Abstract
Hand proteins are evolutionally conserved basic helix-loop-helix (bHLH) transcription factors implicated in development of neural crest-derived tissues, heart and limb. Hand1 is expressed in the distal (ventral) zone of the branchial arches, whereas the Hand2 expression domain extends ventrolaterally to occupy two-thirds of the mandibular arch. To circumvent the early embryonic lethality of Hand1 or Hand2-null embryos and to examine their roles in neural crest development, we generated mice with neural crest-specific deletion of Hand1 and various combinations of mutant alleles of Hand2. Ablation of Hand1 alone in neural crest cells did not affect embryonic development, however, further removing one Hand2 allele or deleting the ventrolateral branchial arch expression of Hand2 lead to a novel phenotype presumably due to impaired growth of the distal midline mesenchyme. Although we failed to detect changes in proliferation or apoptosis between the distal mandibular arch of wild-type and Hand1/Hand2 compound mutants at embryonic day (E)10.5, dysregulation of Pax9, Msx2 and Prx2 was observed in the distal mesenchyme at E12.5. In addition, the inter-dental mesenchyme and distal symphysis of Meckel’s cartilage became hypoplastic, resulting in the formation of a single fused lower incisor within the hypoplastic fused mandible. These findings demonstrate the importance of Hand transcription factors in the transcriptional circuitry of craniofacial and tooth development.
Introduction
Defects in migration, expansion, and differentiation of neural crest cells result in a variety of birth defects affecting craniofacial and cardiovascular structures. The neural crest is a transient and pluripotent population of cells that is formed in the dorsal lip of the neural tube as a result of inductive interactions between the neural plate and the surface ectoderm (Selleck MA et al., 1999). During craniofacial development, neural crest cells migrate ventrolaterally and populate the branchial arches, where they proliferate and form the frontonasal process and discrete swellings that demarcate each branchial arch. Lineage trace studies utilizing a Wnt1-Cre transgene and the ROSA26 conditional reporter (R26R) mouse line have confirmed in vivo that cranial neural crest cells eventually differentiate into bone, cartilage, teeth, cranial ganglia, and connective tissue of the face and neck (Chai et al., 2000; Chai and Maxson, 2006).
Early in embryonic development, the ectomesenchyme of the mandibular arch can be divided along a horizontal axis into two equal parts that have distinct developmental capabilities. The rostral (oral) ectomesenchyme participates in tooth formation via interactions with the overlying oral ectoderm, while the caudal (aboral) mandibular ectomesenchyme gives rise to Meckel’s cartilage (reviewed in Miletich and Sharpe, 2004). Meckel’s cartilage is unique among cartilages in that it exists provisionally prior to permanent mandibular bone formation in the first branchial arch (Sohal et al., 1999).
The mandible first appears as a mesenchymal condensation at E11.5 in mice. At around E13.5, bone begins to form by means of intramembranous ossification, which involves differentiation of mesenchymal precursors to active osteoblasts along the Meckel’s cartilage in a proximo-distal direction (Ramaesh and Bard, 2003). As intramembranous ossification proceeds, BrdU-positive proliferating cells diminish from the body of the mandible except at the proximal and distal symphysis of Meckel’s cartilage where proliferating cells contribute to the lengthening mandible (Ramaesh and Bard, 2003). By E16.5, the body of the mandibular bone is surrounded by a periosteum and the distal region of the mandible encloses the Meckel’s cartilage followed by gradual disintegration of Meckel’s cartilage (Ramaesh and Bard, 2003). In contrast to the posterior (proximal) end of Meckel’s cartilage, which undergoes endochondral ossification to form middle ear ossicles (Kronenberg, 2003), the anterior (distal) and intermediate portions of Meckel’s cartilage degenerate, undergo apoptosis, and are resorbed and replaced by bone (Ishizeki et al., 1999; Trichilis and Wroblewski, 1997). Recently, it was reported that the development and growth of incisors may contribute to osteoclast differentiation and activation, as well as initiation of resorption of Meckel’s cartilage (Sakakura et al., 2005).
Tooth development is regulated by inductive tissue interactions between the oral epithelium and the subjacent mesenchyme of the first branchial arch. Numerous signaling pathways have been implicated in each stage of tooth development (reviewed in Matalova et al., 2004). In particular, Fibroblast growth factor-8 (Fgf8), bone morphogenetic protein-4 (Bmp4), sonic hedgehog (Shh), and Wnt7b are epithelial-derived growth factors that induce an auto-regulatory positive feedback loop as well as mutual inhibitory signals to form incisor-molar boundaries within the epithelium (Hardcastle et al., 1998; Jeong et al., 2004; Sasaki et al., 2005; Stottmann et al., 2001). Tooth initiation becomes morphologically distinguishable between embryonic day (E) 11.5 and E12 when the oral ectoderm thickens in prospective tooth-forming regions of the mandibular and maxillary arches (reviewed in Tucker and Sharpe, 2004). This thickened ectoderm, known as the dental lamina, proliferates and starts to invaginate into the underlying neural crest-derived mesenchyme. Around E13.5 at the bud stage of tooth development, the mesenchyme proliferates and condenses around the developing epithelial bud. During this period, the mesenchyme induces the epithelial enamel knot, which becomes the signaling center of the developing tooth. By E14.5, the tooth bud takes the shape of a cap, and neural crest-derived ectomesenchymal cells are concentrated at the dental papilla. Cytodifferentiation of the tooth occurs after E16.5 at the bell stage, in which the ectoderm gives rise to the enamel- secreting ameloblasts, and the ectomesenchyme to dentine-secreting odontoblasts, pulp, and alveolar bone.
In the present work, we investigated the function of Hand genes in the development of mandibular arch derivatives. The class II bHLH proteins Hand1 (eHand, Thing1, and Hxt) and Hand2 (dHand, Thing2, and Hed) are expressed in partially overlapping domains in postmigratory neural crest cells in the branchial arches. Hand1 expression is confined to the ventral one-third of the mandibular arch and arch two, whereas Hand2 is expressed in the ventral two-thirds of the mandibular arch and arch two (Clouthier et al., 2000; Cserjesi et al., 1995; Srivastava et al., 1995; Srivastava et al., 1997). Hand1−/− embryos die by E8.5 due to yolk sac deficiency and Hand2−/− embryos die around E10.5 because of cardiac insufficiency (Firulli et al., 1998; Srivastava et al., 1997). Although Hand2−/− embryos show retarded branchial arch development and increased apoptosis, it is not clear whether the phenotype is a primary effect of the absence of Hand2 or secondary to heart abnormalities.
Despite early embryonic lethality of homozygous embryos, other genetically manipulated mice offer some insights into the role of Hand genes in the development of branchial arch-derived tissues. Mice lacking the endothelin-1-dependent Hand2 branchial arch enhancer die perinatally and exhibit a spectrum of craniofacial defects, including cleft palate, mandibular hypoplasia and cartilage malformations (Yanagisawa et al., 2003). Lineage analysis using Hand2-Cre:R26R embryos showed Hand2-progeny cells in both mandibular and molar mesenchyme at around E11.5, and later in dental papilla as well as in chondrogenic and osteogenic structures including Meckel’s cartilage (Ruest et al., 2003). In transgenic embryos expressing β-galactosidase under the same 7.4kb upstream region of the Hand2 gene, transgene expression was observed in the broad mesenchyme of the incisor region. It was also reported that treatments of tooth germ explants with Hand2 anti-sense oligodeoxynucleotide resulted in impaired differentiation of ameloblasts and odontoblasts, suggesting a potential role of Hand2 in early tooth formation (Abe et al., 2002).
To further explore roles of Hand genes during craniofacial development, we deleted the Hand1 gene in the branchial arch ectomesenchyme using Cre recombinase controlled by the neural crest cell-specific Wnt1 promoter. We also manipulated the expression domain and the level of both Hand genes in the branchial arches during embryogenesis by generating compound mutants carrying Hand1 conditional alleles and mutated Hand2 alleles. We found that development of the distal midline mesenchyme of the mandibular arch was sensitive to the total Hand gene dosage.
MATERIALS AND METHODS
Mouse strains
Hand1-null allele, referred to as Hand1lacZ or Hand1KO, and Hand1 conditional allele, referred to as Hand1loxP, have been described previously (Firulli et al., 1998; McFadden et al., 2005). Hand2-null allele and Hand2 mutant allele lacking the ventrolateral branchial arch enhancer, referred to as Hand2BA, have been described elsewhere (Srivastava et al., 1997; Yanagisawa et al., 2003). A transgenic mouse line expressing Cre recombinase under the neural crest specific promoter Wnt1 (Wnt1::Cre) has been previously characterized (Jiang et al., 2000). Animals were kept on a 12h/12h light/dark cycle under specific, pathogen-free conditions. Wild-type or heterozygous littermates were used as controls. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committees.
RT-PCR
Mandibular and second branchial arches were collected from wild-type (Hand1loxP/loxP; n=13), mutant (Wnt1::Cre; Hand1lacZ/loxP; n=9, Wnt1::Cre;Hand1loxP/loxP; n=8) and heterozygous (Hand1lacZ/loxP; n=14) embryos at E10.5 and pooled for RNA preparation. Total RNA was isolated using Trizol (Invitrogen) according to the manufacturer’s protocol. Randam hexamer-primed first-strand cDNAs were synthesized from 1 µg of total RNA using Superscript II (Invitrogen). Real-time PCR was performed using Assayson-Demand TaqMan primers and probes for mouse Hand1 and Hand2 (Applied Biosystems) on an ABI PRISM 7000 sequence detection system (Applied Biosystems). Expression levels were normalized to 18S RNA.
Cartilage and bone staining
Euthanized newborn mice were skinned, eviscerated and fixed in 95% ethanol and stained with Alizarin Red and Alcian Blue to examine bone and cartilage formation, respectively (Yanagisawa et al., 1998). Cartilaginous fetal skeletons (E15.5) were prepared and stained with Alcian Blue as previously described (Jegalian and De Robertis, 1992).
β-galactosidase staining
Embryos were harvested and rinsed with PBS and pre-fixed in PBS containing 2% paraformaldehyde and 0.25% glutaraldehyde for 1–3 hours on ice. Staining for β-gal activity was performed as previously described (McFadden et al., 2000).
