Abstract
Several angiogenic factors and extracellular matrix-degrading enzymes that promote invasion and metastasis of cancer are produced by stromal fibroblasts that surround cancer cells. The expression of genes that code for some of these proteins is regulated by the transcription factor NF-κB. In this report, we demonstrate that conditioned medium (CM) from estrogen receptor (ER)-negative but not ER-positive breast cancer cells induces NF-κB in fibroblasts. In contrast, CM from both ER-positive and ER-negative breast cancer cells induces NF-κB in macrophages and endothelial cells. NF-κB activation in fibroblasts was accompanied by induction of interleukin 6 (IL-6) and urokinase plasminogen activator (uPA), both of which promote angiogenesis and metastasis. A survey of cytokines known for their ability to induce NF-κB identified IL-1α as the factor responsible for NF-κB activation in fibroblasts. Analysis of primary breast carcinomas revealed the presence of IL-1α transcripts in majority of lymph node-positive breast cancers. These results along with the known role of IL-1α and IL-6 in osteoclast formation provide insight into the mechanism of metastasis and hypercalcemia in advanced breast cancers.
Breast cancers often progress from a benign, noninvasive, estrogen-dependent growth phenotype to a malignant, estrogen-independent, invasive, and metastatic growth phenotype (1, 2). This progression is usually accompanied by loss or altered function of estrogen receptor (ER) and resistance to antiestrogen therapy (3, 4). In addition, expression of genes that promote invasion and metastasis also is increased. For example, sera from patients with aggressive breast cancers often contain increased level of urokinase plasminogen activator (uPA), a potentiator of metastasis (5). Elevated expression of matrix metalloproteinases 2 and 9 (MMP2 and MMP9) and the angiogenic factor interleukin 6 (IL-6) also has been observed in invasive breast cancers (6–8). Although the majority of these proteins are derived from cancer cells, some of these proteins are made in fibroblasts that surround cancer cells. For example, the matrix metalloproteinase gene stromelysin III is expressed predominantly in fibroblasts that surround breast cancer (9). A cancer cell-derived factor is likely to be the key molecule that regulates stromelysin III expression. Consistent with this possibility, paracrine induction of insulin-like growth factors, gelatinase A, and hepatocyte growth factor (HGF) in fibroblasts by conditioned medium (CM) of breast cancer cells has been reported (10–12).
Breast cancer cells secrete a number of cytokines and growth factors that include leukemia inhibitory factor, pleiotrophin (PTN), basic fibroblast growth factor members (bFGF), vascular endothelial growth factor (VEGF), and epidermal growth factor family members (13–15). Although the targets of these cytokines and growth factors in fibroblasts are not well studied, extracellular signal-activated transcription factors such as activator protein 1 (AP-1), NF-κB, STAT family of transcription factors, and serum response factors are the likely central targets (16). Interestingly, some of these transcription factors, particularly AP-1 and NF-κB, regulate the expression of uPA, MMP9, HGF, and IL-6 (17–20).
In this study, we have focused our attention on the activity of NF-κB in fibroblasts. NF-κB is a heterodimeric transcription factor composed of various combinations of members of the Rel/NF-κB family of polypeptides (20–22). In resting cells, NF-κB usually resides in the cytoplasm because of its association with inhibitory proteins IκBs (20–22). Several IκBs, including IκBα, IκBβ, p105/IκBγ, p100/IκBδ , IκBɛ, cactus, and relish have been identified (20–23). Upon stimulation of cells by inducers such as tumor necrosis factor α (TNFα) and IL-1, IκBs are degraded and NF-κB is translocated to the nucleus where it modulates the expression of genes involved in inflammation, extracellular matrix degradation, cell adhesion, angiogenesis, and prevention of cell death (20–27).
Recently we have demonstrated that the activity of NF-κB is constitutive in ER-negative but not ER-positive breast cancer cells (28). From these results, we hypothesized that constitutive activity of NF-κB is responsible for invasive growth phenotype of ER-negative breast cancer cells (29). To explore the possibility that NF-κB is constitutively active not only in ER-negative breast cancer cells but also in fibroblasts that surround them (because of host–tumor cell interaction), we have compared NF-κB DNA-binding activity in fibroblasts treated with CM from a panel of ER-positive and ER-negative breast cancer cell lines. We show that CM from a subset of ER-negative breast cancer cells induces NF-κB DNA-binding activity and this NF-κB activation is accompanied by induction of IL-6 and uPA in fibroblasts. Furthermore, we demonstrate that IL-1α is the factor responsible for this induction of NF-κB. Finally, an analysis of IL-1α expression in primary breast cancers is presented.
