Abstract
Whitewater Arroyo virus (WWAV) is a North American New World arenavirus, first isolated from rats in New Mexico in 1993, and tentatively associated with three human fatalities in California in 1999-2000. However, it remains unclear whether WWAV was the cause of these, or any other, human infections. One important characteristic of viruses that influences pathogenic potential is the choice of cellular receptor and the corresponding tropism of the virus. In the arenaviruses, these properties are determined largely by the viral glycoprotein (GP). We have previously noted for the New World clade B arenaviruses, which include four severe human pathogens, that the ability to cause human disease correlates with the ability of the GP to use the human transferrin receptor 1 (hTfR1) to enter cells. In addition, pseudotyped retroviral vectors displaying the GPs from pathogenic clade B viruses transduced a range of cell lines in vitro that was distinct from those that could be transduced by non-pathogenic clade B viruses. WWAV was initially classified as a New World clade A virus, based on sequence analysis of its nucleoprotein gene. However, more extensive analyses have revealed that WWAV and the other North American arenaviruses are probably recombinant clade A/B viruses, and that the WWAV GP is more closely related to the clade B GPs. Based on this finding, we sought to understand more about the possible pathogenic potential of WWAV by determining whether its clade B-like GP exhibited the characteristics of a pathogenic or non-pathogenic clade B virus. Our studies found that WWAV GP did not use hTfR1 for entry, and that its overall in vitro tropism was most similar to the GPs from the nonpathogenic clade B viruses. Although many viral factors in addition to GP receptor use and tropism determine whether a virus is able to cause disease in humans, our analysis of the WWAV GP does not support the idea that WWAV is a human pathogen.
Introduction
The arenaviruses are enveloped, single-stranded RNA viruses, of interest because five members of the group can cause severe hemorrhagic fevers in humans, with mortality rates reaching twenty percent (Geisbert and Jahrling, 2004). The family is divided into two groups, Old World and New World viruses, based initially on serologic cross-reactivity and geographic distribution, and later confirmed by genomic sequence analyses (Clegg 1993; Bowen et al., 1996). The vast majority of the arenaviruses are vectored by rodents, in which they cause persistent infections, with the possible exception of Tacaribe virus (TCRV), which was isolated from bats (Downs et al, 1963). The distribution of the viruses is restricted to the areas that are populated by their specific rodent vector, and humans are accidental hosts, being occasionally infected by contact with rodent excreta.
The human pathogens in the Old World arenaviruses comprise Lassa fever virus (LASV) and lymphocytic choriomeningitis virus (LCMV). Lassa fever is a febrile illness, restricted to Western Africa, that in severe cases can lead to pulmonary edema, respiratory distress, bleeding from mucosal surfaces and shock (McCormick et al., 1987). In contrast, LCMV is more widespread, being vectored by the common house mouse Mus Musculus, and is the only Old World arenavirus found in North America. LCMV infection can be asymptomatic or cause febrile CNS disease (Bonthius et al., 2007), and it is also a suspected teratogen (Jamieson et al., 2006; Sheinbergas 1976). It was recently associated with fatal outcomes in immunosupressed patients receiving organ transplants (Fischer et al., 2006).
The New World complex comprises eighteen members, distributed between three clades, A, B and C (Bowen et al. 1996; Charrel and de Lamballerie, 2003) (Fig. 1). Clade B contains four known human pathogens, capable of causing hemorrhagic fevers in South America; Junin (JUNV), Machupo (MACV), Guanarito (GTOV), and Sabia (SABV) viruses, that cause Argentine, Bolivian, Venezuelan and Brazilian hemorrhagic fevers, respectively. Clade B also contains three non-pathogenic members.
Figure 1. Recombinant origin for the North American New World arenaviruses.
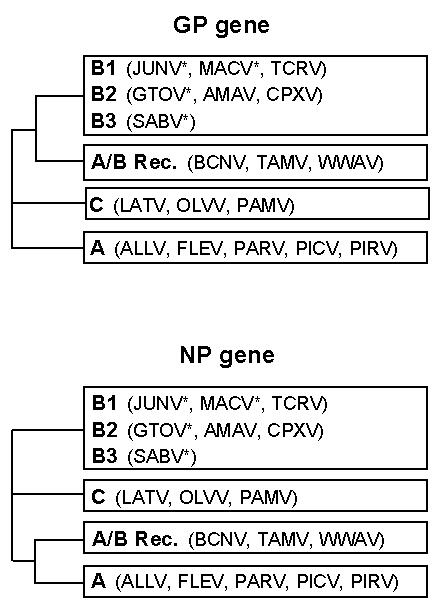
Sequence comparison of the GP and NP genes from the New World arenaviruses revealed three distinct clade, A, B and C. In addition, the North American arenaviruses, that include WWAV, are most closely related to clade A in the NP gene, but clade B in GP. A/B Rec. refers to this putative recombinant lineage. The strains that are pathogenic for humans (*) are found throughout clade B, in each of the three distinct sub-groups, B1, B2 and B3. The pathogenicity of WWAV remains unclear. Adapted from Charrel et al., 2001, 2002)
North America is home to three New World arenaviruses, carried by members of the Cricetidae rodent family. Tamiami virus (TAMV) was isolated from hispid cotton rats (Sigmodon hispidus) in Florida in 1963 (Calisher et al., 1970; Jennings et al., 1970). Whitewater Arroyo virus (WWAV) was isolated from white-throated woodrats (Neotoma albigula) in 1993 (Fulhorst et al., 1996) and has subsequently been found in other Neotoma and Peromyscus species, including deer mice (P. maniculatus). Most recently, Bear Canyon virus (BCNV) was isolated from California mice (P. californicus) in California in 2002 (Fulhorst et al., 2002), and is also carried by large-eared woodrats (N. macrotis) (Cajimat et al., 2007).