Histology
P1 pups were euthanized and heads were harvested and fixed in 10% phosphate-buffered formalin at 4°C. Embryos were harvested and fixed overnight in 4% paraformaldehyde. Paraffin-embedded sections were prepared and stained with hematoxylin and eosin for general histology. β-galactosidase-stained sections were counter-stained with nuclear fast red. Von Kossa staining was used to detect mineralization. Heads from P1 pups were also embedded in LR White acrylic resin, and mineralized bone and tooth tissues were visualized in semi-thin, 1 µm-thick sections by von Kossa staining followed by counterstaining with toluidine blue.
Section in situ hybridization
E10.5 and E14.5 embryos were harvested and fixed in 4% paraformaldehyde overnight at 4°C. Riboprobes for Hand1 and Hand2 (McFadden et al., 2005), Fgf8 (Meyers and Martin, 1999), Shh (Echelard et al., 1993), BMP4 (Jones et al., 1991), Wnt7b (Parr et al., 1993), Collagen α1(II) (Ducy et al., 1997); Osteocalcin (Ducy and Karsenty, 1995); Indian hedgehog and Collagen α1(X) (Akiyama et al., 1999) and Collagen α1(I) (Aigner et al., 1995) were prepared with 35S-UTP (Amersham) using the Maxiscript In Vitro Transcription Kit (Ambion). In situ hybridization was performed as described (Shelton et al., 2000).
Whole mount in situ hybridization
Embryos were harvested at E12.5 and fixed in 4% paraformaldehyde overnight at 4°C. Whole mount in situ hybridization was performed using digoxigenin-labeled riboprobes for Hand1 and Hand2 (Clouthier et al., 2000), Shh (Echelard et al., 1993), Pax9 (Neubuser et al., 1995), Patched (Goodrich et al., 1996), Prx1/MHox (Cserjesi et al., 1992), Prx2/S8 (Opstelten et al., 1991), Msx1 and Msx2 (Thomas et al., 1998) as previously described (Riddle et al., 1993).
TUNEL and immunohistochemistry
Embryos were harvested at E10.5 and E14.5 then fixed overnight in 4% paraformaldehyde and embedded in paraffin. Free DNA ends were detected on transverse sections of embryos using the In situ cell death detection kit (Roche) according to the manufacturer’s protocol. Immunohistochemical detection of phosphorylated Histone H3 was performed as previously described (Shin et al., 2002).
Micro-computed tomography
Mandibular scans were performed on a standard desktop micro-computed tomography (micro-CT) instrument from Skyscan (Model 1072; AartsePaar, Belgium). This instrument has an 80-KeV sealed, air-cooled, microfocused X-ray source with a polychromatic beam derived from a tungsten target and with a spot size of less than 8 um. For these analyses, the X-ray source was operated at maximum power (80 KeV) and at 100 uA. Images were captured using a 12-bit, cooled CCD camera (1024 × 1024 pixels) coupled by a fiber optics taper to the scintillator. Prior to micro-CT analysis, mandibles with their dentition were chemically fixed overnight in 4% paraformaldehyde and 1% glutaraldehyde in 0.1M sodium cacodylate buffer, and then transferred to buffer alone. Mandibles with their dentition were wrapped in thin-film plastic wrap to prevent dessication, and scanned by micro-CT during rotation of the samples for approximately 30 minutes. Cross-sectional x-ray slices were reconstructed using the Skyscan tomography software based on triangular surface rendering to give a three-dimensional distribution of the calcified tissue and soft tissue.
Results
Deletion of Hand1 in neural crest cells
To delete Hand1 specifically in neural crest cells, we crossed Hand1loxP/loxP females to Wnt1::Cre; Hand1lacZ/+ heterozygous males. The Wnt1 promoter-driven Cre is expressed in pre-migratory neural crest cells and deletes efficiently in neural crest cells in vivo (Chai et al., 2000; Jiang et al., 2000). To confirm that the Hand1loxP allele was efficiently recombined in vivo, we analyzed Hand1 expression in E10.5 Wnt1::Cre; Hand1loxP/lacZ (hereafter referred to as Hand1NCKO/KO) embryos by in situ hybridization. In wild-type embryos, Hand1 was expressed in the distal (ventral) portion of the mandibular branchial arch (Fig. 1A, panel a, arrow) and Hand2 was expressed from the distal (ventral) to the ventrolateral portion of the mandibular branchial arch (Fig.1A, panel c). Consistent with a previous report, the Hand1 expression domain was completely included within the Hand2 domain (Clouthier et al., 2000). In contrast, Hand1 expression was absent in the mandibular arch of Hand1NCKO/KO embryos (Fig. 1A, panel b, arrow). The absence of Hand1 did not affect Hand2 expression (Fig. 1A, panel d). As expected, Hand1 expression was unaffected in the heart and limb of Hand1NCKO/KO mice (data not shown).
Figure 1. Neural crest-specific Cre-mediated recombination of Hand1.
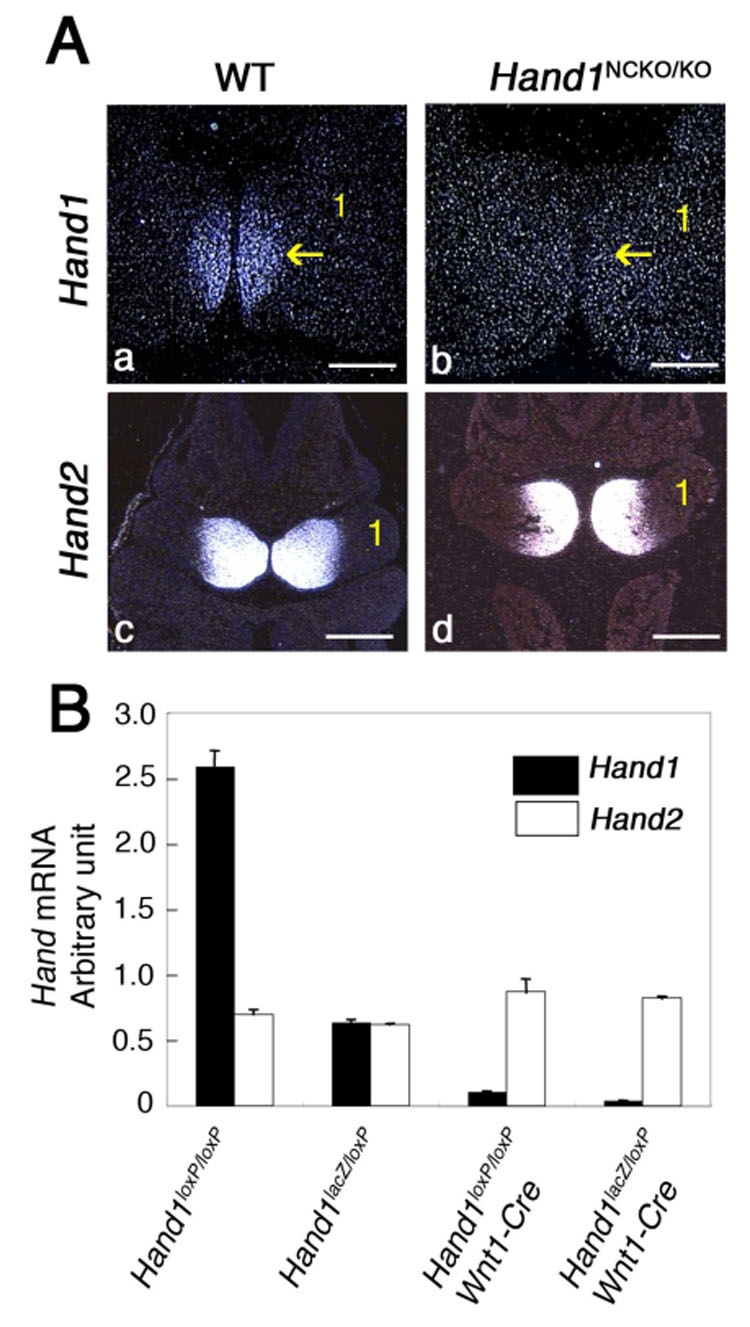
(A). Expression of Hand1 and Hand2 was examined by in situ hybridization of transverse sections of E10.5 wild-type (panels a, c) and Hand1NCKO/KO (panels b, d) embryos. Hand1 expression seen in the distal part of the mandibular arch (panel a, arrow) is absent in the mutant (panel b, arrow); Hand2 expression is unaffected (panel d). Bars indicate 200 µm (panels a, b) and 400 µm (panels c, d). 1; first mandibular arch.
(B). Detection of Hand1 and Hand2 by real-time RT-PCR using RNA extracted from mandibular and second branchial arches of Hand1loxP/loxP, Hand1lacZ/loxp and Hand1NCKO/KO (Hand1loxP/loxP; Wnt1::Cre and Hand1lacZ/loxP;Wnt1::Cre) embryos at E10.5. Values are normalized to 18S expression. Hand1 expression is almost undetectable in the mutants, whereas Hand2 expression is maintained in embryos from all genotypes. Bars are means ± SD.
To quantitate Hand1 expression in Hand1NCKO/KO neural crest cells, real time RT-PCR was performed using RNA extracted from mandibular and second branchial arches of E10.5 embryos. As Fig. 1B shows, Hand1 expression was almost undetectable in the Hand1NCKO/KO branchial arches (both in Wnt1::Cre; Hand1loxP/loxP and Wnt1::Cre; Hand1loxP/lacZ), while the Hand2 level was comparable among the genotypes, indicating that Hand2 was not upregulated to compensate for the loss of Hand1. These data indicate that Hand1 was efficiently deleted in the branchial arches of Hand1NCKO/KO embryos.
Next, we examined Hand1NCKO/KO pups at both postnatal day (P)1 and P28 for defects in craniofacial structures by gross morphology. Unexpectedly, Hand1NCKO/KO pups were anatomically normal and were fertile, and no embryonic lethality was observed (data not shown). These findings suggest that the overlapping expression of Hand2 in the distal region of the rostral mandibular arch can compensate for the loss of Hand1.
