensembles of Eg5 motors must collectively crosslink and slide microtubules in the mitotic spindle. Force-insensitive velocities may allow Eg5 to move under considerable tension while short run lengths prevent interference among neighboring motors. Recent single-molecule fluorescence imaging of GFP-labeled Eg5 tetramers indicates that, in addition to ATP-driven stepping, tetramers also undergo biased diffusion toward the microtubule plus-end, raising the possibility of unique modes of motion in vivo [55]. Extending kinetic and mechanical measurements to Eg5 tetramers, while technically difficult, will be critical to fully understand Eg5 regulation and motility in spindles.
Conclusions
Our understanding of kinesin superfamily members is advancing rapidly, in part because of technological improvements, but also because of the widespread interest in these molecular motors. They are present in every cell of every eukaryotic organism, and are intimately involved in human health and development. Comparisons of conventional kinesin and Eg5 show that in spite of their structural and mechanistic similarities, each has optimized its chemical and mechanical cycle differently for processive stepping along the microtubule. These differences are what generates the diversity for cellular function and fascinates us all.
Acknowledgments
We thank Nick Guydosh, Troy Krzysiak, Andreas Hoenger, and Steve Rosenfeld for helpful discussions and careful reading of the manuscript, and Polly Fordyce for assistance with Figure 1. M.T.V. was supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. S.P.G. was supported by grant GM54141 from NIGMS and Career Development Award K02-AR47841 from NIAMS, National Institutes of Health.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 3.Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 4••.Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loadsin vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. This study is the first single molecule analysis of a processive mitotic kinesin. The paper demonstrates biophysical and biochemical characterization of Eg5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Asbury CL. Kinesin: world’s tiniest biped. Curr Opin Cell Biol. 2005;17:89–97. doi: 10.1016/j.ceb.2004.12.002. A thoughtful and balanced review of Kinesin-1 processivity via asymmetric hand-over-hand walking. [DOI] [PubMed] [Google Scholar]
- 6.Yildiz A, Selvin PR. Kinesin: walking, crawling or sliding along? Trends Cell Biol. 2005;15:112–120. doi: 10.1016/j.tcb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Carter NJ, Cross RA. Kinesin’s moonwalk. Curr Opin Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8•.Cross RA. The kinetic mechanism of kinesin. Trends Biochem Sci. 2004;29:301–309. doi: 10.1016/j.tibs.2004.04.010. A thoughtful and balanced minireview of the ATPase mechanism of Kinesin-1. [DOI] [PubMed] [Google Scholar]
- 9.Block SM. Molecular Motors: point counterpoint. Biophysical Society Discussions; 2006. Kinesin motor mechanics: binding, stepping, tracking, gating, limping… and some newly discovered rotational states. URL: http://www.biophysics.org/discussions/2006/study-book.pdf, Block, p. SP13-A. [Google Scholar]
- 10•.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. An exciting report which shows that purified tetrameric Eg5 can crosslink two parallel microtubules and slide two anti-parallel microtubules relative to one another in vitro. [DOI] [PubMed] [Google Scholar]
- 11••.Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, Gilbert SP, Hoenger A. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. This study shows that AMPPNP promotes an Eg5 two-heads-bound configuration on single microtubule protofilaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 14.Block SM, Goldstein LSB, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 15.Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y-Z, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- 19.Crevel I, Carter N, Schliwa M, Cross RA. Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J. 1999;18:5863–5872. doi: 10.1093/emboj/18.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 21.Rosenfeld SS, Jefferson GM, King PH. ATP reorients the neck linker of kinesin in two sequential steps. J Biol Chem. 2001;276:40167–40174. doi: 10.1074/jbc.M103899200. [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeld SS, Xing J, Jefferson GM, Cheung HC, King PH. Measuring kinesin’s first step. J Biol Chem. 2002;277:36731–36739. doi: 10.1074/jbc.M205261200. [DOI] [PubMed] [Google Scholar]
- 23.Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klumpp LM, Hoenger A, Gilbert SP. Kinesin’s second step. Proc Natl Acad Sci USA. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 27.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 28.Hancock WO, Howard J. Kinesin’s processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schief WR, Clark RH, Crevenna AH, Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-overhand mechanism. Proc Natl Acad Sci USA. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenfeld SS, Fordyce PM, Jeffereson GM, King PH, Block SM. Stepping and stretching: how kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klumpp LM, Brendza KM, Rosenberg JM, Hoenger A, Gilbert SP. Motor domain mutation traps kinesin as a microtubule rigor complex. Biochemistry. 2003;42:2595–2606. doi: 10.1021/bi026715r. [DOI] [PubMed] [Google Scholar]