MATERIALS AND METHODS
Breast Cancer Cell Lines and Collection of CM.
All human breast cancer cell lines were purchased from American Type Culture Collection. HLF-1, MCF-7, MDA-MB-231, MDA-MB-468, SK-BR-3, and ZR-75–1 were grown in MEM + 10% FCS. HBL-100 was grown in MEM + 10% FCS + 10−9 M insulin. MDA-MB-435 and MDA-MB-436 were grown in MEM containing 10% FCS, 1 mM sodium pyruvate, 1 mM MEM nonessential amino acids and vitamins. T47-D and THP-1 cells were grown in RPMI + 10% FCS. Culturing and conversion of RM22 cells from E- to F-phenotype is as described previously (30). Fibroblasts isolated from a skin biopsy of a normal individual were grown in DMEM + 10% FCS as described previously (31). Confluent cultures (≈107 cells) were washed in PBS and incubated with 10 ml serum-free MEM for 24 hr. The medium was collected after 24 hr and used for treating HLF-1 grown in 60-mm plates (≈80% confluent).
Electrophoretic Mobility-Shift Assays (EMSA).
Whole-cell extracts were prepared and subjected to EMSA as described previously (32). Oligonucleotide probes containing consensus sequences for NF-κB, AP-1, and CTF/NF-1 were purchased from Promega. In antibody neutralization experiments, 10 ml CM was incubated with neutralizing antibodies for 1 hr at room temperature. HLF-1 was incubated with antibody-treated CM for 3 hr. Antibodies for this purpose were purchased from R & D Systems.
RNA Preparation and Northern Blotting.
Total RNA from HLF-1 was prepared by using guanidinium isothiocyanate/cesium chloride/ultracentrifugation method (33). RNA was electrophoresed on an agarose-formaldehyde gel and subjected to Northern blot analysis as described previously (34).
Analysis of Primary Breast Cancer for IL-1α Expression.
Tumors were collected immediately after surgery, washed extensively in PBS, and minced into small pieces. Total RNA was prepared by guanidinium isothiocyanate/cesium chloride/ultracentrifugation method (33). Five micrograms of RNA was reverse-transcribed by using oligo(dT) primer and reverse transcription–PCR (RT-PCR) kit (Stratagene). Reverse-transcribed RNA (1/10th volume) was subjected to 20 cycles of PCR by using IL-1α gene-specific primers (5′-ACCAACCAGTGCTGCTGAAGGA-3′, nucleotides 656–677 and 5′-AGACTCCAGACCTACGCC-3′, nucleotides 870–887, GenBank accession no. E04022). Keratin 19 mRNA was amplified similarly by using primers 5′-AGGTGGATTCCGCTCCGGGCA-3′ and 5′-ATCTTCCTGTCCCTCGAGCA-3′ (35). As a control, mRNA corresponding to ribosomal protein gene 36B4 was also amplified by PCR (34). Amplification of 36B4 was linear under this PCR condition. PCR products were visualized by Southern blotting.
RESULTS
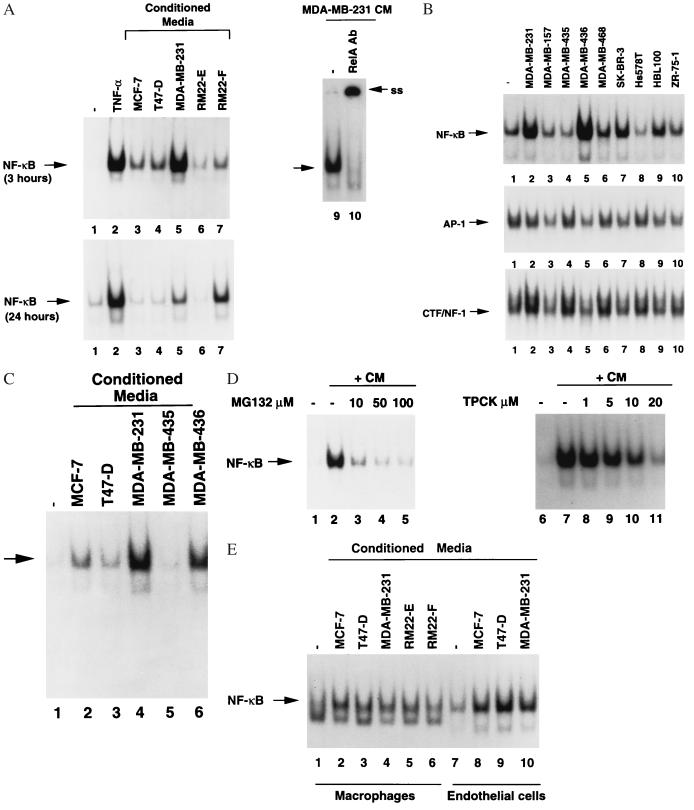
We first examined the effect of CM from ER-negative and ER-positive breast cancer cells on the DNA-binding activity of NF-κB in fibroblasts. Toward this end, human lung fibroblast cell line HLF-1 was treated with CM from four ER-positive (MCF-7, T47-D, ZR-75–1, and RM22-E) and nine ER-negative breast cancer cell lines (MDA-MB-157. MDA-MB-231, MDA-MB-435, MDA-MB-436, MDA-MB-468, SK-BR-3, Hs578T, HBL-100, and RM22-F), and DNA-binding activity of NF-κB was determined by EMSA. The CM from four ER-negative cell lines (MDA-MB-231, MDA-MB-436, HBL-100, and RM22-F) strongly induced NF-κB DNA-binding activity in HLF-1 (Fig. 1 A and B). Induction of NF-κB was evident even after incubation of cells with CM for 24 