Manipulation of Hand genes in the branchial arches during development
To test the possibility that Hand2 compensates for functions of Hand1 during branchial arch development, we manipulated expression levels and domains of Hand2 during embryonic development. We generated Hand1NCKO/KO mice that were heterozygous for the Hand2-null allele (Hand2KO/+), or homozygous for a Hand2-mutant allele lacking the branchial arch enhancer (Hand2BA/BA), to reduce the Hand2 dosage in the branchial arches. Hand2BA/BA mice were previously shown to develop a hypoplastic mandible, deformed tympanic ring and cleft palate (Yanagisawa et al., 2003) attributable to the loss of Hand2 expression specifically in the ventrolateral domain of the mandibular arch. Therefore, Hand1NCKO/KO; Hand2BA/BA mice lack the ventrolateral expression of Hand2 without affecting the distal branchial arch expression. Hand1NCKO/KO; Hand2KO/+ mice were born at full term with expected Mendelian frequency, however, they died within 24 hours after birth because of failure to suckle. Gross examination revealed a hypoplastic jaw with normal proximo-distal patterning and cleft palate, which are similar to, but milder than those of Hand2BA/BA mice. Similarly, all Hand1NCKO/KO; Hand2BA/BA mice exhibited hypoplastic mandible and cleft palate and died within 24 hours of birth.
To further examine craniofacial structures in Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA mice, we performed bone and cartilage staining on P1 pups. In wild-type animals, bilateral palatine processes extended horizontally and fused to form the secondary palate (dashed line in Fig. 2A). In contrast, the palatine processes of Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA mice did not elevate to form the secondary palate (dashed line in Figs. 2B, D), as was observed in Hand2BA/BA mice (Fig. 2C). This causes the underlying presphenoid bone to be visible in the ventral view (arrows in Fig. 2B–D). The size of the mandible in Hand1NCKO/KO; Hand2KO/+ pups was smaller than that of wild-type (compare Fig. 2E and F), and Hand1NCKO/KO; Hand2BA/BA pups developed a much smaller, deformed mandible compared with the rest of the mutants (Figs. 2F, G and H). In both Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA mice, secondary structures derived from the most posterior segments of Meckel’s cartilages developed normally as demonstrated by a lack of noticeable malformations in corresponding middle ear ossicles (data not shown). These data clearly indicate that there is a dosage effect of Hand genes on the development of subsets of cranial neural crest derivatives (Table 1).
Figure 2. Craniofacial defects in P1 mice with compound mutations in Hand1 and Hand2.
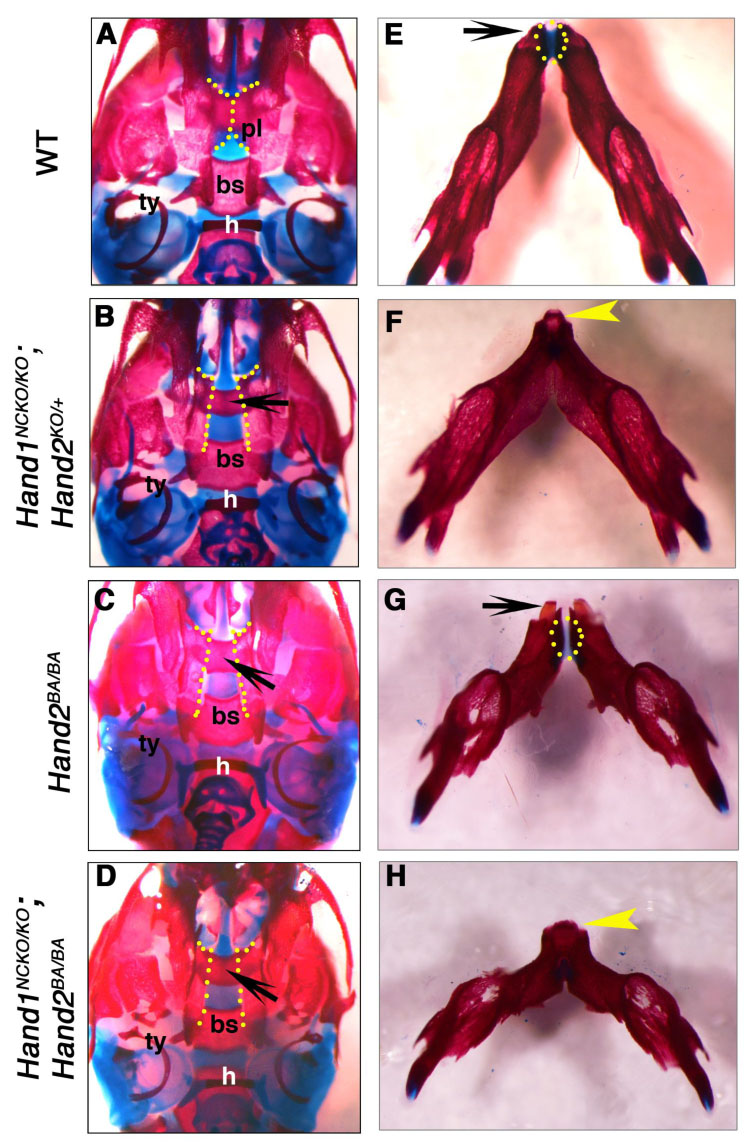
Bone and cartilage staining of wild-type (A, E), Hand1NCKO; Hand2KO/+ (B, F), Hand2BA/BA (C, G) and Hand1NCKO; Hand2BA/BA (D, H) mutant mice are shown. (A–D) Ventral views of the skull. Fusion of the bilateral palatine processes observed in the wild-type mouse (A, dashed line) is absent in the mutants (B-D, dashed line). Underlying presphenoid bone is visible in the mutants (arrows in B–D). (E–H) Ventral views of the mandible. The mandible of the mutants (F–G) is shorter and deformed. The angle between the left and right mandible is wider compared to the wild-type mouse (E). Note that the deformity of the mandible increases as total Hand1 and Hand2 gene dosage decreases. Wild-type (E) and Hand2BA/BA (G) mice show two incisors at the distal tip (arrow), whereas the mandible and lower incisors are fused in Hand1NCKO; Hand2KO/+ (F) and Hand1NCKO; Hand2BA/BA (H) mice (arrowhead). Note that the distal symphysis of the mandible is present in the wild-type and Hand2BA/BA mice (E, G, dotted area) but absent in the compound mutants (F, H).
Table 1.
Summary of mandible staining of Hand mutants at P1
| Genotype | N | Hypoplastic mandible | Fused mandible | Single lower incisor |
|---|---|---|---|---|
| Hand1KO/+ | 10 | 0 | 0 | 0 |
| Hand1NCKO/KO 1) | 14 | 0 | 0 | 0 |
| Hand1KO/+; Hand2KO/+ | 11 | 0 | 0 | 0 |
| Hand1NCKO; Hand2KO/+ | 8 | 8/8 (mild) | 8/8 | 8/8 |
| Hand1NCKO; Hand2BA/+ | 8 | 1/8 | 1/8 | 0/8 |
| Hand2BA/BA | 9 | 9/9 (severe2)) | 0/9 | 0/9 |
| Hand1NCKO; Hand2BA/BA | 8 | 8/8 (severe) | 7/8 | 5/8 |
This genotype includes Wnt1::Cre (+); Hand1loxP/lacZ and Wnt1::Cre (+); Hand1loxP/loxP
Severe mandibular defects include truncation and absence of the angular process.
Hand1 and midline defects
Careful examination of stained mandibular skeletal preparations revealed the distal symphysis of the mandible in wild type and Hand2BA/BA pups (Figs. 2E, G, dotted area). In contrast, Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA pups developed a fused mandible due to the absence of symphysial cartilage (Figs. 2F, H, arrowhead). In addition, a single incisor developed at the midline of the distal tip of the mandible where the two halves were fused. The incisor abnormality was detected with complete penetrance in Hand1NCKO/KO; Hand2KO/+ pups. In Hand1NCKO/KO; Hand2BA/BA pups, two lower incisors were occasionally found within the fused mandible (data not shown). Lower molars and upper teeth were normal in both mutants (data not shown). In addition to the incisor defect, we observed abnormal fusion of the hyoid and thyroid cartilage at the midline in Hand1NCKO/KO; Hand2KO/+ mutants, but not in Hand2BA/BA or Hand1NCKO/KO; Hand2BA/BA pups (data not shown).
To better understand the nature of the lower incisor defect, we performed micro-CT analysis of the whole head from wild-type and Hand1NCKO/KO; Hand2KO/+ mutant pups. In the wild-type pup, two well-developed and separated incisors were detected at the distal (incisal) tip within the mandible (Fig. 3A), whereas a single, somewhat rectangularly-shaped incisor was found within the fused distal tip of the mandible in the mutant (Fig. 3B). Von Kossa staining of P1 wild-type mandible showed two distinct incisors distal-lateral to Meckel’s cartilage surrounded by a thin layer of mineralized matrix and separated by well-developed cartilaginous symphysis (Fig. 3C, asterisk). Two separate incisors were also detected in the Hand2BA/BA pups, although the symphysis was smaller than that of wild-type (Fig. 3D, asterisk). In contrast, a single fused incisor with a common dental pulp was observed in both Hand1NCKO/KO; Hand2KO/+ (Fig. 3E) and Hand1NCKO/KO; Hand2BA/BA pups (Fig. 3F). A markedly reduced symphysis was observed near the base of the incisor, occasionally embedded within the single incisor (Fig. 3H, arrow). H&E staining of transverse sections of P1 embryos revealed that polarized odontoblasts lined the pulp, and dentin and the enamel organ was well formed in wild-type embryos (Fig. 3G, inset). In the mutant, polarized odontoblasts and ameloblasts were observed and a dentin layer was formed, however, the enamel layer was severely reduced or defective (Fig. 3H, inset). In addition, the extrinsic muscle of the tongue, genioglossus, and the geniohyoid muscle were also smaller, possessing fewer myofibers in the mutants than in the wild type mice (compare g and gh in Figs. 3I and J).
Figure 3. Abnormal fusion of the lower incisors.