- 32.Crevel IM, Nyitrai M, Alonso MC, Weiss S, Geeves MA, Cross RA. What kinesin does at roadblocks: the coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci USA. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. A careful and thorough analysis of kinesin backstepping induced by nucleotide analogs. The results provide evidence for a gated front head mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyer ML, Gilbert SP, Johnson KA. Purification and characterization of two monomeric kinesin constructs. Biochemistry. 1996;35:6321–6329. doi: 10.1021/bi960017n. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y-Z, Taylor EW. Kinetic mechanism of a monomeric kinesin construct. J Biol Chem. 1997;272:717–723. doi: 10.1074/jbc.272.2.717. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert SP, Johnson KA. Pre-steady-state kinetics of the microtubule-kinesin ATPase. Biochemistry. 1994;33:1951–1960. doi: 10.1021/bi00173a044. [DOI] [PubMed] [Google Scholar]
- 37.Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, Mandelkow E, Hoenger A. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asenjo AB, Krohn N, Sosa H. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat Struct Biol. 2003;10:836–842. doi: 10.1038/nsb984. [DOI] [PubMed] [Google Scholar]
- 39••.Asenjo AB, Weinberg Y, Sosa H. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat Struct Mol Biol. 2006;13:648–654. doi: 10.1038/nsmb1109. This single molecule study uses fluorescence polarization microscopy to determine the neck linker configuration for the forward and rearward heads of dimeric kinesin. The neck linker region is very mobile during steady walking, but the mobility decreases in the presence of AMPPNP (ATP state) and ADP+AlF4− (ADP•Pi state) with the neck-linker region close to parallel to the microtubule axis. [DOI] [PubMed] [Google Scholar]
- 40.Tomishige M, Stuurman N, Vale RD. Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat Struct Mol Biol. 2006;13:887–894. doi: 10.1038/nsmb1151. [DOI] [PubMed] [Google Scholar]
- 41.Hahlen K, Ebbing B, Reinders J, Mergler J, Sickmann A, Woehlke G. Feedback of the kinesin-1 neck-linker position on the catalytic site. J Biol Chem. 2006;281:18868–18877. doi: 10.1074/jbc.M508019200. [DOI] [PubMed] [Google Scholar]
- 42.Hackney DD. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc Natl Acad Sci USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S. Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci USA. 2002;99:5977–5981. doi: 10.1073/pnas.092546199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Carter NJ. Cross RA: Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. This study shows processive backstepping by kinesin under superstall backward loads at an ATP-dependent rate with no indication of substeps. [DOI] [PubMed] [Google Scholar]
- 45.Auerbach SD, Johnson KA. Alternating site ATPase pathway of rat conventional kinesin. J Biol Chem. 2005;280:37048–37060. doi: 10.1074/jbc.M502984200. [DOI] [PubMed] [Google Scholar]
- 46.Svoboda K, Mitra PP, Block SM. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc Natl Acad Sci USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwok BH, Yang JG, Kapoor TM. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol. 2004;14:1783–1788. doi: 10.1016/j.cub.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 48.Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale R, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hildebrandt ER, Gheber L, Kingsbury T, Hoyt MA. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J Biol Chem. 2006;281:26004–26013. doi: 10.1074/jbc.M604817200. [DOI] [PubMed] [Google Scholar]
- 50.Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- 51••.Krzysiak TC, Gilbert SP. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J Biol Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. This transient-state kinetic analysis identifies Eg5 as the first kinesin motor to have a rate-limiting ATP hydrolysis step. In addition, this study shows a requirement for a slow structural transition upon collision with the microtubule that is required for ATP binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Rosenfeld SS, Xing J, Jefferson GM, King PH. Docking and rolling, a model of how the mitotic motor Eg5 works. J Biol Chem. 2005;280:35684–35695. doi: 10.1074/jbc.M506561200. Fluorescence resonance energy transfer (FRET) is used to measure the structural transitions at the neck linker and other domains of monomeric Eg5 as a function of ATP. This study also documents the requirement for a slow conformational change of the neck linker upon collision with the microtubule and before ATP binding. [DOI] [PubMed] [Google Scholar]
- 53.Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- 54.Yan Y, Sardana V, Xu B, Homnick C, Halczenko W, Buser CA, Schaber M, Hartman GD, Huber HE, Kuo LC. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J Mol Biol. 2004;335:547–554. doi: 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 55.Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 56.Klumpp LM, Mackey AT, Farrell CM, Rosenberg JM, Gilbert SP. A kinesin switch I arginine to lysine mutation rescues microtubule function. J Biol Chem. 2003;278:39059–39067. doi: 10.1074/jbc.M304250200. [DOI] [PMC free article] [PubMed] [Google Scholar]


