Abstract
Fibroblast and macrophage are two dominant cell types respond cooperatively to degrade implanted biomaterials. Using an electrospun Dextran/Poly-lactide-co-glycolide (PLGA) scaffold as a model, an in vitro fibroblast/macrophage co-culture system was developed to investigate the degradability of implantable biodegradable materials. SEM showed that both fibroblasts and macrophages were able to degrade the scaffold, separately or cooperatively. Under the synergistic coordination of macrophages and fibroblasts, scaffolds showed faster degradation rate than their counterparts incubated with a single type of cells as well as in PBS or cell culture medium. Lysozyme, non-specific esterase (NSE), gelatinase, hyaluronidase-1 and α-glucosidase were upregulated in the presence of the scaffold, suggesting their roles in the cell-mediated scaffold degradation. In addition, the expressions of cell surface receptors CD204 and Toll like receptor 4 (TLR4) were elevated one week after cell seeding, implying that these receptors might be involved in scaffold degradation. The results of in vivo subdermal implantation of the scaffold further confirmed the biodegradability of the Dextran/PLGA scaffold. The fibroblast/macrophage co-culture model adequately mimicked the in vivo environment and could be further developed into an in vitro tool for initial biomaterial evaluation.
Keywords: Fibroblasts, Macrophages, Co-culture, Biodegradation, Enzyme, Receptor
1. Introduction
Biomaterials are recognized as foreign and post-implant events involve their integration and/or elimination followed by tissue reconstitution at the implant sites [1]. Ideally, the degradation rates of implants should be engineered to last the intended span of efficacy and to synchronize with the pace of tissue regeneration. In particular, the degradation profile of biomaterials serving as drug delivery vehicles should be controlled precisely for optimal efficacy. Thus, a thorough understanding of material degradation in the biological environment is prudent to their utilization in vivo.
Most in vitro biomaterial degradation studies have been performed in phosphate buffered saline (PBS) [2], enzyme solutions [3], or even non-physiological and very harsh conditions, such as papain and NaOH at 150°C [4]. In contrast, cell mediated material degradation bears a greater resemblance to the physiological environment, which has been exemplified by a recent study demonstrating the importance of intimate biomaterial-cell contact in the degradation of polyethylene carbonate (PEC) [5]. Cells secrete potent hydrolytic and oxidizing agents to mediate or accelerate bond cleavage of materials [6]. Besides, they deposit extracellular matrix (ECM) on biomaterials and the ECM turnover invokes further production of enzymes to accelerate material degradation [7]. The continuous stresses applied by attached cells also hasten polymer degradation [6]. More importantly, different cell types function synergically to modulate the local environments [8], which increase the complexity to material degradation.
Almost all implanted biomaterials are enclosed by collagenous capsules with fibroblasts and macrophages being the two dominant cell types residing inside. They both produce a variety of hydrolases [6] which might degrade materials efficiently. As professional and non-professional phagocytes, respectively, macrophages and fibroblasts are involved in breaking down of many natural macromolecules [9, 10] and could ingest the degraded fragments [6]. In addition, they also produce superoxide anions [11] that could be transformed to more potent oxidants capable of initiating homolytic reactions on polymers [6]. Macrophages have been used to investigate the degradation of many biomaterials such as polyurethanes [12]. However, the effects of fibroblasts on biomaterial degradation are under-explored, which could be due to the partial understanding on fibroblast hydrolases, especially those connecting to synthetic polymer degradation.
In this investigation, using an electrospun Dextran/PLGA scaffold [13] as a model material, we established a fibroblast/macrophage co-culture model to emulate the in vivo environment in order to study the behavior and mechanisms of material degradation. Accordingly, the effects of cells on scaffold morphology, its dry weight lost and changes in medium pH were monitored. The expressions of putative receptors for recognition/clearance of the degraded scaffold were determined by real-time PCR. The activities of some major hydrolytic enzymes during the scaffold degradation were discerned. This co-culture system was further validated by correlating the results with in vivo implanted scaffolds in a mouse subdermal model.