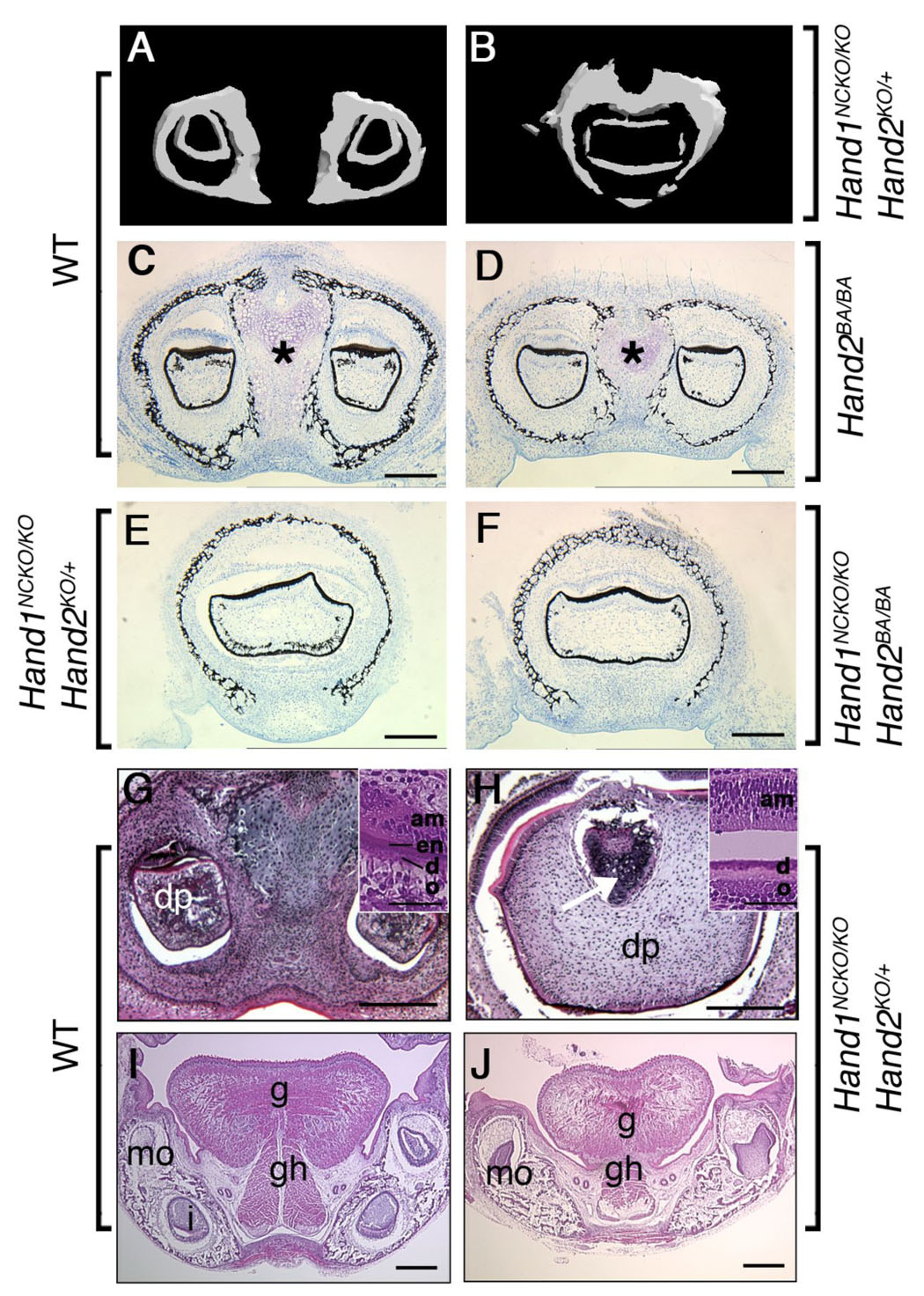
(A, B) Micro-CT analysis of the lower incisors in P1 embryos. In wild-type embryo (A), two lower incisors are located within the mandible. In contrast, a large deformed single incisor is observed within the fused mandible of the mutant (B). (C–F) Von Kossa staining of coronal sections of the mandible in plastic-embedded wild-type (C), Hand2BA/BA (D), Hand1NCKO/KO;Hand2KO/+ (E) and Hand1NCKO/KO;Hand2BA/BA (F) embryos. In wild-type embryos, a well-developed distal symphysis of Meckel’s cartilage is observed (asterisk in C). The symphysis is much smaller in the Hand2BA/BA mutant (asterisk in D) and becomes undetectable in Hand1NCKO/KO;Hand2KO/+ and Hand1NCKO/KO;Hand2BA/BA embryos (E, F). Fused incisors are seen in mutant embryos (E, F). (G–J) H&E staining of coronal paraffin sections of P1 wild-type (G, I) and Hand1NCKO/KO;Hand2KO/+ (H, J) embryos. (G, H) In wild-type embryos (G), two incisors with normal cytodifferentiation, including dental papilla (dp), odontoblasts (o), dentin (d), enamel (en) and ameloblasts (am) are observed (inset in G). In the mutant (H), a large fused incisor is observed, and the enamel layer is absent (inset in H). The distal symphysis is embedded in the dental papilla (H, arrow). (I, J) Well-developed extrinsic muscles (g; genioglossus, gh; geniohyoid) are observed in the tongue of wild-type embryos (I). In the mutant (J), muscle fibers are sparse and hypoplastic. Bars indicate 100 µm (C–J) and 40 µm (inset in G and H). mo; molar, i; incisor.
Changes in transcription factors implicated in tooth development in Hand1NCKO/KO; Hand2KO/+ embryos
To determine whether defects in the patterning of the odontogenic epithelium could account for the incisor fusion in Hand1NCKO/KO; Hand2KO/+ mandibles, we performed section in situ hybridization in E10.5 embryos and evaluated the expression of signaling molecules that induce the early patterning of the first branchial arch. The expression of Shh and Wnt7b, which are essential for specification of the oral epithelium, was unaffected in HAND1NCKO/KO; Hand2KO/+ embryos compared with wild-type embryos (Figs. 4A–D). Bmp4 defines the incisor domain in the oral epithelium and no alteration of the expression domain of Bmp4 was observed in HAND1NCKO/KO; Hand2KO/+ embryos (compare arrows in Figs. 4E, F). These observations suggest that compromized mesenchymal Hand expression does not affect specification of the odontogenic epithelium in mutant embryos.
Figure 4. Normal specification of the odontogenic epithelium in E10.5 mutant embryos.
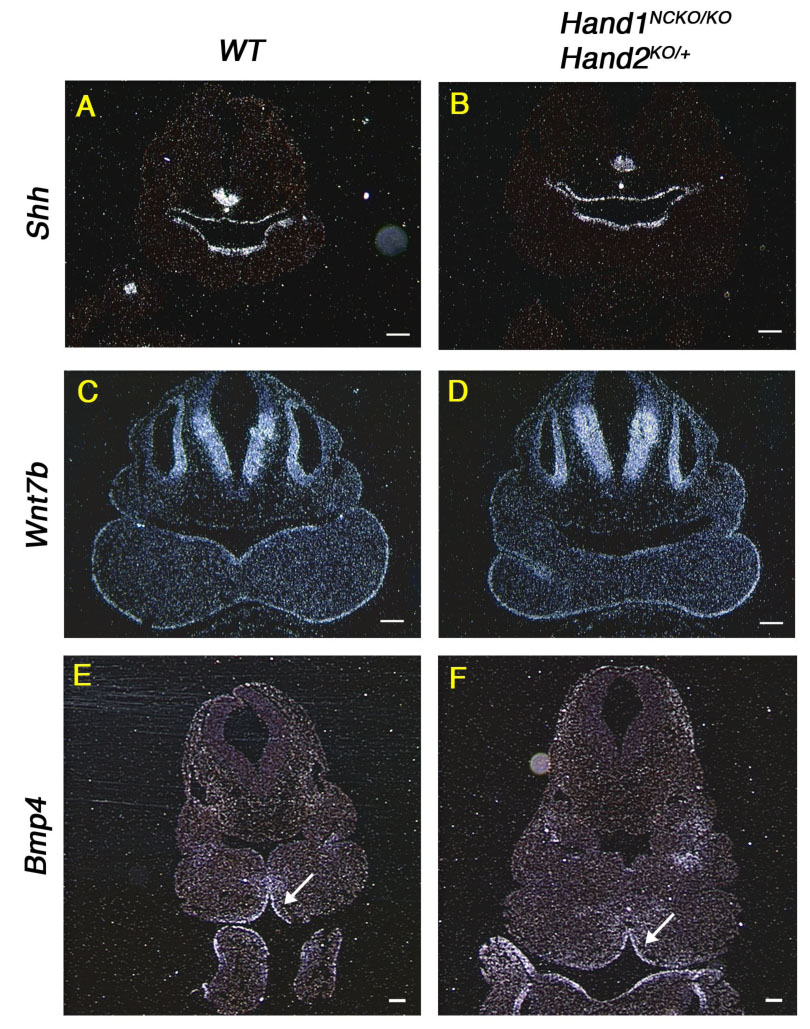
In situ hybridization of transverse sections from E10.5 wild-type (A, C, E) and Hand1NCKO/KO;Hand2KO/+ (B, D, F) embryos probed with Shh (A, B), Wnt7b (C, D) and Bmp4 (E, F). Arrow in E and F indicates that a Bmp4 domain is not altered in the mutant embryo compared with wild-type embryo. Bars indicate 100 µm.
Next, we examined by whole-mount in situ hybridization using E12.5 embryos whether transcription factors implicated in tooth organogenesis are altered or misexpressed in Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA mutants, thus causing abnormal lower incisor development. Since Hand1 and Hand2 are not expressed in the oral epithelium and the defect is restricted to lower incisors, we first focused on genes expressed in the mesenchyme of the distal mandibular arch. Pax9 marks prospective sites of odontogenesis prior to any morphological manifestation (Neubuser et al., 1995). Pax9−/− teeth proceed to the tooth bud stage but eventually arrest and result in agenesis (Peters et al., 1998). In wild-type embryos, Pax9 showed restricted expression in the distal mesenchyme of the mandibular arch and in the maxillary process (Figs. 5A, a). In Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA embryos, Pax9 expression was unaffected in the aboral mesenchyme, however, a small portion of the Pax9 expression domain between the lower incisor tooth buds was reduced (Figs. 5b, c, arrows).
Figure 5. Dysregulation of branchial arch genes in embryos with compound mutations in Hand1 and Hand2.