hr. Interestingly, MDA-MB-231, MDA-MB-436, and RM22-F cells invade basement membrane in in vitro invasion assay and/or form metastatic tumors in nude mice (30, 36, 37). Furthermore, progression of RM22 cell line from ER-positive, epithelial-like phenotype (RM22-E phenotype) to ER-negative, fibrosarcoma phenotype (RM22-F phenotype) (30) is accompanied by secretion of the factor(s) that induce NF-κB (Fig. 1A, compare lane 6 with lane 7). The induction of NF-κB by CM from two ER-positive cell lines (MCF-7 and T47-D) was weak and transient because no NF-κB DNA-binding activity was observed after 24 hr of CM addition (Fig. 1A). The CM from breast cancer cells induce only NF-κB DNA-binding because DNA binding of transcription factors AP-1 and CTF/NF-1 was not affected by any CM (Fig. 1B). The NF-κB:DNA complex contains the NF-κB subunit RelA as RelA-specific antibody supershifted DNA–protein complex (Fig. 1A, compare lane 9 with lane 10). To determine whether NF-κB in normal fibroblasts is induced by CM from breast cancer cells, fibroblasts isolated from a skin biopsy of a normal individual was incubated with CM from MCF-7, T47-D, MDA-MB-231, MDA-MB-435, and MDA-MB-436 cells (Fig. 1C). Strong induction of NF-κB was observed in fibroblasts incubated with CM from MDA-MB-231 and MDA-MB-436. NF-κB activation is accompanied by degradation of both IκBα and IκBβ (data not shown). The induction of NF-κB was inhibited by two known inhibitors of NF-κB activation, tosylphenylalanyl chloromethyl ketone (TPCK) and MG132 (Fig. 1D) (38). These results suggest that NF-κB activation by CM follows a classical pathway involving signal-induced degradation of IκBs by proteasomes (20–22).
Figure 1.
Induction of NF-κB in fibroblasts by hormone-independent breast cancer cell-derived CM. (A) NF-κB activation in fibroblasts. Six micrograms of whole-cell extract from HLF-1 treated with CM from indicated cell lines for 3 hr (Upper) or 24 hr (Lower) was subjected to EMSA by using NF-κB probe. NF-κB activation by TNFα (10 ng/ml) is also shown as a control. Presence of the NF-κB subunit RelA in DNA–protein complex was verified by antibody supershift assay. In this assay, whole-cell extracts from HLF-1 treated with MDA-MB-231 CM was incubated with either NF-κB probe alone (lane 9) or probe with an antibody against RelA (lane 10). The DNA–protein complex supershifted by the antibody is indicated (ss). (B) CM from breast cancer cells do not effect DNA binding of transcription factors AP-1 and CTF/NF-1. HLF-1 was treated with CM from indicated cell lines for 3 hr, and 6 μg of whole-cell extract was subjected to EMSA by using NF-κB probe (Top), AP-1 probe (Middle), and CTF/NF-1 probe (Bottom). (C) Induction of NF-κB in normal fibroblasts. Fibroblasts isolated from a skin biopsy were treated with CM from indicated cell lines for 3 hr and analyzed for NF-κB DNA-binding activity. (D) Protease/proteosome inhibitors prevent NF-κB activation by MDA-MB-231 CM. HLF-1 was incubated with indicated concentrations of TPCK or MG132 for 1 hr before the addition of MDA-MB-231 CM. Whole-cell extract was prepared 3 hr after the addition of CM and subjected to EMSA. (E) Induction of NF-κB in macrophages and endothelial cells by breast cancer cell CM. THP-1 cells (macrophages, lanes 1–6) and bovine heart endothelial cells (lanes 7–10) were incubated with CM from indicated cell lines for 24 hr. Whole-cell extracts were analyzed for NF-κB as above.
