Abstract
A polymerase chain reaction (PCR) assay was combined with a broth-culture enrichment system to detect Salmonella shed in feces from subclinically infected swine. The effectiveness of the broth culture-polymerase chain reaction (BC-PCR) assay to identify pigs shedding Salmonella in feces was compared with a microbiological culture and a commercial enzyme linked immunosorbent assay (ELISA) kit to detect Salmonella-specific serum antibody. A total of 67 pigs were tested by each of the 3 methodologies. Forty-one pigs tested positive for Salmonella by BC-PCR and ELISA identified 6 positives and 23 suspicious samples. It was shown that the BC-PCR assay is a rapid diagnostic tool for detecting of Salmonella shed by asymptomatic swine compared with current diagnostic technologies.
Introduction
Bacteria of the genus Salmonella are gram negative rods, many of which are important human and animal pathogens. Salmonella infections in swine spread rapidly by oral exposure to feces. Organisms multiply rapidly in the intestine and can spread to various internal organs, causing a reduction in growth performance, a risk of infection to people via contaminated pork products, and a potential source of infection for other pigs (1). Detection of Salmonella in swine is difficult because infection may not result in clinical symptoms. Shedding of Salmonella in asymptomatic, carrier-swine is intermittent with bacterial cells generally being shed in numbers below the detection limit for standard culture methods resulting in an under-estimation of herd prevalence (2). Therefore, it is recommended that evaluation of the Salmonella infection status of a herd or individual animal requires repeated testing (3).
A rapid, reliable tool to assist disease control management within barns should aim to reduce the number of carrier-swine, thereby reducing the incidence of salmonellosis in both people and animals. For this purpose, a number of assays have been developed to decrease the time required to identify Salmonella in food, feces, and other clinical samples (4,5,6). However, careful examination of factors affecting detection of Salmonella, showed that detection is significantly increased when multiple enrichment-broths or plating media are utilized (7,8), and while large numbers (1 × 105/g feces) of bacteria can be shed by carriers, more typically, only smaller numbers (less than 50/g feces) are shed (3,9,10).
The objective of this study was to compare current diagnostic methodologies for detecting asymptomatic carrier-swine shedding Salmonella bacteria in their feces.
Materials and methods
Experimental design
Salmonella bacteria were identified in fecal samples (n = 67) using 3 diagnostic methodologies; microbial culture (single- and double-broth enrichment), polymerase chain reaction (PCR) (direct and broth culture-PCR), and a commercial enzyme linked immunosorbent assay (ELISA) kit, to determine serological status. Results from each assay were compared using the observed proportion of agreement and kappa statistics to ensure that agreement exceeds chance levels.
Serum and fecal samples were collected from 57, 5 to 6 mo old healthy pigs, from 3 farms in southeastern Saskatchewan that were known to have experienced sporadic cases of enteric salmonellosis. Additional serum and fecal samples were collected from 10 age-matched healthy pigs at a 4th farm, known to be free from clinical salmonellosis for the past 2 to 3 y. None of the animals used in this experiment showed clinical signs of salmonellosis.
Microbial culture
Single-enrichment culture for Salmonella was performed as follows: fecal samples (0.5 g) were inoculated into tetrathionate (9 mL) and selenite (9 mL) broths for incubation at 37°C for 24 h. Fecal samples (0.2 g) were also inoculated into Rappaport-Vassiliadis (9 mL) broth and incubated at 42°C for 24 h. After incubation, each broth was then plated onto 4 selective, solid medias (Xylose-Lysine-Tergitol-4 [XLT-4], Salmonella/Shigella (SS), Hektoen, and MacConkey) and incubated at 37°C. After 24 h growth, suspected Salmonella colonies were subcultured onto blood-agar and MacConkey-agar and incubated at 37°C for a further 24 h. Presumptive Salmonella isolates were confirmed using conventional biochemical tests and an agglutination assay (Bacto-Salmonella O antisera; Difco Laboratories, Detroit, Michigan, USA). Isolates determined to be Salmonella by these procedures were sent to Health Canada, Laboratory for Foodborne Zoonoses, Guelph, Ontario, for serotyping.
Double-enrichment microbial culturing involved subculturing 1 mL of each of the initial enrichment-broths into fresh broth (9 mL) after an incubation period of 5 d at room temperature, as previously published (11). Subculture of each 2nd broth to selective, solid media was subsequently performed, as outlined above.