2. Materials and methods
Metharylated Dextran/PLGA scaffolds were prepared following the methodology established by us [13].
2.1 Macrophage/Fibroblast co-culture model
RAW 264.7 macrophages (ATCC, Manassas, VA, USA) were maintained in DMEM (Gibco Grand Island, NY, USA) supplemented with inactivated fetal bovine serum (Hyclone, Logan, UT, USA) and 1% Pen/Strep (Gibco Grand Island, NY, USA). M.DUNNI mouse dermal fibroblasts (CRL-2017) (ATCC, Manassas, VA, USA) were cultured in DMEM with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 1% Pen-Strep. Passages two to twenty were used.
Scaffolds (~0.8 cm2) were rinsed in ethanol and deposited in 48-well plates. Equal numbers of fibroblasts and macrophages were seeded (2,000 cells, each) on the scaffolds. The controls included co-cultured cells (2,000 cells, each) without scaffolds; macrophages and fibroblasts, individually (2,000 cells, each) with and without scaffolds. All samples (n=8) and controls (n=8) were incubated in DMEM with inactivated fetal bovine serum and the media were changed every two days. Samples were collected at day 3, 7, and 21, respectively, for enzyme activity assays and receptor studies. Two days before sample collection, phenol red-free media were used for all fluorescent signal assays and serum-free media were used for all enzyme and receptor studies, respectively. For quantitative results, data were normalized with cell numbers or with total protein concentrations determined by BCA assay (Pierce, Rockford, IL, USA). Images were acquired with an inverted light microscope (Axiovert 200M, Zeiss, Munich, Germany) and analyzed by Axiovision 4 imaging software (Zeiss, Jena, Germany).
2.2 Scaffold morphological change and dry weight lost
Cell-laden scaffolds were retrieved at weeks 4 and 8, respectively, rinsed twice in PBS, and incubated with trypsin solution at 37 °C under agitation for 10 min. After further agitation on a vortexer, the detached cells were removed. Subsequently, the scaffolds were incubated in proteinase K solution (0.5 mg/mL) (Promega, Madison, WI, USA) for 15 min to remove all cell-secreted proteins and the proteins in cell culture media adsorbed to the scaffold fibers. Pristine scaffolds subjected to the same treatments were used as controls. Scaffolds were rinsed in water extensively and air dried. The dry weights lost of the degraded scaffolds were monitored and their morphological changes (both the surface and interior) were examined under a scanning electron microscope (LEO/Zeiss 1550, Zeiss, Munich, Germany) following a method described by us [13]. For comparison, scaffolds incubated in PBS and cell culture medium under the same incubation conditions, respectively, were used as controls.
2.3 pH value variation
Cell culture media were collected from each sample group (n=20) at day 3 and day 21, respectively, after seeding and their pH values were measured by a micro pH electrode (Lazar, Los Angeles, CA). The pH value of pure cell culture media incubated under the same conditions as other samples was set as the baseline (arbitrarily set as 0). All data were validated with the pH decline of the corresponding scaffold-free controls.
2.4 Enzyme activity assays
Non-specific esterase
The experiment was performed utilizing a modified manufacturer’s protocol for α-Naphthyl Acetate Non-Specific Esterase assay (Sigma, St. Louis, MO, USA). Briefly, 1 ml of sodium nitrite was mixed with 1 ml of fast blue BB base solution, and the admixture was blended with 40 ml of pre-warmed water (37°C). Thereafter, 5 ml of TRIZMAL buffer and 1 ml of naphthyl acetate solution were added. Formaldehyde pre-fixed samples were immersed in the reagent mixture and incubated for 30 min at 37°C in dark. The absorbances of all supernatants at 540 nm were recorded. The stained specimens were evaluated semi-quantitatively under a microscope. The color intensity of stained samples was scored by another observer and numerical rating of 1 to 5 was assigned according to the following criteria: 1 = negative, 2 = sporadic detection, 3 = sparse but consistent, 4 = uniformly present and 5 = intense and widespread.