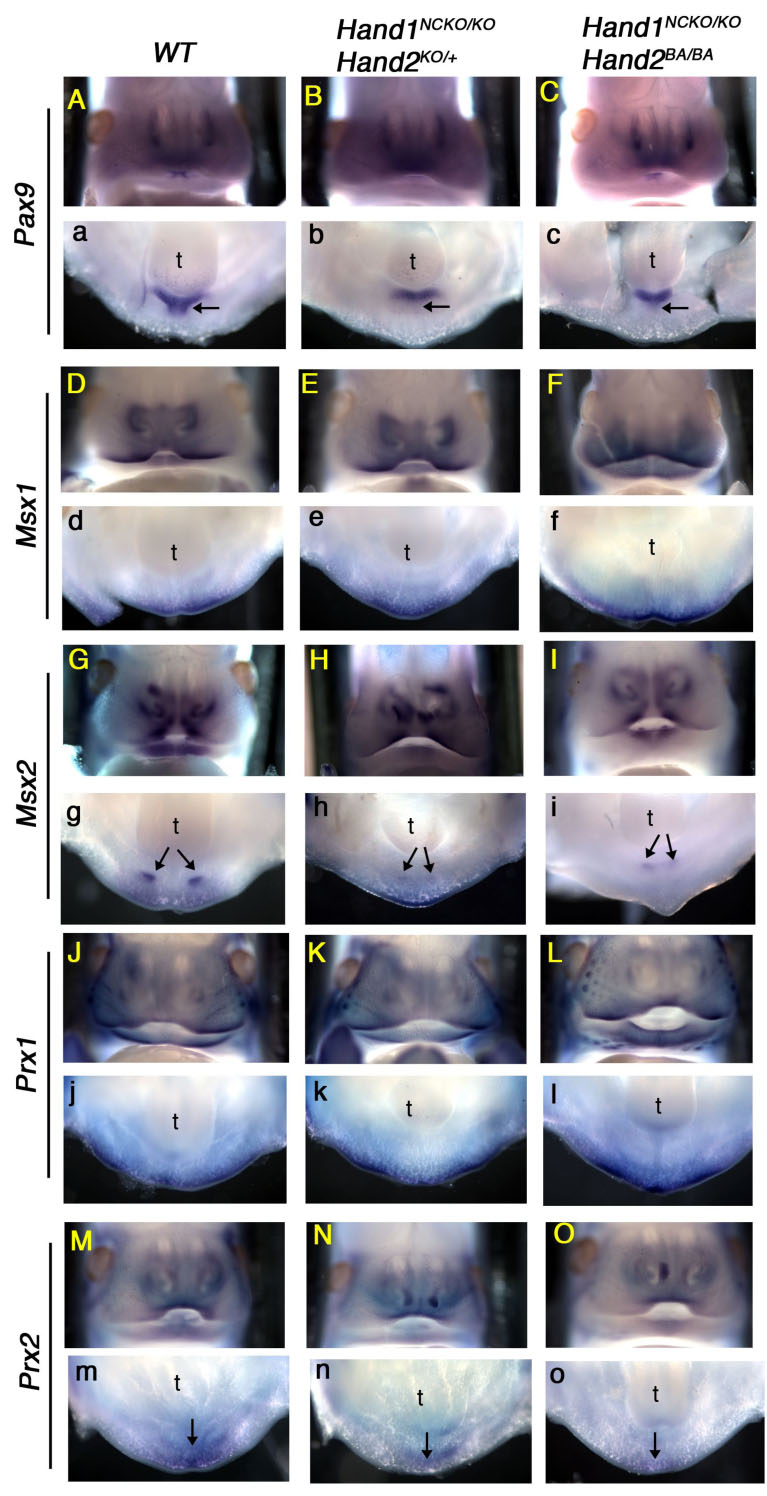
Whole-mount in situ hybridization was performed in E12.5 wild-type (A, a, D, d, G, g, J, j, M, m), Hand1NCKO/KO;Hand2KO/+ (B, b, E, e, H, h, K, k, N, n) and Hand1NCKO/KO;Hand2BA/BA (C, c, F, f, I, i, L, l, O, o) embryos. Frontal views (upper case) and transverse oral views (lower case) are shown. Pax9 is expressed in the dental and inter-dental (arrow in a) mesenchyme of the wild-type embryos, however, inter-dental expression is absent in both mutant embryos (arrows in b, c). Msx2 is expressed in the incisors (arrows in g) and Prx2 is expressed in the distal midline mesenchyme of wild-type embryos (arrow in m). In both mutants, Msx2 and Prx2 are down-regulated (arrows in h, i, arrow in n, o). Other candidate marker genes, Msx1 and Prx1, showed no remarkable changes.
Msx1 and Msx2 are well-known BMP target genes and Msx1 is downstream of Hand2 in the ectomesenchyme of the mandibular arch (Semba et al., 2000; Thomas et al., 1998). In wild-type embryos, Msx1 was expressed in the mesenchyme of the distal mandibular arch, maxillary arch and in the nasal process, and mutant embryos exhibited a similar pattern (Figs. 5D–F, d–f). Msx2 was restricted to the region where prospective incisors form in the mandibular arch in the wild-type embryo (Fig. 5g, arrow). In mutant embryos, Msx2 expression was significantly weaker, and the reduced distance between the two lower incisors was readily visible (Figs. 5h, I, arrows). These data suggest that Hand1 and Hand2 may be upstream of Msx2 in the mandibular arch mesenchyme (Figs. 5G–I).
The position- and gene dosage-dependent lower incisor and mandible phenotype in Hand mutant mice share a striking similarity with that of Prx1−/−; Prx2−/− mice (Lu et al., 1999; ten Berge et al., 1998; ten Berge et al., 2001). Prx1 and Prx2 belong to the paired-related homeobox gene family expressed in the craniofacial primordium, branchial arches and limb buds during embryogenesis (Cserjesi et al., 1992; Leussink et al., 1995). Prx1−/−; Prx2−/− double homozygous neonates exhibit some novel defects that are not observed in single knockout mice, including a hypoplastic mandible with absent or only one lower incisor. Therefore, to determine if Hand1 and Hand2 are upstream of Prx1 and Prx2, we examined expression of Prx genes in our mutant embryos. Prx1 was expressed in the maxillary arch, vibrissae and the distal mesenchyme of the mandibular arch, and no difference was detected between the wild-type and mutant embryos (Figs. 5J–L, j–l). Prx2 was expressed in more distal regions of the maxillary and mandibular arches compared to Prx1 in the wild-type embryos (Fig. 5m). While normal maxillary expression of Prx2 was maintained in mutant embryos (Figs. 5N, O), the mandibular expression was reduced (Figs. 5n, o).
These results suggest that initial tooth organogenesis occurred normally in the mutant embryos; however, a reduction in total Hand gene dosage may have caused down-regulation of Pax9, Msx2 and Prx2 genes and lead to the developmental defect of the midline mesenchyme and/or lower incisor development. The possibility still exists that a loss of appropriate cells between the lower incisors altered the expression domains of these genes within the mandibular arch.
Defect of the distal midline mesenchyme in Hand1NCKO/KO; Hand2KO/+ mutant mice
To determine the onset of the fusion defect of mandible and incisors with respect to distal midline mesenchyme development in Hand1NCKO/KO; Hand2KO/+ and Hand1NCKO/KO; Hand2BA/BA mutants, we focused on the phenotype of Hand1NCKO/KO; Hand2KO/+ embryos and performed whole-mount lacZ staining followed by serial sectioning. At E10.5, both Hand1KO/+ control and Hand1NCKO/KO; Hand2KO/+ embryos contained lacZ-positive cells in the distal part of the mandibular arch and no difference was observed between the genotypes (data not shown). Next, we harvested mutant embryos between E11.5 and E14.5, the critical period of dental lamina formation through cap stage tooth formation, and compared them with Hand1KO/+ control embryos. At E11.5, strong lacZ expression was observed in the midline mesenchyme extending from the mandibular component of the first branchial arch in the Hand1KO/+ embryos (Fig. 6A). A slight reduction of the lacZ-positive mesenchyme was observed in Hand1NCKO/KO; Hand2KO/+ mutant (Fig. 6B). At E12.5, the dental lamina had proliferated and invaginated into the underlying mesenchyme in Hand1KO/+ embryos (Fig. 6C, de). A lacZ-positive mesenchyme was well-developed and demarcated two prospective incisor tooth buds. In contrast, the invagination of the dental lamina was less distinct and lacZ-positive mesenchyme did not extend between the prospective incisor tooth buds in Hand1NCKO/KO; Hand2KO/+ embryos (Fig. 6D, arrow). At E13.5, a more localized thickening of the dental lamina and formation of the corresponding dental mesenchyme were observed in Hand1KO/+ embryos (Fig. 6E). Strong lacZ expression was observed in the mesenchyme between the tooth buds and between and above the genioglossus muscles in the tongue. The dental mesenchyme was negative for lacZ (Fig. 6E, arrow). In mutant embryos, the two incisor tooth buds were fused and the lacZ-positive mesenchyme was no longer evident except in a small area aboral to the tooth bud (Fig. 6F). The lacZ staining was also significantly reduced in the mutant tongue (data not shown). At E14.5, cap-shaped incisor tooth buds were visible in control embryos (Fig. 6I, dotted area). The lacZ expression was most intense at the symphysis (Fig. 6G, arrow) and persisted along the distal-proximal axis (Figs. 6G, I, K). The site of fusion of Meckel’s cartilage, as well as its surrounding perichondrium, was composed of both lacZ-positive and negative cells (Fig. 6I). The inter-dental mesenchyme, which extends toward the oral epithelium and separates the two incisors in control embryos was strongly positive for lacZ (Fig. 6I, arrow). Neural crest-derived dental mesenchyme where Hand2 is expressed (Abe et al., 2002) was negative for lacZ (Fig. 6I, double arrows). In mutant embryos, the symphysis of Meckel’s cartilage was markedly reduced in size and located aborally as indicated by the lacZ-positive area (Fig. 6H, arrow), and the two incisor primordia were fused near the oral base, giving rise to a fused incisor (Fig. 6J, I, dotted area). Abnormal blunting of the tip of Meckel’s cartilage was also observed in the mutant (Fig. 6L, arrow).
Figure 6. Development of Hand1-positive distal midline mesenchyme during embryogenesis.