A tumor microenvironment contains infiltrating macrophages and endothelial cells. To examine whether CM from breast cancer cells contain factor(s) that induce NF-κB in macrophages and endothelial cells, human macrophage cell line THP-1 and bovine heart endothelial cells were incubated with CM for 24 hr and tested for NF-κB DNA-binding activity. In contrast to our findings in fibroblasts, CM from both ER-positive (MCF-7 and T47-D) and ER-negative (MDA-MB-231) cells induced NF-κB activity in THP-1 and endothelial cells (Fig. 1E). CM from both MCF-7 and MDA-MB-231 cells induced NF-κB in murine macrophage cell line RAW-264.7 (data not shown). These results indicate that although all breast cancer cells secrete factor(s) that induce NF-κB in macrophages and endothelial cells, only a subset of hormone-independent breast cancer cells secrete factor(s) that induce NF-κB in fibroblasts.
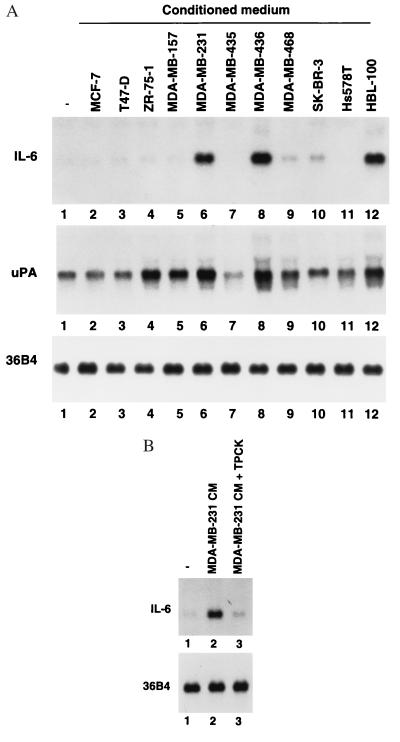
To determine the consequences of NF-κB activation, the expression of two NF-κB-regulated genes, IL-6 and uPA, were examined in CM-treated HLF-1. Interestingly, CM from the cell lines that activated NF-κB (MDA-MB-231, MDA-MB-436, and HBL-100) also induced IL-6 mRNA (Fig. 2A, lanes 6, 8, and 12). IL-6 induction by MDA-MB-231 CM was inhibited by TPCK, which suggests the requirement of NF-κB activation in IL-6 induction (Fig. 2B). Induction of uPA also was observed with the CM from MDA-MB-231, MDA-MB-436, and HBL-100 cells (Fig. 2A). Unlike in the case of IL-6, CM from ZR-75–1 also induced uPA. Because uPA gene expression depends on a number of regulatory elements including NF-κB, AP1, and PEA3 (17, 39), specific cytokine/growth factor secreted by ZR-75–1 may induce uPA independent of NF-κB.
Figure 2.
CMs that activate NF-κB induce IL-6 and uPA expression in HLF-1. (A) Ten micrograms of total RNA from HLF-1 treated with CM from indicated cell lines for 4 hr was subjected to Northern blotting and probed with IL-6 (Upper) and uPA (Lower) cDNA. Integrity of the RNA was examined by reprobing the blot with the ribosomal protein gene 36B4 (34). (B) TPCK inhibits IL-6 induction by CM. HLF-1 was incubated with 50 μM TPCK for 1 hr before the addition of CM from MDA-MB-231 cells. RNA was prepared 4 hr after CM addition and analyzed as above.