There has been considerable interest in WWAV since 2000, when a preliminary report implicated it in the deaths of three people in California (CDC, 2000; Enserink 2000). The patients presented with febrile illness and respiratory distress, with two developing hemorrhagic symptoms and liver failure. However, only one of the victims reported a possible contact with rodent droppings before becoming ill, and no subsequent reports have appeared confirming these initial findings. Therefore, the association of WWAV with these or any other human infections remains to be established.
The arenavirus genome consists of two RNA strands, a large (L) strand of about 7200 nucleotides and a small (S) strand of about 3500 nucleotides. Each codes for two proteins, arranged in non-overlapping open reading frames of opposite (ambisense) orientation. The S segment codes for the glycoprotein (GP) and nucleoprotein (NP), whereas the L segment carries the viral polymerase (L) and a zinc-finger-like protein (Z).
The phylogeny of the arenaviruses has been traditionally based on a 613-649 nucleotide sequence of the NP gene (Bowen et al., 1997). Based on such analyses, WWAV was initially classified as a clade A virus. Subsequently, a more extensive analysis of a 3178 nucleotide sequence of the S strand revealed that the GP gene had closer homology to the GP of clade B viruses (Archer and Rico-Hesse, 2002; Charrel et al., 2001;). Similar findings were obtained upon analysis of the S strands from TAMV and BCNV, but not in any South American arenaviruses (Charrel et al., 2002), leading to the suggestion that the three North American arenaviruses represent a phylogenetic lineage that originated by recombination between clade A and B viruses, and is separate from the other South American viruses (Figure 1)
Receptor use by viruses has a significant influence on viral tropism and pathogenicity. The arenaviruses are known to infect a wide range of cell types across many different species (Reignier et al., 2006; Rojek et al., 2006), suggesting that they may be able to use different cellular receptors. To date, two distinct arenavirus receptors have been identified; α-dystroglycan (α-DG), which is used by LASV, certain isolates of LCMV, and the New World clade C viruses Oliveros and Latino (Cao et al., 1998; Kunz et al., 2005; Reignier et al., 2006; Spiropoulou et al., 2002), and human transferrin receptor 1 (hTfR1), which is used by certain clade B viruses for entry into human cells (Radoshitzky et al., 2007; Flanagan et al., submitted for publication).
Using GP pseudotyped retroviral vectors as surrogates for arenavirus entry, we have recently demonstrated that hTfr1 is not the only receptor to be used by clade B viruses. Although the pathogenic members of the clade B lineage can use hTfR1 to enter cell lines in vitro, the non-pathogenic members TCRV and Amapari (AMAV) use entry pathways that are independent of either α–DG or hTfR1 (Flanagan et al., submitted for publication). We have also observed marked differences in the ability of clade B GPs to direct entry into human and rodent lymphocyte cell lines, and which also segregate between the pathogenic and non-pathogenic clade B viruses (Oldenburg et al., 2007), and tropism differences between GTOV and AMAV have also previously been noted (Rojeck et al., 2006). Taken together, these studies demonstrate that receptor use and tropism characteristics of the GP can distinguish human pathogens from non-pathogens in the clade B lineage. Since sequence comparisons have revealed that the WWAV GP is closely related to the clade B GPs, and because the ability of this virus to cause human disease is unclear, we undertook the present study to investigate whether the WWAV GP exhibited the same characteristics as the GPs from the clade B human pathogens.
Results
Generation of WWAV GP pseudotyped retroviral vectors
Retroviral vectors pseudotyped with arenaviruses GPs have proved to be useful tools with which to study arenavirus entry as they exhibit entry characteristics and tropism patterns similar to the parental arenaviruses (Oldenburg et al., 2007; Reignier et al., 2006, Rojek et al., 2006). We generated retroviral vectors carrying the GP from the prototype WWAV strain AV 9310135, originally isolated from the kidney of an infected white-throated woodrat (Fulhorst et al., 1996). Since the vectors encode a GFP reporter gene, the ability of the WWAV GP to direct entry into cells could be assessed by measuring GFP expression in a target population by FACS analysis at 48 hours post-transduction.
The titer of the WWAV vectors was initially measured on human 293A cells and compared to the titers obtained with a panel of GP pseudotyped vectors, including LASV, MACV and TCRV. As a control, we used vectors pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G), which confers a very broad tropism to retroviral vectors and is used to control for the transduction efficiency of retroviral vectors on all the cell lines tested. Unconcentrated supernatant stocks of the WWAV vectors produced a titer of approximately 2.5 × 104 transducing units per ml (Figure 2A), which is a similar to the titers we have previously observed for other clade B vectors (Oldenburg et al., 2007).
Figure 2. Titers of GP pseudotyped retroviral vectors on various cell lines.