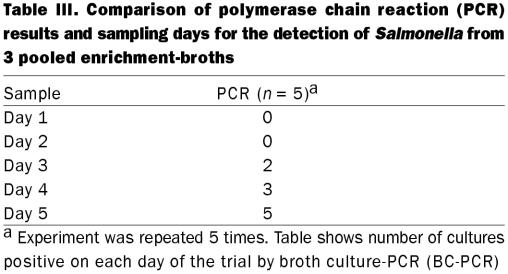
Enrichment culture prior to BC-PCR was performed by adding feces to tetrathionate (0.5 g, 9 mL), selenite (0.5 g, 9 mL), and Rappaport-Vassiliadis (0.2 g, 9 mL) broths and incubating for 24 h at the appropriate temperature, and then moving the cultures to room temperature for 5 d. The 5-day culture system was optimized in a preliminary experiment wherein Salmonella-positive feces were used and each day 500 μL of each broth was pooled and tested for the presence of Salmonella by BC-PCR until samples showed positive. This experiment was repeated 5 times.
DNA extraction and PCR
DNA was extracted from porcine fecal samples using the method described by Cohen (12). Approximately 0.2 g of feces was suspended in 1 mL of lysis buffer (5M guanidine thiocyanate [GuSCN], 22 mM EDTA, 0.05M Tris-HCl [pH 6.4], 0.65% Triton X-100) and incubated at room temperature for 1 h. After centrifugation (15 000 × g, 30 s), the supernatant was transferred to a clean tube containing 50 μL diatomaceous earth (DE) suspension (20% diatomaceous earth in 0.17 M HCl). After vortex and centrifugation (15 000 × g, 30 s), the pellet was washed twice with 1 mL GuSCN-Tris buffer (5.5 M guanidine thiocyanate, 0.05 M Tris-HCl [pH 6.4] and 80% ethanol) and once with acetone. The DE was vacuum dried and DNA was eluted by the addition of 50 μL of water.
The DNA was extracted from bacteria grown in the pre-PCR enrichment-broths through a phenol/chloroform extraction (13). After 5 d of enrichment, 500 μL aliquots of each broth (tetrathionate, selenite, Rappaport-Vassiliadis) were pooled into 1 tube and centrifuged (15 000 × g, 5 min). The pellet was resuspended in 600 μL of bacterial lysis buffer (100 mM NaCl, 500 mM Tris [pH 8.0], 10% sodium dodecyl sulfate [SDS]) containing 6 μL proteinase K (20 mg/mL). After a 2 h incubation at 65°C, DNA was isolated by adding and mixing an equal volume of Tris-buffered phenol-chloroform (50:50 v/v, pH 8.0) followed by centrifugation at 15 000 ×g for 5 min. The aqueous phase was further purified by repeating the phenol/chloroform extraction process. The DNA was isolated by precipitation with salted 95% ethanol (0.3 M Na acetate) and washed with 80% ethanol. The DNA was vacuum dried and resuspended in approximately 30 μL of water.
A set of PCR primers were developed for a 427 base pairs (bp) region of S. enterica subspecies enterica serovar Typhimurium that included the 3' end of the Fim A gene (Salm-F; CGCAGGTGC CTTTCTCCAT, bases 152 to 486) and the 5' end of the Fim I gene (Salm-R; CGCGCCTTTCCTTATCATCT, bases 1-17). The PCR reaction mixture included 1 × PCR buffer (Invitrogen, Burlington, Ontario), MgCl2+ (2 mM), dNTP's (0.25 mM), 40 pMol each primer, Taq polymerase (1.2 U/mL) in a 48 μL reaction volume, and mixed with 2 μL of sample DNA. A thermal cycler (PTC 200; MJ Research Inc., Waltham, Massachusetts, USA) program consisted of a denaturation step (3 min, 94°C), then 40 cycles (30 s at 94°C, 60 s at 55°C, and 60 s at 72°C) of amplification, and finished with a prolonged extension phase (5 min, 72°C). Amplified products were electrophoresed through a 1.2% agarose gel, stained with ethidium bromide and visualized with a ultra violet (UV) transilluminator.