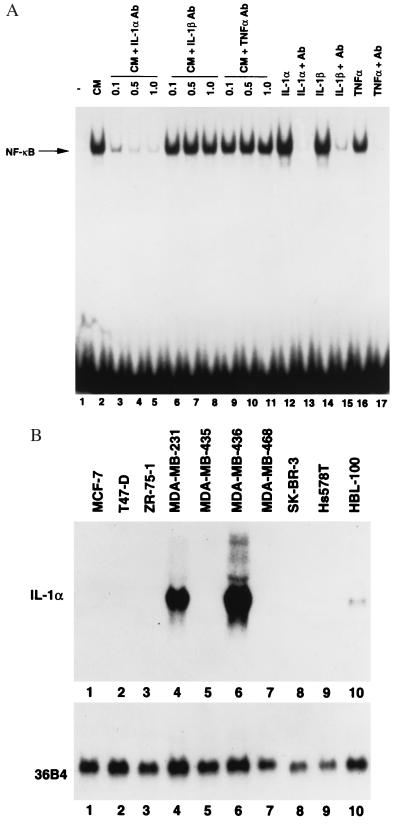
Cytokines/growth factors that can induce NF-κB include TNFα, IL-1α, IL-1β, and leukemia inhibitory factor (20–22, 40). To determine whether any of these factors present in CM is responsible for NF-κB activation in fibroblasts, HLF-1 cells were exposed to MDA-MB-231 CM that had been pretreated with neutralizing antibodies against TNFα, IL-1α, IL-1β (Fig. 3A), and leukemia inhibitory factor (data not shown). Neutralizing antibodies against IL-1β, TNFα, and leukemia inhibitory factor did not effect NF-κB activation (Fig. 3A, compare lane 2 with lanes 6–11, data not shown). In contrast, CM pretreated with antibody against IL-1α failed to activate NF-κB, suggesting that IL-1α is responsible for NF-κB activation in fibroblasts.
Figure 3.
IL-1α is responsible for NF-κB activation in fibroblasts. (A) Neutralizing antibodies against IL-1α prevent NF-κB activation in fibroblasts. MDA-MB-231 CM was incubated with neutralizing antibodies against IL-1α (lanes 3–5), IL-1β (lanes 6–8), or TNFα (lanes 9–11) at indicated concentrations (μg/ml) for 1 hr at room temperature. HLF-1 was incubated with antibody-treated CM for 3 hr, and NF-κB DNA-binding activity was measured by EMSA. Efficiency of antibodies to neutralize respective cytokines also was tested simultaneously to ensure the quality of antibodies (lanes 12–17). (B) IL-1α is expressed only in cell lines whose CM induce NF-κB in fibroblasts. Twenty micrograms of total RNA from indicated cell lines was subjected to Northern blotting with IL-1α probe. Integrity of RNA was examined by reprobing the blot with the ribosomal protein gene 36B4.
CM from only MDA-MB-231, MDA-MB-436, and HBL-100 induced NF-κB. If IL-1α in their CM is responsible for NF-κB activation, expression of IL-1α mRNA should be restricted to only these cell lines. As expected, IL-1α transcripts were detected only in MDA-MB-231, MDA-MB-436, and HBL-100 cells (Fig. 3B). These results further support the possibility that IL-1α is responsible for NF-κB activation.
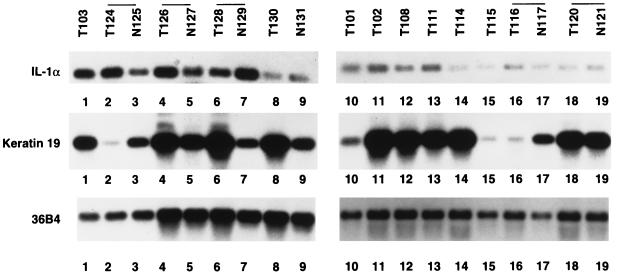
To assess the possibility that IL-1α is expressed in primary breast cancers, total RNA from freshly obtained breast cancer samples was prepared. Whenever available, RNA from normal tissues adjoining the tumor also was prepared. RNA from these tumors was reverse-transcribed and subjected to 20 cycles of PCR by using the ribosomal protein gene 36B4-specific primers. Amplification of abundantly expressed 36B4 mRNA was linear under these PCR conditions. The IL-1α mRNA was similarly amplified, and amplified product was identified by Southern blotting. Keratin 19 mRNA also was amplified to ensure that the tumor samples are composed of mammary epithelial cells. A representative Southern blot is shown in Fig. 4. A comparative analysis of IL-1α expression, ER, progesterone receptor (PR) (as determined by immunohistochemistry), and lymph node metastasis is shown in Table 1. A semiquantitative estimate of IL-1α was calculated by considering the lowest level of expression in a particular series of experiments as +. A low level (1+) of IL-1α expression was observed in tumor and normal tissues of three patients (T116, N117, T120, N121, T130, and N131). Three other patients displayed elevated the level of IL-1α in both normal and tumor tissues (T124, N125, T126, N127, T128, and N129). Note that the tumor has metastasized to lymph nodes in these patients. A marked increase in IL-1α expression was observed in one of seven patients where only tumor samples were examined (T103). All lymph nodes (22 of 22) were positive in the patient where expression of IL-1α was highest. It is also interesting that three of four tumors with elevated IL-1α are either ER-negative or heterogeneous with ER-positive and ER-negative cells. Note the abundant expression of keratin 19 in most of the tumor samples, particularly in IL-1α positive tumors. These preliminary studies in primary breast cancer along with results obtained with breast cancer cell lines suggest that IL-1α is expressed preferentially in ER-negative breast cancers and IL-1α in these cancers may contribute to metastasis by inducing the expression of NF-κB-regulated genes in fibroblasts.