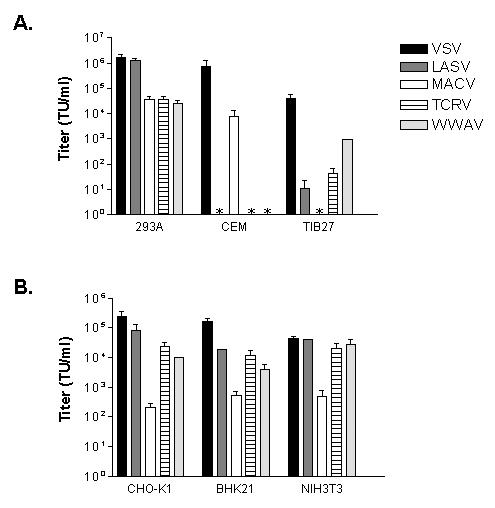
Titers were measured on (A) human kidney endothelial cells (293A, human T lymphocytes (CEM) and murine T lymphocytes (TIB27), and (B) rodent cell lines. Values shown are mean +/− SE for 2-8 independent experiments. All vector/cell combinations that gave no titer (*) were confirmed using 10 X concentrated stocks of vectors.
WWAV tropism on human and rodent cell lines
We have previously characterized the in vitro tropism of GPs from different arenaviruses by measuring their ability to transduce a panel of cell lines. The GPs were derived from the Old World viruses LASV and LCMV, and the New World clade B viruses JUNV, MACV, GTOV, TCRV and AMAV. These initial screens identified specific cell lines that revealed differences in the tropism of the different GPs. For example, we found that lymphocyte cell lines produced three distinct patterns of entry: LASV and LCMV vectors were unable to efficiently transduce either human or rodent lymphocytes, pathogenic clade B vectors such as MACV could transduce human CEM lymphocytes but not mouse TIB27 lymphocytes, while nonpathogenic clade B vectors such as TCRV could infect TIB27 but not CEM cells (Oldenburg et al., 2007). In addition, we observed differences in the relative efficiency with which different clade B vectors could transduce rodent cell lines (Oldenburg et al., 2007). Together these observations provided a tropism profile that could distinguish between the pathogenic and non-pathogenic clade B GPs.
We first examined the ability of WWAV pseudotyped vectors to transduce human CEM T lymphocytes and murine T1B27 T lymphocytes. We observed that they were unable to transduce CEM cells, even when concentrated stocks of vectors were used. In contrast, T1B27 cells were susceptible to the WWAV vectors (Figure 2A). Comparison to MACV and TCRV vectors, which are representative pathogenic and non-pathogenic clade B viruses respectively, revealed that the WWAV GP properties are more similar to the non-pathogenic clade B GP.
Next, we examined the ability of the WWAV vectors to transduce three different rodent cells lines, NIH 3T3, CHO-K1 and BHK21 cells, in comparison to VSV-G, LASV, MACV and TCRV GP vectors (Figure 2B). In agreement with the results from the lymphocyte studies, we found that the WWAV vectors were more similar to the TCRV vectors than the MACV vectors. Together, these data show that the WWAV GP has entry characteristics that are most similar to the non-pathogenic clade B GPs.
Low pH requirement for WWAV entry
Upon cell entry, some viruses require the acidic environment of the endosome in order to trigger virus-cell fusion. We have previously noted that within the clade B1 viruses, this requirement is virus- and cell-type specific (Oldenburg et al., 2007). Specifically, while all clade B1 GPs tested were sensitive to inhibition by NH4Cl on 293A cells, TCRV entry was markedly more sensitive than either JUNV or MACV entry when tested on K562 cells. We therefore examined the pH-sensitivity of WWAV entry using both 293A and K562 cells (Figure 3). The data revealed that the characteristics of WWAV entry are most similar to TCRV entry, being pH-sensitive on both cell lines.
Figure 3. NH4Cl sensitivity.
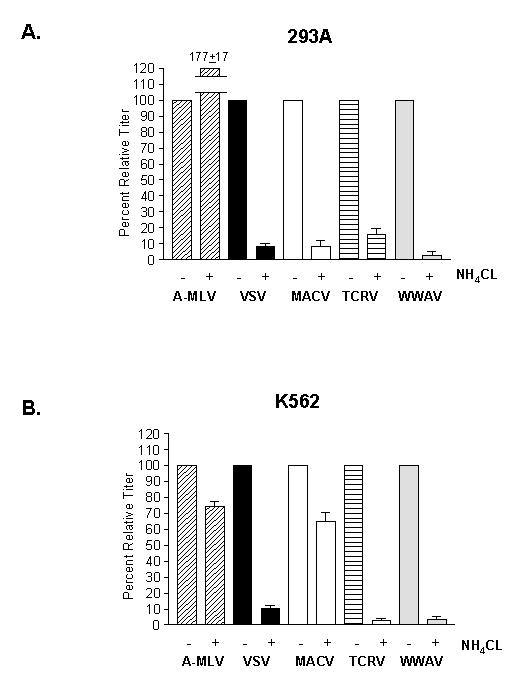
Effect of NH4Cl treatment on titer of indicated vectors on (A) 293A and (B) K562 cells. In each case, the titers were made relative to the values obtained in the absence of drug. Control vectors were pseudotyped with the amphotropic murine leukemia virus (A-MLV) Env, which is pH-independent and VSV-G, which is pH-dependent.
WWAV GP does not useα-DG
α-DG was the first receptor described for the arenavirus family, and can be utilized by LASV, certain strains of LCMV and clade C viruses (Cao et al., 1998; Kunz et al., 2005; Reignier et al., 2006; Spiropoulou et al., 2002). We have also shown that it is not required for entry by the clade B viruses, since both pathogenic and non-pathogenic GP pseudotyped vectors transduce equally well murine embryonic stem (mES) cells with and without DG (Reignier et al., 2006; Oldenburg et al., 2007). We therefore determined the titers of WWAV vectors on DG +/− and −/− mES cells. The WWAV vectors gave equivalent titers on the two cell lines when normalized to control VSV-G vectors, showing no requirement for α-DG for entry (Figure 4).