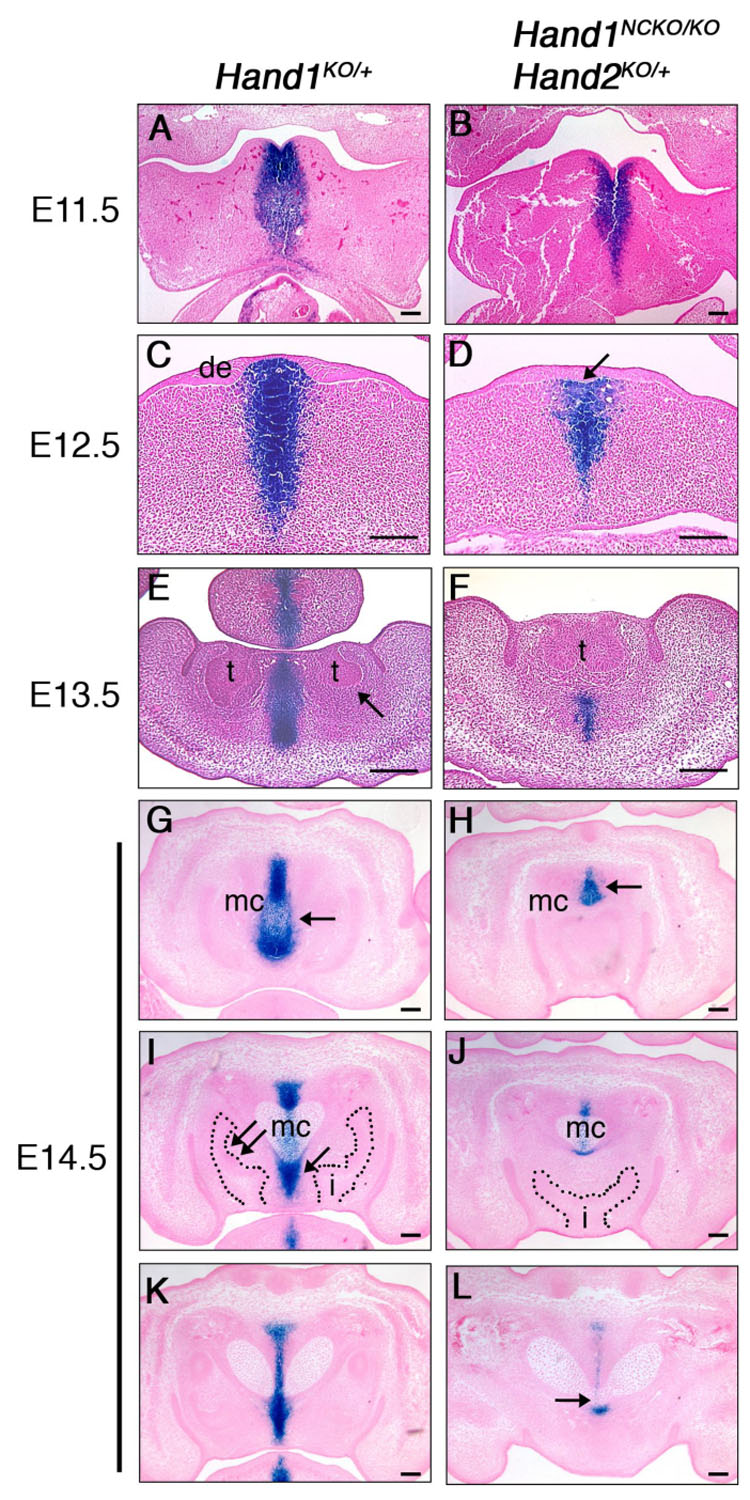
LacZ staining of Hand1KO/+ (A, C, E, G, I, K) and Hand1NCKO/KO;Hand2KO/+ (B, D, F, H, J, L) embryos. Transverse sections (A–F) and coronal sections (G–L) are shown. (A, B) LacZ staining is observed in the distal tip of the mandibular arch in both wild-type and mutant embryos at E11.5. (C, D) At E12.5, two distinct thickenings of the dental epithelium (de) corresponding to the future incisors are observed in wild-type embryo (C), whereas one linear thickening of the epithelium was observed in the mutant (D, arrow). (E, F) In wild-type embryos, two well-developed incisor buds are observed. Strong lacZ staining was observed in the distal midline mesenchyme between the tooth buds and in the center of the tongue in wild-type embryo (E). Dental mesenchyme is negative for lacZ (E, arrow). In the mutant, lacZ-positive mesenchyme is markedly reduced and tooth buds are fused (F). (G–L) Sections were taken in a distal (G, H) to proximal (K, L) direction in E14.5 embryos. In wild-type embryos, strong lacZ staining is detected in the distal symphysis of the Meckel’s cartilage (G, arrow) and the inter-dental mesenchyme (I, arrow). Two cap-stage incisors are observed (i in I). Dental mesenchyme is negative for lacZ (I, double arrow). In the mutant, the distal symphysis is significantly reduced in size (H, arrow), and a single incisor is observed (i in J). The distal end of Meckel’s cartilage is tapered and fused (arrow in L). t; tooth bud, i; incisor, mc; Meckel’s cartilage. Bars indicate 100 µm.
To investigate the cellular mechanism of retarded growth of the distal midline mesenchyme in mutant embryos, we performed proliferation and apoptosis assays on E10.5 and E14.5 sections using anti-phosphorylated histone H3 immunostaining and TUNEL assay, respectively. No difference was observed in proliferation or apoptosis of mesenchymal cells between wild-type and Hand1NCKO/KO; Hand2KO/+ embryos at E10.5 (Supplemental Fig. 1A, B). At E14.5, we found a trend for increased apoptosis in the distal incisor portion of the mutant mandible, however, it was not statistically significant (p=0.053, Supplemental Fig. 1B). No difference in proliferation was detected in the distal or proximal portion of the mandible at E14.5. These data indicate that Hand genes are not responsible for maintenance of cell numbers in the distal midline mesenchyme at early or late developmental stages, although the assessment of cell cycle length at E10.5 may reveal the underlying cause of growth defect of the midline mesenchyme.
Distal midline mesenchyme defect leads to premature ossification of the mandible in Hand1NCKO/KO;Hand2KO/+ mice
In the mandibular arch, the absence of ventrolateral Hand2 expression or the reduction of Hand2 in the entire Hand2 expression domain, in combination with the loss of Hand1, resulted in the distal mesenchyme defect and eventually led to fusion of the hemi-mandibles and lower incisors. To gain mechanistic insight into the relationship between the distal mesenchyme and the abnormal fusion event during late embryosgenesis, we compared the development of the mandible in mutant embryos and wild-type embryos. Bone and cartilage staining of wild-type E14.5 embryos showed a well-developed distal symphysis of Meckel’s cartilage (Fig. 7A, arrow), and mineralization of the membranous bone was observed along the body of the Meckel’s cartilage (Fig. 7A, double arrows). In contrast, mutant embryos showed a proportionally small and flat distal tip of the Meckel’s cartilage (Fig. 7B, arrow), and bone mineralization reached beyond the anterior part of the Meckel’s cartilage (Fig. 7B, double arrows). Von Kossa staining of E14.5 wild-type and Hand1NCKO/KO; Hand2KO/+ embryos were consistent with bone staining; while the distal portions of Meckel’s cartilage showed no signs of calcified matrix (Fig. 7C) and mineralization was evident only around the proximal portion of the Meckel’s cartilage in wild-type embryos (Fig. 7E, arrow), deposition of calcified matrix was already observed lateral to the Meckel’s cartilages throughout the proximal-distal axis in Hand1NCKO/KO; Hand2KO/+ embryos (Figs. 7D and F, arrows). Since Hand2BA/BA embryos, which showed more severe hypoplastic mandible (see Fig. 2G, Table 1), did not exhibit deposition of calcified matrix at the distal portion of the mandible (Supppledental Figure 2) and did not develop fused mandible, increased deposition of calcified matrix in Hand1NCKO/KO; Hand2KO/+ embryos was not simply a consequence of the shortening of Meckel’s cartilage. Rather, it was indicative of accelerated membranous ossification of the mandible.
Figure 7. Premature mineralization of the mandible in Hand1NCKO/KO; Hand2KO/+ embryos.
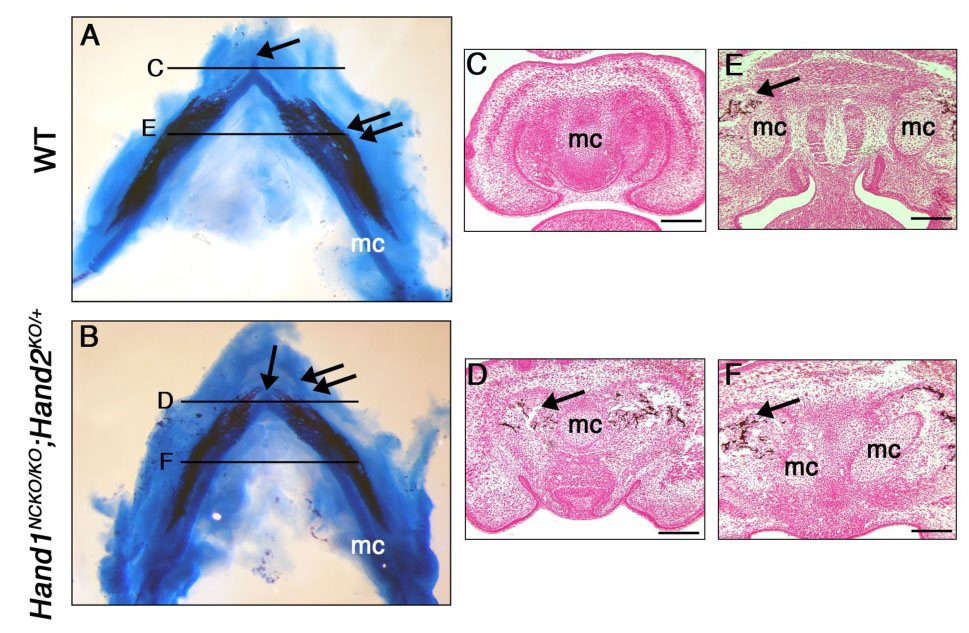
(A, B) Alcian blue and alizarin red staining of E14.5 wild-type (A) and Hand1NCKO/KO;Hand2KO/+ (B) embryos. In wild-type embryo (A), a well-developed distal symphysis of the Meckel’s cartilage (arrow) and mineralization of the mandible (double arrow) is observed along the body of Meckel’s cartilage. In the mutant (B), the distal symphysis is absent (arrow) and mineralization is accelerated at the distal portion of Meckel’s cartilage (double arrows). (C–F) Coronal sections of the mandible from E14.5 wild-type (C, E) and mutant (D, F) embryos stained with Von Kossa. Levels are indicated in A and B. Mineralization is observed around the proximal portion of Meckel’s cartilage in both wild-type and mutant embryos (arrow in E and F). In contrast, mineralization in the distal tip of the Meckel’s cartilage is only observed in the mutant (arrow in D, compare C and D). Bars indicate 100 µm. mc; Meckel’s cartilage.