Figure 4.
IL-1α transcripts are present in primary breast cancer. Total RNA from breast tumor samples (T) and normal tissues that surround the tumor (N) was subjected to RT-PCR by using primers specific to IL-1α (Top), keratin 19 primers (Middle), or ribosomal protein gene 36B4 (Bottom). RT-PCR products were visualized by Southern blotting.
Table 1.
IL-1α, ER/PR, and lymph node metastasis in primary breast cancers
| Tumor no. | IL-1 | ER | PR | Node status |
|---|---|---|---|---|
| T101 | 2+ | — | — | 0/21 |
| T102 | 2+ | — | — | 1/17 |
| T103 | 15+ | — | +/− | 22/22 |
| T108 | + | 50% | — | 0/1 |
| T111 | 2+ | 70% | 80% | 1/14 |
| T114 | + | + | + | 0/20 |
| T115 | + | 80% | 80% | 7/17 |
| T116 | + | — | — | 0/19 |
| N117 | + | — | — | |
| T120 | + | 30% | 60% | 0/18 |
| N121 | + | |||
| T124 | 10+ | — | NA | 2/20 |
| N125 | 3+ | |||
| T126 | 6+ | + | + | 13/20 |
| N127 | 3+ | |||
| T128 | 2+ | — | — | 1/1 |
| N129 | 7+ | |||
| T130 | + | + | + | 0/20 |
| N131 | + |
Quantitation is based on densitometric scanning of autoradiograms, and the values were corrected for the difference in 36B4 level. A value of + was assigned to the sample that displayed least IL-1α-amplified product. Percentage of cells that stained positive for ER and PR is given for tumors that are heterogeneous. Tumor that stained weakly for PR is indicated as +/−. Positive lymph node versus number of lymph nodes examined is also shown. NA, not available.
DISCUSSION
In this report, we show that ER-negative breast cancer cells secrete IL-1α and secreted IL-1α induces NF-κB in fibroblasts. In contrast, both ER-positive and ER-negative breast cancer cells secrete a factor(s) that can induce NF-κB in endothelial cells and macrophages.
That IL-1α is expressed only in a subset of ER-negative breast cancer cells suggests that ER somehow modulates the expression and activity of IL-1α. In this regard, preliminary studies revealed that CM from MDA-MB-231 cells transduced with ER have partially lost their ability to induce NF-κB and IL-6 in fibroblasts (data not shown). Unexpectedly, IL-1α expression in these cells was only marginally effected by ER (data not shown). It is likely that ER modulates IL-1α activity either by directly inhibiting IL-1α expression or by increasing the expression of factors that antagonize the binding of IL-1 to its active receptors (41).
The present study has provided a link between ER-negative phenotype and overexpression of two cytokines, IL-1α and IL-6. These two cytokines collectively influence the expression of a number of genes in cancer cells as well as in stromal cells. Two recent studies have indicated that increased expression of E-selectin and collagenase 3 in endothelial cells and stromal fibroblasts, respectively, is a result of breast cancer cell-derived IL-1 (42, 43). Induction of these two genes could be a result of NF-κB activation in stromal cells. E-selectin promoter contains a NF-κB-binding site (44). Collagenase 3 promoter binds to AP-1, which can synergize with NF-κB through protein–protein interaction (45, 46). Thus, IL-1α alone could account for overexpression of four genes in ER-negative breast cancers. These genes are involved in angiogenesis (E-selectin, IL-6) and metastasis (collagenase 3, uPA and IL-6). In addition, IL-1α and IL-6 may contribute to other clinical characteristics of breast cancers. For example, both IL-1α and IL-6 can contribute to chemotherapeutic resistance by decreasing penetration of drugs and/or by inhibiting activation of proteases involved in apoptosis (47, 48). IL-6 may alter the gene expression pattern in breast cancer cells by activating JAK/STAT pathways and Ras/MAPK pathways (49). Finally, both IL-1α and IL-6 promote osteoclast formation and hypercalcemia, which frequently is observed in advanced breast cancer even in the absence of bone metastasis (41, 50).