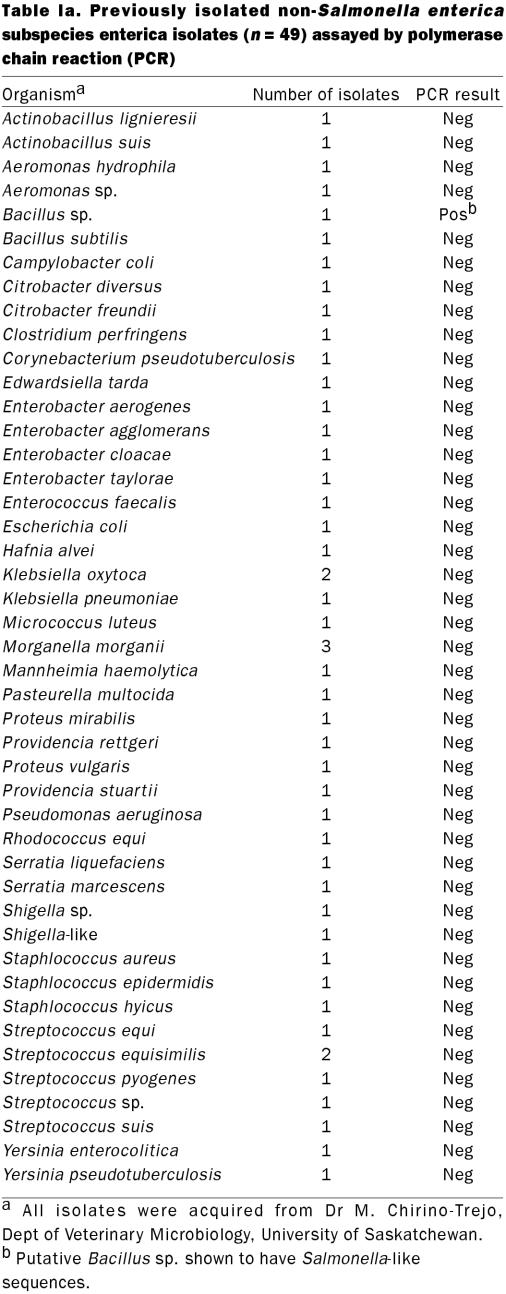
To demonstrate the specificity of the primers, 111 archived Salmonella isolates and 45 other bacterial species (49 isolates) were tested (Table Ia and Ib) and confirmed using a previously published assay (14).
Table Ia.
Table Ib.
To determine the detection limit, fecal samples identified as negative for Salmonella by microbial culture and BC-PCR, were spiked with 10-fold serial dilutions (neat to 1 × 10−10 ) of log phase S. enterica subspecies enterica serovar Infantis culture. Dilutions of bacteria were also plated on blood-agar to establish the number of colony forming units (CFU). Each sample was tested by BC-PCR and microbial culture for the presence of Salmonella bacteria. This experiment was repeated 3 times.
Salmonella-ELISA
Serum samples (n = 67) were tested for the presence of Salmonella-specific antibodies using a commercial Salmonella ELISA kit (Vetsign; GuildHay, Guildford, Surrey, United Kingdom), as recommended by the manufacturer. Serum-to-positive ratios (SP%) of 10 or lower were interpreted as negative, while SP% above 25 were interpreted as positive for Salmonella. Samples with an SP% between 10 and 25 were considered suspicious.
Results
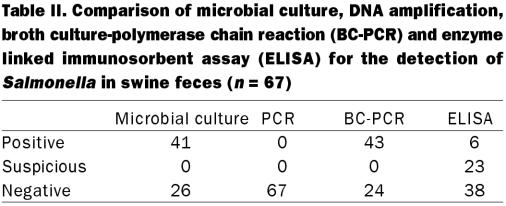
Results of the 3 assays performed to identity asymptomatic Salmonella-carrier swine are shown in Table II. Single-enrichment microbial culture identified 37 (55.2%) animals shedding Salmonella sp. Combining the results of single-and double-enrichment broth culture systems identified a total of 41 (58%) shedding pigs. Suspected Salmonella isolates (n = 121) were sent for serotyping. Fifteen pigs (37.6%) were shown to be infected with S. enterica subspecies enterica serovar California and 10 (24.4%) with S. enterica subspecies enterica serovar Infantis. Seven (17%) of the infected pigs had mixed infections consisting of both serovar Infantis and serovar California, and 1 (1.4%) had the non-motile serotype (antigen I:6,7,14:-:-). Three (7.3%) pigs had infections which were apparently incorrectly identified as Salmonella, but may have been Citrobacter amalonaticus, and samples from 5 pigs (12.2%) were non-viable at the time of serotyping.
Table II.