During intramembranous bone development, proliferation of mesenchymal cells is followed by their commitment to become osteoprogenitor cells, which differentiate into pre-osteoblasts and then into bone matrix-forming mature osteoblasts. To determine if the premature deposition of bone matrix is attributable to defects in the differentiation of mutant osteoblasts, sections of E14.5 wild-type and Hand1NCKO/KO; Hand2KO/+ embryos were probed by radioactive in situ hybridization for markers of osteoblasts. In both wild-type and mutant mandibles, collagen α1(I) expression was detected surrounding the Meckel’s cartilage as well as in the adjacent incipient intramembranous bone (Figs. 8A, B, arrow). On the other hand, osteocalcin expression, a late marker of osteoblast differentiation (Ducy et al., 1997), was markedly reduced in the mandibles of Hand1NCKO/KO; Hand2KO/+ embryos (compare Fig. 8C and D). The osteoblasts in the mutant bone were not terminally differentiated, yet seemed to be inappropriately active leading to a premature deposition of the bone matrix.
Figure 8. Expression of bone and cartilage differentiation markers in E14.5 wild-type and Hand1NCKO/KO;Hand2KO/+ embryos.
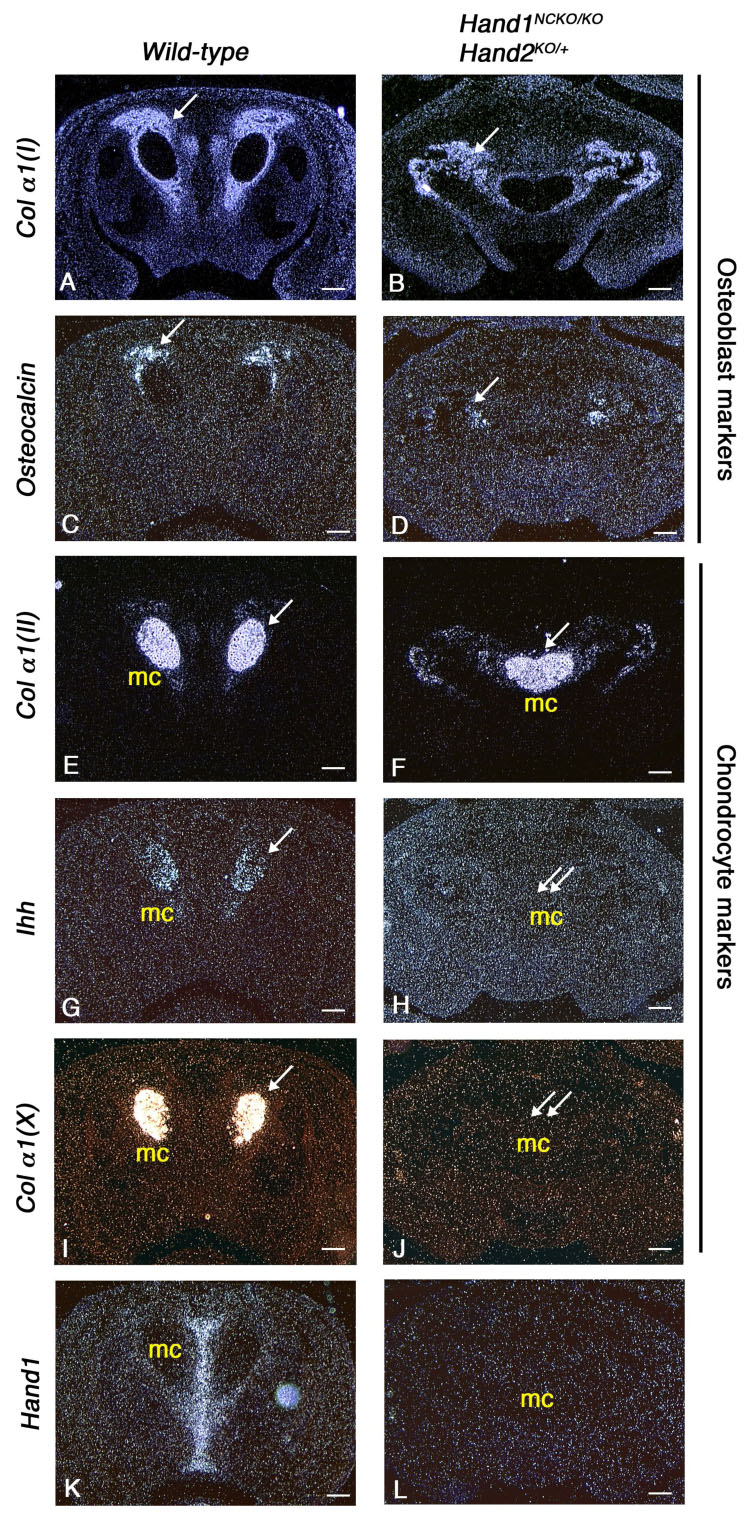
Coronal sections of the mandible from wild-type (A, C, E, G, I, K) and mutant embryos (B, D, F, H, J, L) are hybridized with osteoblast differentiation markers (A–D) or chondrocyte differentiation markers (E–J). Membranous bone ossification takes place around Meckel’s cartilage (arrows in A–D). While collagen αI (I) is expressed in both wild-type (A) and mutant (B) embryos, osteocalcin, a mature osteoblast marker, is down-regulated in the mutant (D). Note that Ihh and collagen α1 (X) expressed in the wild-type Meckel’s cartilage (arrows in G, I) are absent in the mutant embryos (double arrows in H, J). Hand1 expression performed as a control is absent in the mutant embryo (K, L). Bars indicate 100 µm.
It is known that at the distal symphysial region of the mandible, chondrocytes within Meckel’s cartilage undergo terminal differentiation and the cartilage is eventually replaced by bone tissues (Ishizeki et al., 1999). To determine if the hypoplasia of the distal symphysis of Meckel’s cartilage observed in Hand1NCKO/KO; Hand2KO/+ embryos lead to a delay in chondrocyte maturation, we examined the expression of chondrocyte maturation markers. Collagen α1(II), a marker of proliferating and prehypertrophic chondrocytes, was expressed in both wild-type and mutant embryos (Figs. 8E, F). However, Ihh (Indian hedgehog) and collagen α1(X), a specific marker of pre-hypertrophic chondrocytes and hypertrophic chondrocytes, respectively, was absent in mutant embryos (Figs 8G–J). Considering that Hand1 was mainly expressed in the midline between the Meckel’s cartilages in wild-type embryos (Fig. 8K), and both Hand1 and Hand2 were expressed only in subsets of cells in Meckel’s cartilage symphysis (Ruest et al., 2003), the defect may be secondary to a retarded growth of midline mesenchyme at an earlier developmental stage. However, a direct role of Hand genes in terminal differentiation of Meckel’s chondrocytes needs to be examined.
Discussion
In the present study, we performed tissue specific ablation in mice of the Hand1 gene in combination with various mutant alleles of Hand2. We show that growth of the distal midline mesenchyme of the first branchial arch, which develops into the interdental mesenchyme and distal symphysis of Meckel’s cartilage, is highly sensitive to dosage of the Hand genes. Secondary to the loss of distal midline mesenchyme, terminal differentiation of chondrocytes in Meckel’s cartilage symphysis is affected and membranous ossification of the mandible becomes dysregulated, which together impair the coordinated growth of lower incisors.
Redundant roles of Hand genes in development of the distal midline mesenchyme of the mandibular arch
In the branchial arches, the Hand1 domain is completely included in the Hand2 domain, and deletion of Hand1 in the first and second branchial arches in Hand1NCKO/KO mice was not sufficient to cause developmental defects. The overlapping expression domains of Hand1 and Hand2 and a highly conserved sequence in the HLH region of the two genes strongly suggest that Hand2 compensates for loss of Hand1. Indeed, removing one copy of Hand2 or deleting the ventrolateral branchial arch expression of Hand2 in conjunction with branchial arch deletion of Hand1 evoked a novel phenotype, perturbing the development of distal midline mesenchyme. The synergistic effect of Hand1 and Hand2 has been observed during cardiac development (McFadden et al., 2005). In cardiac-specific deletion mutants of Hand1, expansion of the left ventricle where Hand1 is predominantly expressed, is perturbed in a Hand2 gene dose-sensitive manner, causing a more severe phenotype than seen in single-knockout embryos. The requirement of Hand gene dosage for proper development of the distal craniofacial mesenchyme suggests that Hand1 and Hand2 redundantly regulate the same sets of target genes in the branchial arches. Knock-in studies in which the Hand2 locus is replaced with Hand1 and vise versa will be required to establish redundant roles of Hand in vivo.
Our findings shed light on the previously unrecognized role of Hand genes in the regulation of inter-dental mesenchyme development. In contrast to a previous report showing that increased apoptosis was responsible for hypoplastic branchial arch in Hand2-null embryos (Thomas et al., 1998), we failed to detect significant differences in proliferation or apoptosis between wild-type and Hand1/2 compound mutants at E10.5 and E14.5. However, this does not exclude the possibility that a slight decrease of proliferation and/or a gradual increase of apoptosis that was not detected during these time windows, may be responsible for compromised distal midline mesenchyme growth. It is recently reported that Hand1 regulates proliferation of cardiomyocytes by controlling cyclinD1 and cyclin dependent kinase 4 (Risebro et al., 2006). In the absence of Hand1, proliferation of cardiomyocytes in the distal outflow tract was decreased and myocytes undertook differentiation pathway. Therefore, it is plausible that Hand genes are involved in the initial proliferation of specific ectomesenchymal cell populations at the distal midline. Second possibility is that loss of Hand expression in the distal midline alters the boundaries of transcription factors expressed in adjacent domains of the branchial arch. For instance, Hand2 restricts dorsal expression of eng2 and dorsal-lateral expression of bapx1 (=Nkx3.2) to specify ventral branchial cartilages during zebrafish development (Miller et al., 2003). Therefore, the spatial relationship of HAND expression domain with that of other transcription factors may be critical for regulation of transcriptional circuitry during midline mesenchyme development.
In Hand compound mutants, Shh, Wnt7b, Fgf8, Patched and Msx1 were expressed normal levels and a Bmp4-positive incisor domain was not disrupted in E10.5 mutant embryos, suggesting that Hand1 and Hand2 are not involved in specification of the oral epithelial boundary between incisors and molars. Rather, it seems that lower incisors were fused due to hypoplastic inter-dental mesenchyme. It has been reported that several key molecules known to regulate development of the distal midline of the mandibular arch, including Hand2, Dlx3, Alx4, Pitx1 and BMP7, are markedly down-regulated in Dlx5−/−;Dlx6−/− embryos (Depew et al., 2002). In these mutants, Meckel’s cartilage and the mandible are absent and transformation of the mandible to maxilla is observed. Lower incisors do form, but, they are not associated with each other and occasionally accompanied by a remnant of the distal midline cartilaginous structure. In Hand mutant embryos, it seems that the growth of the distal mesenchyme affects normal positioning of the incisors by allowing a direct contact of two apposing dental buds. Finally, it is possible that the inter-dental mesenchyme produces inhibitory signals that prevent fusion of the incisor buds.