In summation, it is likely that constitutive activation of NF-κB in cancer cells (28, 51) as well as in fibroblasts that surround them may contribute to chemotherapy-resistant and metastatic growth of ER-negative breast cancers. Interestingly, analysis performed with primary breast cancers reveals a positive correlation between increased IL-1α expression and lymph node metastasis (Table 1). Although we were unable to detect IL-1α in plasma of 40 breast cancer patients by using enzyme-linked immunoassays (data not shown), further studies with a large number of samples using RT-PCR or in situ hybridization with riboprobes will be required to clarify whether IL-1α expression can predict clinical characteristics of breast cancer. In addition, it will be interesting to test whether inhibition of IL-1α-mediated NF-κB activation can reduce the growth and metastasis of ER-negative breast cancers. This can be achieved by the use of either soluble receptors for IL-1, which antagonize IL-1 action (49), IL-4, which up-regulates the expression of soluble IL-1 receptors (52), or specific inhibitors that can inhibit the activity of IκB kinases (53).
Acknowledgments
We thank P. Chambon, K. Cornetta, A. King, A. Passaniti, D. F. Spandau, X.-L. Sun, G. Sledge, and Y. C. Yang for various reagents, cell lines, and advice. This work is supported by Grant IRG-161k from the American Cancer Society, RGK Foundation, and Gustavus and Louise Pfeiffer Research Foundation (to H.N.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HLF, human lung fibroblasts; EMSA, electrophoretic mobility-shift assay; IL-1α, interleukin 1α; IL-6, interleukin 6; CM, conditioned medium; ER, estrogen receptor; TNFα, tumor necrosis factor α; TPCK, tosylphenylalanyl chloromethyl ketone; uPA, urokinase plasminogen activator.
References
- 1.Clarke R, Dickeson R B, Brunner N. Ann Oncol. 1990;1:401–407. doi: 10.1093/oxfordjournals.annonc.a057790. [DOI] [PubMed] [Google Scholar]
- 2.Horwitz K B. J Steroid Biochem Mol Biol. 1994;49:295–302. doi: 10.1016/0960-0760(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 3.Johnston S R D, Saccani-Jotti G, Salter S J, Coppen M, Ebbs S R, Dowsett M. Cancer Res. 1995;50:3331–3338. [PubMed] [Google Scholar]
- 4.Dumont J A, Bitonti A J, Wallace C D, Baumann R J, Cashmann E A, Cross-Doersen D E. Cell Growth Differ. 1996;7:351–359. [PubMed] [Google Scholar]
- 5.Duffy M J, Reilly D, McDermott E, O’Higgins N, Fennelly J J, Andreasen P A. Cancer. 1994;74:2276–2280. doi: 10.1002/1097-0142(19941015)74:8<2276::aid-cncr2820740811>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Montegudo C, Menino M J, San-Jaun J, Liotta L A, Stetler-Stevenson W G. Am J Pathol. 1990;136:585–592. [PMC free article] [PubMed] [Google Scholar]
- 7.D’Errico A, Garbisa S, Liotta L A, Castronovo V, Stetler-Stevenson W G, Grigoni W. Mod Pathol. 1991;4:239–246. [PubMed] [Google Scholar]
- 8.Basolo F, Fiore L, Fontanini G, Conaldi P G, Calvo S, Falcone V, Toniolo A. Cancer Res. 1996;56:3118–3122. [PubMed] [Google Scholar]
- 9.Basset P, Bellocq J P, Wolf C, Stoll I, Hutin P, Limacher J M, Podhajcer O L, Chenard M P, Rio M C, Chambon P. Nature (London) 1990;348:699–704. doi: 10.1038/348699a0. [DOI] [PubMed] [Google Scholar]
- 10.Singer C, Rasmussen A, Smith H S, Lippman M E, Lynch H T, Cullen K J. Cancer Res. 1995;55:2448–2454. [PubMed] [Google Scholar]
- 11.Noel A C, Polette M, Lewalle J M, Munaut C, Emonard H P, Birembaut P, Foidart J M. Int J Cancer. 1994;56:331–336. doi: 10.1002/ijc.2910560306. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T, Matsumoto, Kiritoshi A, Tano Y, Nakamura T. Cancer Res. 1997;57:3305–3313. [PubMed] [Google Scholar]
- 13.Kellokumpu-Lehtnen P, Talpaz M, Harris D, Van Q, Kurzrock R, Estrov Z. Int J Cancer. 1996;66:515–519. doi: 10.1002/(SICI)1097-0215(19960516)66:4<515::AID-IJC15>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Riegel A T, Wellstein A. Breast Cancer Res Treat. 1994;31:309–314. doi: 10.1007/BF00666163. [DOI] [PubMed] [Google Scholar]
- 15.Harris, A. L., Fox, S., Bicknell, R., Leek, R., Relf, M., Lejeune, S. & Kaklamanis, L. (1994) Cancer 74 (Suppl.), 1021–1025. [DOI] [PubMed]