Polymerase chain reaction amplification using S. enterica-specific primers yielded a single, 427 bp DNA fragment. Feces from 43 out of 67 (64.2%) pigs were positive for Salmonella by BC-PCR. A positive PCR result could be obtained with as little as 4.8 pg of total genomic DNA or approximately 3 CFU/g feces as determined by parallel microbiological culture. All archived Salmonella isolates (n = 111) tested positive by PCR (Table Ia and Ib). Forty-eight of 49 isolates of 45 other bacterial species were negative by PCR. One possible cross reaction was identified; a single archived field-isolate of a putative Bacillus species. The optimal time for the detection of Salmonella in culture broth was 5 d after inoculation into porcine feces. The BC-PCR, performed on DNA from pooled culture broth, was inconsistently positive for the presence of Salmonella on days 1 to 4 and consistently positive day 5 (Table III). None of the samples tested positive when DNA was extracted directly from feces.
Table III.
Only 6 animals (9.0%) had positive serological evidence of systemic infection with Salmonella at the time of collection. A further 23 (34.3%) were identified as suspicious and the remaining 38 (56.7%) were found negative by ELISA.
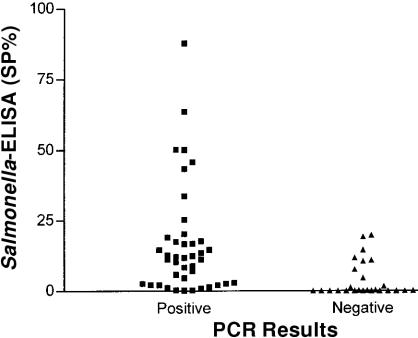
The BC-PCR and microbial culture (combined single- and double-broth enrichment culture systems) identified similar numbers of shedding and non-shedding animals (observed proportion of agreement = 97%, Kappa = 0.94). Two additional fecal samples were positive for Salmonella by BC-PCR. None of the culture positive samples were negative by BC-PCR. Based on this comparison, the BC-PCR test had a sensitivity of 95.3% and specificity of 93.3%. The Salmonella-specific ELISA identified fewer positive and suspicious samples than did microbial culture (observed proportion of agreement = 82%, Kappa=0.65) or BC-PCR (observed proportion of agreement = 75%, Kappa=0.52). Only samples resulting in an SP% above 50 appeared to be in agreement with both BC-PCR and enrichment culture positive results (Figure 1).
Figure 1. Comparison of polymerase chain reaction (PCR) results for detection of Salmonella sp. in swine feces with serological evidence of systemic anti-Salmonella sp. immune response measured by enzyme linked immunosorbent assay (ELISA) (SP%, serum sample to positive ratio expressed as a percent).
Discussion
The DNA amplification technology has proven to be a necessity when working with numbers of bacteria that are too low to be detected by standard culture techniques or when specimens contain bacteria that cannot be cultured. In this study, it was possible to detect S. enterica serovars within 5 d of receiving fecal samples using an enrichment-broth PCR technique (BC-PCR). This assay significantly reduces the expense associated with the biochemical identification of Salmonella commonly used during routine microbiological testing, while maintaining the benefits of high sensitivity provided by enrichment-broth culturing systems. Amplification could be observed with as few as 3 CFU/g feces; a result consistent with other studies that use PCR to detect Salmonella (15,16) and other bacteria (17,18).
In this study, the use of a double-enrichment microbial culture, in addition to the single-enrichment microbial culture, increased the sensitivity and resulted in the detection of 4 additional positive animals. Others have made similar observations regarding double broth-enrichment systems (8). The BC-PCR, performed from pooled broths after 5 d of enrichment, identified 2 more positive samples than did the microbial culture without the need for repeated subculturing on selective media and biochemical tests. The BC-PCR had a low detection limit and detected slightly more positive samples than did the microbial culture. These results are comparable to the findings of other studies (15,19,20).
The fact that Salmonella DNA could not be detected in extracts taken directly from fecal samples, is likely due to low numbers of bacteria and the presence of inhibitors coextracted with DNA from the samples. It has been demonstrated previously that the PCR is sensitive to the presence of inhibitory substances that may reside in certain types of samples, such as food (21,22) and human fecal samples (23,24). Similar inhibitors may also be present in porcine fecal samples. Culture-enrichment, wherein there was either an increased number of bacteria or deterioration of fecal-derived inhibitors was necessary to overcome a dramatic lack of sensitivity in testing fecal samples directly.