Lysozyme
Lysozyme activities were determined with EnzChek® Lysozyme Assay Kit (Molecular Probes; Eugene, OR, USA). Collected media were diluted in 50 μL of assay buffer and were mixed with 50 μL of fluorescein-conjugated Micrococcus lysodeikticus for 30 min at 37°C. The fluorescence intensities were measured at 494/518 nm and the lysozyme activity of experimental samples were determined from the standard curve.
α-glucosidase
10 μL of cell culture supernatant was mixed with 100 μL of 6 mM 4-methylumbelliferyl-alpha-D-glucoside (Sigma, St. Louis, MO, USA) (in 0.1 M citric acid, 0.2 M Na2HPO4, pH6.0) and incubated at 37°C for 1h. The reaction was terminated with 2 mL of glycine buffer (0.2 M, pH 10.5) and the fluorescent intensity was recorded by a cytofluor (Perspective Biosystems, Framingham, MA, USA) at 365/445 nm [14].
β-glucosidase
After the samples were rinsed with 80 μL of PBS, 80 μL of acetate buffer (0.2 M, pH 4) were added to each well. Then, 100 μL of 6 mM 4-methylumbelliferyl-beta-D-glucoside (Sigma, St. Louis, MO, USA) were incubated with each sample as a substrate at 37°C for 1 hour. The assay was terminated by adding 2 mL of glycine buffer (0.2 M, pH 10.5) and the fluorescent signal was measured at 365/445 nm [15].
Collagenases
Detection of total collagenases/gelatinases activities with an EnzChek® Gelatinase/Collagenase Assay Kit (Molecular Probes; Eugene, OR, USA) was performed according to manufacturer’s instructions. In brief, cell culture supernatants prepared in the substrate buffer at different dilutions (100 μL), were incubated with 20 μL of fluorescent-labeled gelatin substrate in 80 μL of buffer for 24 hours at room temperature in dark. The fluorescent intensity was quantified at 495/515 nm. The enzyme activities in the samples were determined by comparing them with the activities of collagenase standards.
Hyaluronidases
0.5 mL of potassium hyaluronate (4 mg/mL in PBS, pH 5.3) was added to 0.3 mL of cell culture supernatant and incubated for 30 min at room temperature. The reactions were stopped by 5 mL of acid albumin (pH 3.75) at 37 °C for 10min. Reduction in liquid turbidity at 600 nm was measured and the enzyme activity was calculated by referencing a hyaluronidase standard plot (Sigma, St. Louis, MO, USA) [16].
2.5 Real-time PCR
Cells were re-suspended in cell lysis buffer (Stratagen, La Jolla, CA, USA). Real-time PCR was performed with a LightCycler (Roche, Mannheim, Germany) using the Brilliant® 2 step QRT-PCR Kit (Stratagen, La Jolla, CA, USA). PCR primers were summarized in Table 1. At least three replicates were performed on each sample and the housekeeping gene GAPDH was used as a reference. Scaffold-free samples were used to normalize gene expressions. The relative gene expression ratios were analyzed with the software REST©. Expression ratios above zero were considered as up-regulated, while those below zero as down-regulated. If the gene expression was unaltered, the ratio should be 1 or −1. Any change in ratio > 2 or < −2 was regarded as regulated expression [13].
Table 1.