Figure 4. Effect of DG expression in murine ES cells on vector titers.
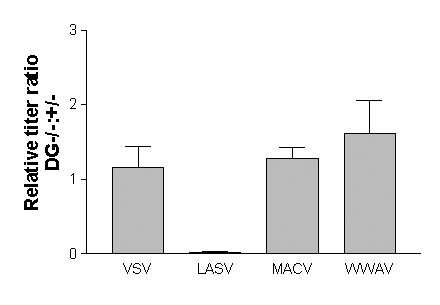
The titers of GP pseudotyped vectors were measured on DG −/− and +/− murine ES cells and the ratios determined. Values were normalized to the ratio obtained with VSV-G vectors, to control for any differences between the two cell lines in their ability to support retroviral vector transduction. LASV GP vectors were highly dependent on DG for entry, while MACV and WWAV GP vectors were not affected by the loss of DG. Values shown are mean +/− SE for 2-3 independent experiments.
Human transferrin receptor 1 use by WWAV GP
Human transferrin receptor 1 (hTfR1) was recently identified as a cellular receptor that can be used by clade B viruses to gain entry into human cells (Radoshitzky et al., 2007). Our subsequent studies have shown that hTfR1 can only be used by the pathogenic clade B GPs (JUNV, MACV and GTOV), but not by the non-pathogenic GPs from TCRV or AMAV (Flanagan et al., submitted for publication). Accordingly, the ability to use hTfR1 for entry can be considered to be a property that distinguishes the pathogenic and non-pathogenic clade B GPs.
We examined whether WWAV GP entry into cells utilized hTfR1. First, we compared the titers of the panel of pseudotyped vectors on CHO-K1 cells and TRVb-1 cells. TRVb-1 cells are CHO-K1 derivatives, which do not express functional endogenous TfR1, but stably express human TfR1 (McGraw et al., 1987). It has been shown that the presence of hTfR1 in CHO-K1 cells increases the titers of MACV vectors but not TCRV (Flanagan et al., submitted for publication; Radoshitzky et al., 2007).
We first confirmed the presence of hTfR1 on the surface of TRVb-1 cells by FACS analysis using an anti-CD71 antibody, which specifically recognizes the human form of TfR1 (Figure 5A). Examination of the titers of the vectors on the two cell lines confirmed that the MACV vector titers were increased by the presence of hTfR1 by approximately 2 orders of magnitude, while the VSV, LASV and TCRV vectors were not significantly different between the two cell lines (Figure 5B). This suggests that WWAV GP does not use hTfR1 as a receptor.
Figure 5. Effect of hTfR1 on vector titers on CHO-K1 cells.
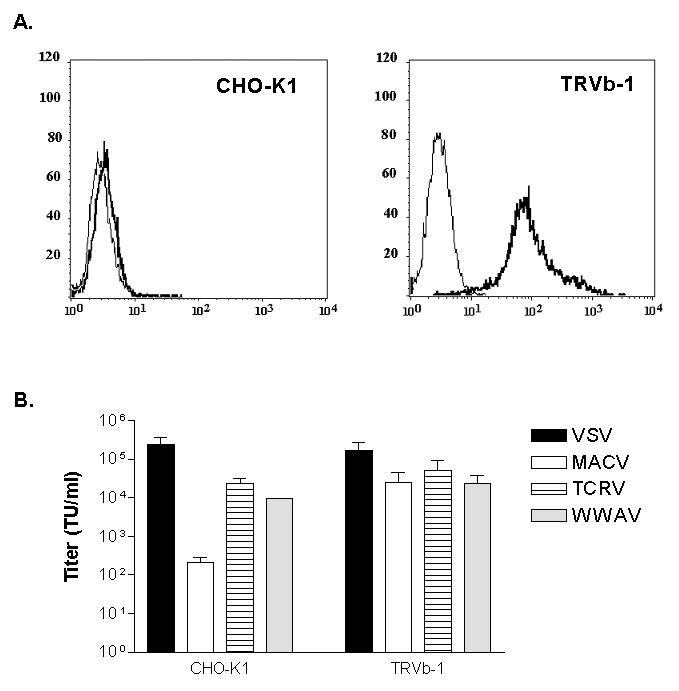
(A) Flow cytometric analysis of CHO-K1 and TRVb-1 cells was performed using an anti-hTfR1 antibody. TRVb-1 cells do not express endogenous CHO-K1 TfR1 but are stably transfected with hTfR1. Narrow lines represent secondary antibody only. (B) Comparison of vector titers on CHO-K1 versus TRVb-1 cells. Values shown are mean +/− SE for of 3-8 independent experiments.
To confirm this result, we performed antibody inhibition studies by pre-treating human 293A and TRVb-1 cells with an anti-hTfR1 antibody before adding pseudotyped vectors. We observed a significant reduction in titer for the MACV vectors on both cell lines, even at the lowest dose of antibody used (5 nM). In contrast, the titers of the VSV, TCRV and WWAV vectors were only slightly inhibited at the higher dose (50 nM) (Figure 6A, B). Finally, we transiently expressed hTfR1 in CHO-K1 cells and showed that only the titer of the MACV vectors was increased (Figure 6C). Together, these data show that the WWAV GP does not require hTfR1 to enter cells. Combined with the observations of tropism on the panel of human and rodent cell lines, we conclude that WWAV GP behaves more like the non-pathogenic members of clade B than the pathogenic members.