- 16.Hill C S, Treisman R. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 17.Hansen S K, Nerlov C, Zabel U, Verde P, Johnsen M, Baeuerle P A, Blasi F. EMBO J. 1992;11:205–213. doi: 10.1002/j.1460-2075.1992.tb05043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato H, Seiki M. Oncogene. 1993;8:395–405. [PubMed] [Google Scholar]
- 19.Miyazawa K, Kitamura A, Kitamura N. Biochemistry. 1991;30:9170–9176. doi: 10.1021/bi00102a007. [DOI] [PubMed] [Google Scholar]
- 20.Siebenlist U, Franzoso G, Brown K. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 21.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 22.Verma I M, Stevenson J K, Schwarz E M, Antwerp D V, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 23.Whiteside S T, Epinat J, Rice N R, Israel A. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 25.Wang C-Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 26.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z-G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 28.Nakshatri H, Bhat-Nakshatri P, Martin D A, Goulet R J, Sledge G W. Mol Cell Biol. 1997;17:3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson E W, Paik S, Brunner N, Sommers C L, Zugmaier G, Clarke R, Shima T B, Torri J, Donahue S, Lippman M E, Martin G R, Dickson R B. J Cell Physiol. 1992;150:534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi H, Taylor R M, Chrest F J, Masui T, Utsumi K, Tatematsu M, Passaniti A. Cancer Res. 1995;55:399–407. [PubMed] [Google Scholar]
- 31.Spandau D F. Oncogene. 1994;9:1861–1868. [PubMed] [Google Scholar]
- 32.Nakshatri H, Bhat-Nakshatri P. Ann Biochem. 1997;249:103–104. doi: 10.1006/abio.1997.2154. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Nakshatri H, Broullet P, Bhat-Nakshatri P, Chambon P. Gene. 1996;174:79–84. doi: 10.1016/0378-1119(96)00391-5. [DOI] [PubMed] [Google Scholar]
- 35.Noguchi S, Aihara T, Motomura K, Inaji H, Imaoka S, Koyama H. Cancer. 1996;78:1235–1240. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1235::AID-CNCR10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 36.Sommers C L, Byers S W, Thompson E, Torri J A, Gelmann E P. Breast Cancer Res Treat. 1994;31:325–335. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- 37.Price J E, Polyzos A, Zhang R D, Daniels L M. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- 38.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 39.Nerlov C, Rorth P, Blasi F, Johnsen M. Oncogene. 1991;6:1583–1592. [PubMed] [Google Scholar]
- 40.Gruss H, Brach M A, Herrmann F. Blood. 1992;80:2563–2570. [PubMed] [Google Scholar]
- 41.Pacifici R. J Bone Mineral Res. 1996;11:1043–1051. doi: 10.1002/jbmr.5650110802. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen M, Corless C L, Kraling B M, Tran C, Atha T, Bischoff J, Barsky S H. Am J Pathol. 1997;150:1307–1314. [PMC free article] [PubMed] [Google Scholar]
- 43.Uria J A, Stahle-Backdahl M, Seiki M, Fueyo A, Lopez-Otin C. Cancer Res. 1997;57:4882–4888. [PubMed] [Google Scholar]
- 44.Min W, Pober J S. J Immunol. 1997;159:3508–3518. [PubMed] [Google Scholar]
- 45.Pendas A M, Balbin M, Llano E, Jimenez M G, Lopez-Otin C. Genomics. 1997;40:222–233. doi: 10.1006/geno.1996.4554. [DOI] [PubMed] [Google Scholar]
- 46.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St. Croix B, Florenes V A, Rak J W, Flanagan M, Bhattacharya N, Slingerland J M, Kerbal R S. Nat Med. 1996;2:1204–1210. doi: 10.1038/nm1196-1204. [DOI] [PubMed] [Google Scholar]
- 48.Lotem J, Sachs L. Proc Natl Acad Sci USA. 1997;94:9349–9353. doi: 10.1073/pnas.94.17.9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 50.Kanis J A, Powles T, Paterson A H G, McCloskey E V, Ashley S. Bone. 1996;19:663–667. doi: 10.1016/s8756-3282(96)00285-2. [DOI] [PubMed] [Google Scholar]
- 51.Sovak M A, Bellas R E, Kim D W, Zanieski G J, Rogers A E, Traish A M, Sonenshein G E. J Clin Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohmori Y, Smith M F, Jr, Hamilton T A. J Immunol. 1996;157:2058–2065. [PubMed] [Google Scholar]
- 53.Stancovski I, Baltimore D. Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]