Real-time PCR primer sequences.
| Gene | Access number | Forward primer sequence | Reverse primer sequence |
|---|---|---|---|
| Alpha-glucosidase | NM_008064 | 5′ CTCCTACCCAGGTCCTTTCC 3′ | 5′ ACAGCTCTCCCATCAGCAGT 3′ |
| Beta-glucosidase | NM_172692 | 5′ ACCCTGGAATGTACCAGCAC 3′ | 5′ GCTCCAAGGACAGAACTTGC 3′ |
| Hyaluronidase-1 | BC021636 | 5′ CATGCACTGGCTTAGATCA 3′ | 5′ GGATGCCGTCTATGTCGTCT 3′ |
| Gelatinase | D12712 | 5′ AATTGGGCACCTACCCCTAC 3′ | 5′ TCCTGGAATGTGTGAGCAAG 3′ |
| TLR-4 | NM_021297 | 5′ TTCTTCTCCTGCCTGACACC 3′ | 5′ TGTCATCAGGGACTTTGCTG 3′ |
| CD204 | AF203781 | 5′ GACGCTTCCAGAATTTCAGC 3′ | 5′ CCAGTGAATTCCCATGTTCC 3′ |
| MARCO | NM_010766 | 5′ AGGGAGACAAAGGGGACCTA 3′ | 5′ CTGGTTTTCCAGCATCACCT 3′ |
| CD 44 | NM_009851.2 | 5′ CGTCCAACACCTCCCACTAT 3′ | 5′ TCCATCGAAGGAATTGGGTA 3′ |
| Endo180 | NM_008626 | 5′ GTCTGGCCAGCTATGAGGAG 3′ | 5′ CTAGGGTCTCTGCGGTTCAG 3′ |
|
| |||
| GAPDH | BC083080 | 5′ ACCAACTG TTAGCCC 3′ | 5′ CTTCCCGTTCAGCTCT 3′ |
2.6 In vivo evaluation with a mice subdermal implant model
Male mice (25–30 g) (n=4) were anesthetized with isoflurane (2–4%) and a small dorsal incision was created. After being pre-sterilized by ethanol, scaffolds were placed into the pockets and the incisions were sutured. All animals received humane care in compliance with a protocol approved by the SUNY-Stony Brook University IACUC (protocol number 2006-1286). Animals were euthanized by CO2 at 3, 7, and 21 days post-surgery and the tissues were sectioned, stained with H&E. Likewise, explanted scaffolds were processed and their morphological changes were evaluated by SEM.
2.7 Statistics
All experimental results were presented as mean ± standard deviation. Whenever relevant, Student’s t-test was used to discern the statistical difference between groups. The significant level was set as p<0.05.
3. Results and discussion
3.1 Scaffold morphological change and dry weight lost
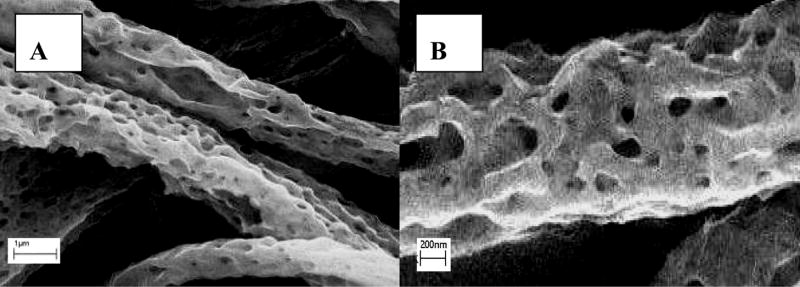
Scaffold morphological changes were monitored by SEM over 8 weeks (Fig. 1). Comparing with the smooth fibers of the pristine scaffold (Fig. 1J), the scaffolds incubated with cells for 4 weeks exhibited abundance of pores and an increase in fiber surface roughness signifying material loss (Fig. 1A: fibroblasts, 1C: macrophages, 1E: co-culture). Moreover, the fibers on the scaffold surface as well as its interior (Fig. 1I: fibroblasts) showed similar appearance suggesting the uniform degradation pattern. The interior of scaffolds incubated with macrophages and co-cultures, respectively, showed patterns (not shown) similar to those incubated with fibroblasts. Since PLGA is more resistant to degradation than dextran, it could be inferred that the pores on the fibers were caused by preferential degradation of dextran with the remnant composed mostly of PLGA. Scaffold degradation inevitably led to some extent of loss of crosslinking contributing to scaffold structural stabilization, which could explain the slight shrinkage (Fig. 1I). In contrast, after incubated in PBS for 4 weeks, the scaffolds showed very moderate change in morphology (Fig. 1H) (scaffold incubated in culture medium showed identical morphology, not shown), suggesting that the degradation rate in PBS was slower, and PLGA is known to undergo auto-hydrolysis in aqueous environment [6]. Initial breakdown of scaffold fibers resulted in the creation of surface defects leading to entrance of water, salts and enzymes into the bulk, thereby, hastening their degradation [6]. After eight weeks, the fiber pore sizes increased considerably due to further erosion of the bulk (Fig. 1B, D and F), suggesting an exponential nature of scaffold degradation.