Figure 6. Role of hTfR1 in viral entry.
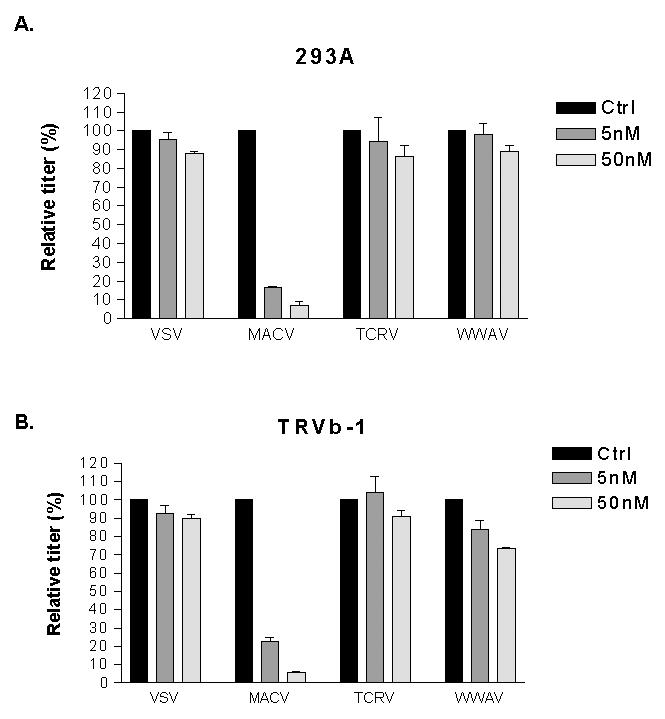
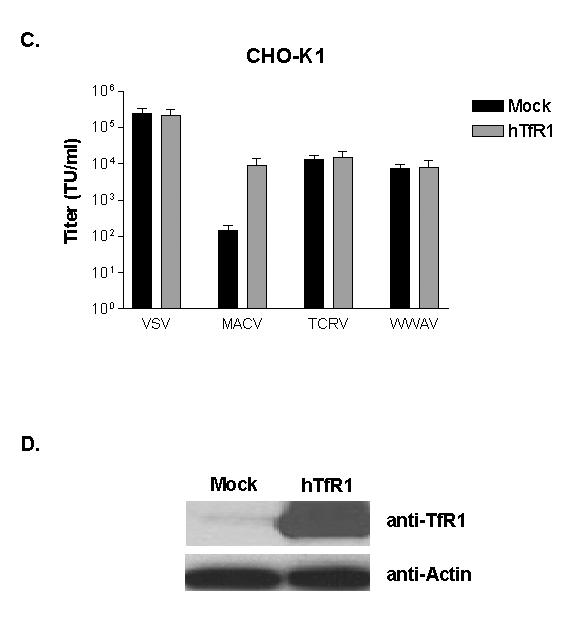
Human 293A (A) and rodent TRVb-1 (B) cells were pretreated with anti-hTfR1 antibody at two different doses, then challenged with the indicated pseudotyped vectors. Titers were converted to percent of control (titers obtained with no antibody pretreatment). Data are reported as mean ± SE of 2 independent experiments. (C) Titers of pseudotyped retroviral vectors on mock transfected CHO-K1 cells, or cells transiently transfected with an expression plasmid for hTfR1. Values shown are mean +/− SE for 3 independent experiments. (D) Western blotting confirming expression of hTfR1.
Discussion
The possibility that WWAV is a new, rodent-borne human pathogen was first suggested in 2000, when a report from the CDC speculated that the virus had caused three fatalities in California in 1999-2000. However, no follow-up information has appeared since that initial report, and there have been no further cases of human infection. Therefore, the status of WWAV as a human pathogen remains unclear.
WWAV is part of a distinct lineage in the New World arenaviruses that also includes BCNV and TAMV. Sequence analysis indicates a recombinant origin for these North American viruses, with the S strand of the virus containing clade A-like sequences in the NP gene, but clade B-like sequences in the GP gene. A proposed crossover point has been mapped to the 3' end of the GP gene (Charrel et al., 2001).
The fact that WWAV has a clade B-like GP is of interest because the clade B arenaviruses include four highly pathogenic viruses that cause severe hemorrhagic fevers in South America. In addition, three clade B viruses have been identified to date that are not associated with human disease. We have described marked differences in the properties of the GPs from pathogenic and non-pathogenic members of clade B that do not align with the phylogenetic relationships in this clade (B1, B2 and B3 viruses) but, instead, correlate with the ability of the viruses to infect humans. (Flanagan et al., submitted for publication). We speculate that these differences are likely due to differences in receptor use and entry pathways and, as such, will represent important determinants of in vivo pathogenicity.
Since WWAV has a clade B-like GP, we hypothesized that its human pathogenic potential could be revealed by an analysis of the characteristics of its GP. Using WWAV GP pseudotyped retroviral vectors, we analyzed several properties, including the ability to transduce human and rodent cell lines, the requirement for low pH during entry, and the ability of the GP to use hTfR1. The results of these analyses suggested that the WWAV GP has properties that are most similar to the non-pathogenic members of clade B. In addition, we found that as well as not requiring hTfR1 to enter cells, WWAV GP also did not use α–DG. Therefore, similar to the findings for TCRV and AMAV (Flanagan et al., submitted; Rojek et al, 2006), WWAV seems to use an additional, unidentified receptor.