Loss of Hand genes affects terminal differentiation of the Meckel’s cartilage symphysis and ossification of the mandible
In Hand1NCKO/KO; Hand2KO/+ embryos, hypoplasia of the distal midline mesenchyme was evident as early as E11.5. Later at E14.5, chondrocytes within the distal symphysis of Meckel’s cartilage failed to express the pre-hypertrophic chondrocyte marker Ihh and the hypertrophic chondrocyte marker Collagen α1(X) (Kronenberg, 2003). Secretion of Ihh tightly controls chondrocyte hypertrophy and differentiation, and Ihh-null mice exhibit initial delay in the formation of hypertrophic chondrocytes (St-Jacques et al., 1999; Vortkamp et al., 1996). An overlap in the expression of Collagen α1 (II), Ihh and Collagen α1(X) in wild-type Meckel’s cartilage indicates that these chondrocytes are in a transition between the proliferating and hypertrophic stages. In contrast, lack of expression of Ihh and Collagen α1(X) in mutant chondrocytes suggests that the terminal differentiation of chondrocytes is affected.
We also found that osteoblasts surrounding Meckel’s cartilage symphysis were prematurely active in Hand mutants, indicated by accelerated deposition of calcified matrix, the presence of Collagen α1(I), an early marker for osteoblast differentiation, and the absence of Osteocalcin, a marker for mature osteoblasts. Production of type I collagen is one of the earliest events associated with osteoblast differentiation (Franceschi and Iyer, 1992). Osteocalcin, a marker of terminally differentiated osteoblasts (Ducy and Karsenty, 1995), inhibits the function of osteoblasts, as osteocalcin mutant mice exhibit an increase in bone formation (Ducy et al., 1996). Thus, in Hand mutant embryos, although prematurely active osteocytes did not undergo excessive proliferation nor exhibit resistance to apoptosis during the formation of the mandible, they contributed to premature deposition of bone matrix.
Taken together, these findings suggest the possibility that impaired chondrocyte differentiation within the Meckel’s symphysis induces a compensatory increase in intramembranous bone formation in the mandible at the distal tip, leading to the fused mandibles and a single incisor. Alternatively, Hands may regulate the activity of mandibular osteoblasts. It is also possible that secreted factor(s) from the symphysis may regulate the timing and the degree of intramembranous ossification of the mandible.
In Hand2-null mutant zebrafish, while dorsal cartilages were well-patterned, anterior ventral cartilages were almost absent, and upper jaw joints were connected by a cartilaginous bridge at the midline (Miller et al., 2003). Excess and ectopic expression of Msx genes (Msxe and Msxb), which depend on upstream ET-1 signaling, was thought to prevent ventral arch neural crest cell differentiation into chondrocytes (Miller et al., 2000; Miller et al., 2003). It is therefore interesting to consider whether the defect of chondrocyte differentiation reflects a direct role of Hand genes, or whether it is mediated through an inhibitory factors that are suppressed by Hand genes during chondrocyte differentiation.
Implications
Craniofacial tissue boundaries are established by specific expression of transcription factors and growth factors within the developing branchial arches. The present study defines a Hand-sensitive distal sub-domain within the mandibular arch demarcated by overlapping expression of Hand1 and Hand2. Our study also demonstrates that Hand genes are critical for the development of distal midline mesenchyme derivatives including the distal symphysis of Meckel’s cartilage and inter-dental mesenchyme. Identification of direct targets of Hand genes as well as enhancer(s) that drives Hand expression in the distal mesenchyme will facilitate dissection of the genetic network involved in craniofacial development.
Supplementary Material
Acknowledgments
We thank members of the Pathology Core Laboratory at the University of Texas Southwestern Medical Center for excellent histological preparations, Mei Xin for technical assistance and Drs. Eric Small and David Clouthier for a critical reading of the manuscript. We also thank Lydia Malynowsky and Jean-Sebastien Binette of McGill University for their excellent technical assistance with the histology and the micro-computed tomography, respectively. This work was supported by the NIH (E.N.O.), the Donald W. Raynolds Clinical Cardiovascular Research Center (E.N.O.), the March of Dimes Birth Defect Foundation (H.Y.), and the Canadian Institutes of Health Research (M.D.M.). Noriko Funato was supported by the JSPS Postdoctoral Fellowships for Research Abroad 2005.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe M, Tamamura Y, Yamagishi H, Maeda T, Kato J, Tabata MJ, Srivastava D, Wakisaka S, Kurisu K. Tooth-type specific expression of dHAND/Hand2: possible involvement in murine lower incisor morphogenesis. Cell Tissue Res. 2002;310:201–212. doi: 10.1007/s00441-002-0611-2. [DOI] [PubMed] [Google Scholar]
- Aigner T, Dietz U, Stoss H, von der Mark K. Differential expression of collagen types I, II, III, and X in human osteophytes. Lab Invest. 1995;73:236–243. [PubMed] [Google Scholar]
- Akiyama H, Shigeno C, Iyama K, Ito H, Hiraki Y, Konishi J, Nakamura T. Indian hedgehog in the late-phase differentiation in mouse chondrogenic EC cells, ATDC5: upregulation of type X collagen and osteoprotegerin ligand mRNAs. Biochem Biophys Res Commun. 1999;257:814–820. doi: 10.1006/bbrc.1999.0494. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Lyons GE, Olson EN. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115:1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Franceschi RT, Iyer BS. Relationship between collagen synthesis and expression of the osteoblast phenotype in MC3T3-E1 cells. J Bone Miner Res. 1992;7:235–246. doi: 10.1002/jbmr.5650070216. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui CC, Sharpe PT. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- Ishizeki K, Saito H, Shinagawa T, Fujiwara N, Nawa T. Histochemical and immunohistochemical analysis of the mechanism of calcification of Meckel's cartilage during mandible development in rodents. J Anat. 1999;194(Pt 2):265–277. doi: 10.1046/j.1469-7580.1999.19420265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegalian BG, De Robertis EM. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71:901–910. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Jones CM, Lyons KM, Hogan BL. Involvement of Bone Morphogenetic Protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development. 1991;111:531–542. doi: 10.1242/dev.111.2.531. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Leussink B, Brouwer A, el Khattabi M, Poelmann RE, Gittenberger-de Groot AC, Meijlink F. Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- Lu MF, Cheng HT, Lacy AR, Kern MJ, Argao EA, Potter SS, Olson EN, Martin JF. Paired-related homeobox genes cooperate in handplate and hindlimb zeugopod morphogenesis. Dev Biol. 1999;205:145–157. doi: 10.1006/dbio.1998.9116. [DOI] [PubMed] [Google Scholar]
- Matalova E, Tucker AS, Sharpe PT. Death in the life of a tooth. J Dent Res. 2004;83:11–16. doi: 10.1177/154405910408300103. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Miletich I, Sharpe PT. Neural crest contribution to mammalian tooth formation. Birth Defects Res C Embryo Today. 2004;72:200–212. doi: 10.1002/bdrc.20012. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Koseki H, Balling R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Opstelten DJ, Vogels R, Robert B, Kalkhoven E, Zwartkruis F, de Laaf L, Destree OH, Deschamps J, Lawson KA, Meijlink F. The mouse homeobox gene, S8, is expressed during embryogenesis predominantly in mesenchyme. Mech Dev. 1991;34:29–41. doi: 10.1016/0925-4773(91)90089-o. [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaesh T, Bard JB. The growth and morphogenesis of the early mouse mandible: a quantitative analysis. J Anat. 2003;203:213–222. doi: 10.1046/j.1469-7580.2003.00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Risebro CA, Smart N, Dupays L, Breckenridge R, Mohun TJ, Riley PR. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development. 2006;133:4595–4606. doi: 10.1242/dev.02625. [DOI] [PubMed] [Google Scholar]
- Ruest LB, Dager M, Yanagisawa H, Charite J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakura Y, Tsuruga E, Irie K, Hosokawa Y, Nakamura H, Yajima T. Immunolocalization of receptor activator of nuclear factor-kappaB ligand (RANKL) and osteoprotegerin (OPG) in Meckel's cartilage compared with developing endochondral bones in mice. J Anat. 2005;207:325–337. doi: 10.1111/j.1469-7580.2005.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Ito Y, Xu X, Han J, Bringas P, Jr, Maeda T, Slavkin HC, Grosschedl R, Chai Y. LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev Biol. 2005;278:130–143. doi: 10.1016/j.ydbio.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Selleck MA, García-Castro MI, Artinger KB, Bronner-Fraser M. Effects of Shh and Noggin on neural crest formation demonstrate that BMP is required in the neural tube but not ectoderm. Development. 1999;125:4919–4930. doi: 10.1242/dev.125.24.4919. [DOI] [PubMed] [Google Scholar]
- Semba I, Nonaka K, Takahashi I, Takahashi K, Dashner R, Shum L, Nuckolls GH, Slavkin HC. Positionally-dependent chondrogenesis induced by BMP4 is co-regulated by Sox9 and Msx2. Dev Dyn. 2000;217:401–414. doi: 10.1002/(SICI)1097-0177(200004)217:4<401::AID-DVDY7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- Sohal GS, Ali MM, Ali AA, Dai D. Ventrally emigrating neural tube cells contribute to the formation of Meckel's and quadrate cartilage. Dev Dyn. 1999;216:37–44. doi: 10.1002/(SICI)1097-0177(199909)216:1<37::AID-DVDY6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Anderson RM, Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Reijnen MJ, van Raaij EJ, Verbeek F, Gaffield W, Meijlink F. Prx1 and Prx2 are upstream regulators of sonic hedgehog and control cell proliferation during mandibular arch morphogenesis. Development. 2001;128:2929–2938. doi: 10.1242/dev.128.15.2929. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Trichilis A, Wroblewski J. Expression of p53 and hsp70 in relation to apoptosis during Meckel's cartilage development in the mouse. Anat Embryol (Berl) 1997;196:107–113. doi: 10.1007/s004290050083. [DOI] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charite J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