The enrichment-step consisted of an incubation period in 3 different Salmonella-permissive broth media, which increases the number of bacteria. Since some strains of Salmonella grow better in some Salmonella-selective-enrichment broths than others (25), an aliquot of each was pooled prior to the extraction of DNA. Salmonella enterica isolates could be consistently detected by BC-PCR in enrichment-broths after 5 d and inconsistently after 1 to 4 d.
The ELISA assays have been reported to be an efficient and rapid means to detect Salmonella infections in pigs (26,27). In this study, however, the ELISA assay only identified approximately half the number of pigs shedding Salmonella that could be identified by microbial culture or BC-PCR, which suggests that ELISA may have limited use in a control program to eliminate salmonellosis in western Canada. It is known that immune responses are not frequently generated in pigs that shed detectable amounts of bacteria, so the observed discrepancy may be the result of differences between intestinal colonization and systemic infection. The manufacturer has claimed that the assay is able to detect S. enterica subspecies enterica serovars Typhimurium, and Choleraesuis and shows cross-reactivity to serovars Infantis, Derby, and Panama. The 2 serovars grown from the feces of pigs in this study included Infantis and California suggesting that the ELISA may not detect all serotypes present in Saskatchewan.
The primers designed for this study did not cross-react with any of 48 isolates representing 26 genuses of bacteria, but did show a weak cross-reaction with a single Bacillus sp. field isolate. Bacillus species are considered saprophytes, found commonly in soil and water. These organisms are not commonly isolated from porcine feces in routine culture conditions (28,29) and this, combined with the selective methods described in this study which favour the growth of Gram negative rather than Gram positive bacteria, would make the detection of false positives unlikely. To characterize this isolate, it was tested with a set of Salmonella invA-specific primers, which also yielded a positive result. Sequencing of the FimA-specific PCR product from this isolate showed a 100% match with the 427 bp to Salmonella FimA gene (data not shown).
This study has demonstrated that BC-PCR is a rapid, efficient diagnostic tool, which may aid in the control of Salmonella in carrier-swine and, therefore, decrease the incidence of disease in both humans and animals.
Footnotes
Acknowledgments
The authors thank B. Andres (Prairie Swine Centre) for the generous contributions of material and C. A. Muckle (Health Canada, Laboratory for Foodborne Zoonoses) for serotyping all of the isolates in this study. This research was funded by the Agriculture Development Fund of the Saskatchewan, Department of Agriculture, Food and Rural Revitalization.
Address all correspondence and reprint requests to Dr. G. Appleyard; telephone: (306) 966-7213; fax: (306) 966-7244; e-mail: greg.appleyard@usask.ca
Received October 18, 2001. Accepted January 23, 2003.
References
- 1.Wood RL, Pospischil A, Rose R. Distribution of persistent Salmonella typhimurium infection in internal organs of swine. Am J Vet Res 1989;50:1015–1021. [PubMed]
- 2.Hurd HS, Schlosser W, Ebel ED. The effect of intermittent shedding on prevalence estimation in populations. Proc 3rd Intl Symp Epidemiology and Control of Salmonella in Pork 1999:57–58.
- 3.Gray JT, Stabel TJ, Fedorka-Cray PJ. Effect of dose on the immune response and persistence of Salmonella choleraesuis infection in swine. Am J Vet Res 1996;57:313–319. [PubMed]
- 4.Bennet AR, Greenwood D, Tennant C, Banks JG, Betts RP. Rapid and definitive detection of Salmonella in foods by PCR. Lett Appl Micro 1998;26:437–441. [DOI] [PubMed]
- 5.Bailey JS. Detection of Salmonella cells within 24 to 36 hours in poultry samples with polymerase chain reaction BAX system. J Food Prot 1998;61:792–795. [DOI] [PubMed]
- 6.Gouws PA, Visser MA, Brozel VS. A polymerase chain reaction procedure for the detection of Salmonella spp. within 24 hours. J Food Prot 1998;61:1039–1042. [DOI] [PubMed]
- 7.Davies PR, Funk JA, Nichols MG, et al. Effects of some methodological factors on detection of Salmonella in swine feces. Proc 3rd Intl Symp Epidemiology and Control of Salmonella in Pork 1999:30–32.