WWAV was originally isolated in 1993 from Neotoma albigula white-throated woodrats from New Mexico (Fulhorst et al., 1996) and has now been found in seven Western states (Fulhorst et al., 1996). However, human infections attributed to a North American arenavirus have been limited to the 2000 CDC report. A survey of 1,094 sigmodontine and 112 murine rodents trapped in Southern California between 1995 and 1998 showed that antibodies to WWAV were found in N. fuscipes, N. lepida, Reithrodontomys megalotis, as well as four other species of Peromyscus rodents. In particular, P. maniculatus, the deer mouse, was found to carry WWAV-specific antibodies (Bennett et al., 2000). This may be significant for human health since this rodent is known to invade human habitations. However, only a very low incidence of seropositivity has been found amongst individuals who are in frequent contact with various wild rodents, with no reported cases of infection or death from arenaviruses in this vulnerable population (Fulhorst et al., 2007). Therefore, the likelihood of a widespread outbreak of an North American arenavirus infection in the general population seems to be small.
Although our data indicate that it is unlikely that WWAV is a human pathogen, we caution that an obvious limitation of any study using GP pseudotyped vector systems is that the pathogenicity of a virus will depend on factors other than just receptor use. Receptor binding and viral entry are just one part of the life cycle of a virus and the complex host-virus interactions that occur downstream of the entry step remain to be described for a complete understanding of the potential threat to human health that WWAV represents.
Material and methods
Cell lines
293T, NIH3T3, CHO-K1, CEM, TIB27, and BHK21 cells were obtained from the American Type Culture Collection and 293A cells were obtained from QBiogene (Irvine, CA). The cells were maintained in D10 medium: Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and 2 mM glutamine (Gemini Bio-Products,West Sacramento, CA) except CEM cells, which were maintained in R10 medium: RPMI (Mediatech) supplemented with 10% FBS and 2 mM glutamine. TRVb-1 cells (McGraw et al., 1987) were a gift from Dr. Timothy McGraw and were cultured in D10. Dystroglycan knockout (−/−) and heterozygous control (+/−) R1 murine embryonic stem cells (Williamson et al., 1997) were generously provided by Dr. Stefan Kunz (The Scripps Research Institute), and Dr. Kevin Campbell (University of Iowa), respectively, and cultured in DMEM supplemented with 20% FBS, 2 mM glutamine, 1 mM non-essential amino acids (Chemicon, Temecula, CA), 0.001% β-mercaptoethanol (Sigma) and 103 U/ml of murine leukemia inhibiting factor (ESGRO, Chemicon). All cells were maintained in 5% CO2, except TIB27 cells, which were maintained in 10% CO2.
Retroviral vector production and titer determination
Retroviral vectors displaying arenavirus GPs were produced by transient transfection of 293T cells using plasmid pCgp, which expresses murine leukemia virus Gag-Pol,, plasmid pMND-eGFP which is a retroviral vector genome expressing the enhanced green fluorescent protein (eGFP) reporter gene, and the appropriate GP expression plasmid, essentially as described (Reignier et al., 2006). The viral GPs that were used were derived from VSV, MACV (Carvallo strain, accession no. AY129248), LASV (Josiah strain, accession no. AY628203) and TCRV (Reignier et al., 2006). The GP from WWAV (strain AV 9310135, accession no AF228063) was chemically synthesized as a codon-optimized version for improved expression in human cell lines and was cloned into the expression vector pCAGGS (Niwa et al., 1991). Retroviral vector supernatants were harvested 48 h post-transfection filtered through a 0.45 μm filter (Millipore Corp., Bedford, MA), and aliquots were stored at −80 °C. Concentrated (10X) vector stocks were generated by ultrafiltration using Centricon Plus-20 30 kD MWCO columns (Millipore). The titer of the vectors on specific target cells was obtained by incubation of cells with vector stocks, followed by FACS analysis on a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ), for eGFP expression 48 hrs later. Titers were expressed as transducing units per ml (TU/ml), calculated by multiplying the percentage of eGFP positive cells in a sample by the number of cells present at the time of transduction, and taking into account any dilution factors.
pH-dependence assay
Five × 105 K562 or 293A cells were pre-treated for 1 hr with 50mM NH4Cl (Sigma), then vector supernatant containing 50mM NH4Cl was added to the cells and incubated for a further 3 hours. The cells were then washed with PBS and incubated in 2ml fresh D10 media for 48 hrs., when the vector titers were determined by FACS analysis as described above. Control transductions were performed in the absence of NH4Cl, and the percent titer for the drug treatment arm compared to the control cells was calculated. Control vectors were included that were pseudotyped with the pH sensitive glycoprotein from VSV, and the pH-insensitive amphotropic murine leukemia virus Env protein.
Human TfR1 cell surface expression
Five × 105 CHO-K1 orTRVb-1 cells were washed and incubated in phosphate buffered saline (PBS) containing 1% bovine serum albumin for 10 mins. The cells were kept on ice throughout the procedure. The cells were incubated with a 1:50 dilution of the mouse anti-human CD71 antibody (clone M-A712, BD Biosciences, San Jose, CA) for 30 minutes, washed, and incubated with a 1:200 dilution of fluorescein isothiocyanate FITC-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA) for 25 mins. The cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson) after a final PBS wash.
Transient transfection of hTfR1 and Western blot analysis
CHO-K1 cells were plated at 75% confluency in 10-cm plates, 16 hrs. before transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. Briefly, a standard transfection mixture containing 24 μg of a plasmid expressing hTfR1 and 60 μl of Lipofectamine in serum-free media was added to each plate, the cells were incubated for 4 hrs. and cultured overnight in D10. The following day, the cells were trypsinized and dispensed into 6-well plates at 40% confluency. The next day, the cells were transduced with vectors pseudotyped with test GPs, and titers determined 48 hrs. later, as described above.
A cell sample was collected at the time of transduction and analyzed by Western blot to determine the levels of expression of hTfR1. Cells were lysed in lysis buffer (20 mM Tris–HCl [pH 7.5], 1% Triton X-100, 0.05% sodium dodecyl sulfate [SDS] containing 5 mg/ml sodium deoxycholate, 150 mM NaCl and 1 mM phenylmethylsulfonyl fluoride [Sigma]) at 4 °C for 10 min, centrifuged in an Eppendorf microfuge at 16,000 × g for 10 min and the cleared supernatants were diluted 1:1 in 2 × SDS gel loading buffer (Biorad, Hercules, CA) plus 5% 2- mercaptoethanol, boiled for 10 min and electrophoresed in 8–16% polyacrylamide gels (Biorad). The proteins were transferred to an Immobilon P polyvinylidifluoride fluoride transfer membrane (Millipore Corp., Bedford, MA) and blocked overnight at 4 °C with blocking buffer (5% dried milk in PBST [PBS {pH 7.4}, 0.1% Tween 20]). HTfR1 was detected using the mouse anti-human Tfr1 antibody clone H68.4 (Invitrogen, Carlsbad, CA), diluted 1:500 in blocking buffer, followed by horseradish-peroxidase-conjugated goat anti-mouse IgG (1:10,000) (Pierce, Rockford, IL). Specific proteins were visualized using the enhanced chemiluminescence detection system (Amersham Biosciences Corp., Piscataway, NJ).
Anti-CD71 antibody pretreatment assay
293A or TRVb-1 cells were seeded in 12-well plates, 1 day prior to transduction. Cells were pretreated with 5 nM or 50 nM of mouse anti-human CD71 Ab (clone M-A712, BD Biosciences, San Jose, CA), or medium alone, for 30 min in a 37°C, 5% CO2 incubator. GP pseudotyped vectors were then added to the mixture for an additional 4 hrs. incubation. The antibody/vector mixtures were then replaced with fresh D10 media and the cells allowed to recover for 48 - 72 hrs. before FACS analysis to determine vector titers, as described above.
Acknowledgements
This work was supported by PHS grant 1U54 AI065359 to the Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (PMC) and a Saban Research Institute Career Development Award (TR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer AM, Rico-Hesse R. High genetic divergence and recombination in Arenaviruses from the Americas. Virology. 2002;304:274–81. doi: 10.1006/viro.2002.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SG, Milazzo ML, Webb JP, Jr, Fulhorst CF. Arenavirus antibody in rodents indigenous to coastal southern California. Am. J. Trop. Med. Hyg. 2000;62:626–30. doi: 10.4269/ajtmh.2000.62.626. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Wright R, Tseng B, Barton L, Marco E, Karacay B, Larsen PD. Congenital lymphocytic choriomeningitis virus infection: Spectrum of disease. Ann. Neurol. 2007 doi: 10.1002/ana.21161. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Nichol ST. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996;219:285–290. doi: 10.1006/viro.1996.0248. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol. Phylogenet. Evol. 1997;8:301–16. doi: 10.1006/mpev.1997.0436. [DOI] [PubMed] [Google Scholar]
- Cajimat MN, Milazzo ML, Hess BD, Rood MP, Fulhorst CF. Principal host relationships and evolutionary history of the North American arenaviruses. Virology. 2007 doi: 10.1016/j.virol.2007.05.031. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Tzianabos T, Lord RD, Coleman PH. Tamiami virus, a new member of the Tacaribe group. Am. J. Trop. Med. Hyg. 1970;19:520–6. doi: 10.4269/ajtmh.1970.19.520. [DOI] [PubMed] [Google Scholar]
- Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MBA. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Fatal illnesses associated with a New World arenavirus—California, 1999–2000. Morb. Mort. Wkly. Rep. 2000;49:709–711. [PubMed] [Google Scholar]
- Charrel RN, de Lamballerie X, Fulhorst CF. The Whitewater Arroyo virus: natural evidence for genetic recombination among Tacaribe serocomplex viruses (family Arenaviridae) Virology. 2001;283:161–6. doi: 10.1006/viro.2001.0874. [DOI] [PubMed] [Google Scholar]
- Charrel RN, Feldmann H, Fulhorst CF, Khelifa R, de Chesse R, de Lamballerie XK. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 2002;296:1118–1124. doi: 10.1016/s0006-291x(02)02053-3. [DOI] [PubMed] [Google Scholar]
- Charre l R.N., de Lamballerie X. Arenaviruses other than Lassa virus. Antiviral Res. 2003;57:89–100. doi: 10.1016/s0166-3542(02)00202-4. [DOI] [PubMed] [Google Scholar]
- Charrel RN, Lemasson JJ, Garbutt M, Khelifa R, De Micco P, Feldmann H, de Lamballerie X. New insights into the evolutionary relationships between arenaviruses provided by comparative analysis of small and large segment sequences. Virology. 2003;317:191–6. doi: 10.1016/j.virol.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Clegg JCS. In: The Arenaviridae. Salvato MS, editor. Plenum; New York: 1993. pp. 175–187. [Google Scholar]