- 8.O'Carroll JM, Davies PR, Correa MT, Slenning BD. Effects of sample storage and delayed secondary enrichment on detection of Salmonella spp in swine feces. Am J Vet Res 1999;60:359–362. [PubMed]
- 9.Clarke RC, Gyles CL. Salmonella. In: Gyles CL, Thoen CO, eds. Pathogenesis of Bacterial Infections in Animals. 2nd ed. Ames: Iowa State University Press, 1993:133–153.
- 10.World Health Organization. Salmonellosis control: the role of animal and product hygiene: a report of a WHO committee. WHO Technical Report Series No. 744. 1988. World Health Organization, Geneva, Switzerland. [PubMed]
- 11.Chirino-Trejo JM. A more sensitive procedure for the detection of Salmonella carriers in swine. Proc Western Can Assoc Swine Pract 1999:50–53.
- 12.Cohen ND, Martin LJ, Simpson RB, Wallis DE, Neibergs HL. Comparison of polymerase chain reaction and microbiological culture for detection of salmonellae in equine feces and environmental samples. Am J Vet Res 1996;57:780–786. [PubMed]
- 13.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press. 1989.
- 14.Rahn K, De-Grandis SA, Clarke RC, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 1992;6:271–279. [DOI] [PubMed]
- 15.Cohen ND, McGruder ED, Neibergs HL, Behle RW, Wallis DE, Hargis BM. Detection of Salmonella enteritidis in feces from poultry using booster polymerase chain reaction and oligonucleotide primers specific for all members of the genus Salmonella. Poult Sci 1994;73:354–357. [DOI] [PubMed]
- 16.Cohen ND, Wallis DE, Neibergs HL, Hargis BM. Detection of Salmonella enteritidis in equine feces using the polymerase chain reaction and genus-specific oligonucleotide primers. J Vet Diagn Invest 1995;7:219–222. [DOI] [PubMed]
- 17.Pao CC, Yen TS, You JB, Maa JS, Fiss EH, Chang CH. Detection and identification of Mycobacterium tuberculosis by DNA amplification. J Clin Microbiol 1990;28:1877–1880. [DOI] [PMC free article] [PubMed]
- 18.Frankel G, Riley L, Giron JA, et al. Detection of Shigella in feces using DNA amplification. J Infect Dis 1990;161:1252–1256. [DOI] [PubMed]
- 19.Soumet C, Ermel G, Rose V, et al. Identification by a multiplex PCR-based assay of Salmonella typhimurium and Salmonellaenteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol 1999;29:1–6. [DOI] [PubMed]
- 20.Thisted-Lamberts S, Ballagi-Pordány A, Nilsson A, Norberg P, Danielsson-Tham ML. A comparison between a PCR method and a conventional culture method for detecting pathogenic Yersinia enterocolitica in food. J Appl Bacteriol 1996;81:303–308. [DOI] [PubMed]
- 21.Dickinson JH, Kroll RG, Grant KA. The direct application of the polymerase chain reaction to DNA extracted from foods. Lett Appl Microbiol 1995;20:212–216. [DOI] [PubMed]
- 22.Rossen L, Norskov P, Holmstrøm K, Rasmussen OF. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA extraction systems. Intl J Food Microbiol 1992;17:37–43. [DOI] [PubMed]
- 23.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol 1990;28:1300–1307. [DOI] [PMC free article] [PubMed]
- 24.Monteiro L, Bonnemaison D, Vekris A, et al. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol 1997;35:995–998. [DOI] [PMC free article] [PubMed]
- 25.Bager F, Petersen J. Sensitivity and specificity of different methods for the isolation of Salmonella from pigs. Acta Vet Scand 1991;32:473–481. [DOI] [PMC free article] [PubMed]
- 26.Proux K, Houdayer C, Humbert F, et al. Development of a complete ELISA using Salmonella lipopolysaccharides of various serogroups allowing to detect all infected pigs. Vet Res 2000;31:481–490. [DOI] [PubMed]
- 27.Møller K, Ahrens P. Comparison of toxicity neutralization-, ELISA- and PCR tests for typing of Clostridium perfringens and detection of the enterotoxin gene by PCR. Anaerobe 1996;2:103–110.
- 28.Salanitro JP, Blake IG, Muirhead PA. Isolation and identification of fecal bacteria from adult swine. Appl Environ Microbiol 1977;33:79–84. [DOI] [PMC free article] [PubMed]
- 29.Robinson IM, Allison MJ, Bucklin JA. Characterization of the cecal bacteria of normal pigs. Appl Environ Microbiol 1981;41:950–955. [DOI] [PMC free article] [PubMed]