- Downs WG, Anderson CR, Spence L, Aitken THG, Greenhall AM. Tacaribe virus, a new agent isolated from Artibeus bats and mosquitoes in Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1963;12:640–646. doi: 10.4269/ajtmh.1963.12.640. [DOI] [PubMed] [Google Scholar]
- Enserink M. Emerging diseases. New arenavirus blamed for recent deaths in California. Science. 2000;289:842–3. doi: 10.1126/science.289.5481.842. [DOI] [PubMed] [Google Scholar]
- Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A, Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR. LCMV in Transplant Recipients Investigation Team. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 2006;354:2235–49. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- Flanagan ML, Oldenburg J, Reignier T, Hamilton GA, Martin VK, Cannon PM. New World clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and independent entry pathways, but human pathogenicity correlates with use of human TfR1. J. Virol. 2007 doi: 10.1128/JVI.01397-07. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF, Bowen MD, Ksiazek TG, Rollin PE, Nichol ST, Kosoy MY, Peters CJ. Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology. 1996;224:114–20. doi: 10.1006/viro.1996.0512. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Bennett SG, Milazzo ML, Murray HL, Jr, Webb JP, Jr, Cajimat MN, Bradley RD. Bear Canyon virus: an arenavirus naturally associated with the California mouse (Peromyscus californicus) Emerg. Infect. Dis. 2002;8:717–21. doi: 10.3201/eid0807.010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF, Milazzo ML, Armstrong LR, Childs JE, Rollin PE, Khabbaz R, Peters CJ, Ksiazek G. Hantavirus and arenavirus antibodies in persons with occupational rodent exposure. Emerg. Infect. Dis. 2007;13:532–8. doi: 10.3201/eid1304.061509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Jahrling PB. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10(12 Suppl):S110–121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ, Kourtis AP, Bell M, Rasmussen SA. Lymphocytic choriomeningitis virus: an emerging obstetric pathogen? Am. J. Obstet. Gynecol. 2006;194:1532–6. doi: 10.1016/j.ajog.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Jennings WL, Lewis AL, Sather GE, Pierce LV, Bond JO. Tamiami virus in the Tampa Bay area. Am. J. Trop. Med. Hyg. 1970;19:527–36. doi: 10.4269/ajtmh.1970.19.527. [DOI] [PubMed] [Google Scholar]
- Kunz S, Borrow P, Oldstone MBA. Receptor structure, binding, and cell entry of arenaviruses. Curr. Top. Microbiol. Immunol. 2002;262:111–37. doi: 10.1007/978-3-642-56029-3_5. [DOI] [PubMed] [Google Scholar]
- Kunz S, Sevilla N, Rojek JM, Oldstone MBA. Use of alternative receptors different than α-dystroglycan by selected isolates of lymphocytic choriomeningitis virus. Virology. 2004;325:432–445. doi: 10.1016/j.virol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Kunz S, Rojek JM, Kanagawa M, Spiropoulou CF, Barresi R, Campbell KP, Oldstone MBA. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J. Virol. 2005;79:14282–14296. doi: 10.1128/JVI.79.22.14282-14296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 1987;155:437–44. doi: 10.1093/infdis/155.3.437. [DOI] [PubMed] [Google Scholar]
- McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J. Cell Biol. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Oldenburg J, Reignier T, Flanagan ML, Hamilton GA, Cannon PM. Differences in tropism and pH dependence for glycoproteins from the Clade B1 arenaviruses: implications for receptor usage and pathogenicity. Virology. 2007;364:132–9. doi: 10.1016/j.virol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters CJ, Buchmeier M, Rollin PE, Ksiazek TG. Arenaviruses. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman R, Straus SE, editors. Fields Virology. 3rd ed. Lippincott-Raven Publishers; Philadelphia, PA: 1996. pp. 1521–1551. [Google Scholar]
- Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn H, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–6. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reignier T, Oldenburg J, Noble B, Lamb E, Romanowski V, Buchmeier MJ, Cannon PM. Receptor use by pathogenic arenaviruses. Virology. 2006;353:111–120. doi: 10.1016/j.virol.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Rojek JM, Spiropoulou CF, Kunz S. Characterization of the cellular receptors for the South American hemorrhagic fever viruses Junin, Guanarito, and Machupo. Virology. 2006;349:476–491. doi: 10.1016/j.virol.2006.02.033. [DOI] [PubMed] [Google Scholar]
- Sheinbergas MM. Hydrocephalus due to prenatal infection with the lymphocytic choriomeningitis virus. Infection. 1976;4:185–91. doi: 10.1007/BF01638922. [DOI] [PubMed] [Google Scholar]
- Smelt SC, Borrow P, Kunz S, Cao W, Tishon A, Lewicki H, Campbell KP, Oldstone MBA. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J. Virol. 2001;75:448–57. doi: 10.1128/JVI.75.1.448-457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulou CF, Kunz S, Rollin PE, Campbell KP, Oldstone MBA. New World arenaviruses clade C, but not clade A and B viruses, utilizes α-dystroglycan as its major receptor. J. Virol. 2002;76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RA, Henry MD, Daniels KJ, Hrstka RF, Lee JC, Sunada Y, Ibraghimov-Beskrovnaya O, Campbell KP. Dystroglycan is essential for early embryonic development: disruption of Reichert's membrane in Dag1-null mice. Hum. Mol. Genet. 1997;6:831–841. doi: 10.1093/hmg/6.6.831. [DOI] [PubMed] [Google Scholar]