Figure 1.
SEM of degraded Dextran/PLGA scaffolds. (A) 4 weeks, and (B) 8 weeks after seeding fibroblasts; (C) 4 weeks, and (D) 8 weeks after seeding macrophages; (E) 4 weeks, and (F) 8 weeks after seeding macrophages and fibroblasts; (G) a typical cell-mediated degradation sample at a higher magnification after 4 weeks, and (H) a PBS-mediated degradation control after 4 weeks. (I) a cross-section of fibroblast-mediated scaffold sample after 4 weeks; (J) pristine scaffold (scale bar: 8.569μm).
The results demonstrated that both fibroblasts and macrophages were capable of degrading the scaffolds. Due to the phagocytic nature of macrophages and the broader arrays of enzymes and oxidants they produced, macrophages had greater capacity in degrading the scaffold than fibroblasts, which was exemplified by the general disparity in both the sizes and distribution of pores on the scaffold fibers in concert with the fiber diameters (Fig. 1A and C vs. Fig. 1B and D). Fibroblasts secrete many hydrolases and function as non-professional phagocytes [9, 10]. However, to our knowledge, it has not been reported that fibroblasts are capable of digesting synthetic biomaterials, particularly, fibrous electrospun scaffolds. It was previously demonstrated by us that fibroblast could produce and deposit ECM inside the Dextran/PLGA scaffold [13]. The turnover of ECM could evoke production of enzymes thus, accelerating scaffold degradation. Attached fibroblasts induced scaffold contraction by exerting stress on the fibers [13], further hastening scaffold degradation. The patterns of fibroblast and macrophage mediated scaffold degradation were comparable (Fig. 1 A–D), suggesting that the two cell types degraded the scaffold through similar mechanisms. Fibroblasts and macrophages act synergistically in degrading scaffold. 8 weeks after cell seeding (Fig. 1F), the scaffold had generally lost its fibrous network structure with an obvious decrease in pore sizes and the diameters of remnant fiber strands were smaller. The scaffolds incubated in the co-culture samples lost approximately 75% of their original dry mass with increase in brittleness, whereas the dry weight loss of scaffold in PBS was about 40%. The higher rates of cell-mediated scaffold degradation further underscored the roles of cells in scaffold degradation.
3.2 pH value drop in the cell culture media
PLGA degrades into lactic acid (pKa 3.86) and glycolic acid (pKa 3.83) [6]. Altered local pH could further accelerate the polymer degradation [8]. Therefore, it is important to determine the extent of pH drop that could potentially be induced by the degradation of the scaffold. In the presence of scaffold, 3 days after cell seeding (Fig. 2), the relative pH drop in the co-culture sample was 0.061±0.004, whereas the relative pH drop of the samples cultured with macrophages and with fibroblasts alone were 0.035±0.003 and 0.009±0.002, respectively. Three weeks after cell seeding, in the presence of scaffold, the pH drop in the co-culture sample was 0.129±0.003, while in the samples cultured with macrophages or fibroblasts alone were, 0.286±0.001 and 0.112±0.002, respectively. Evidently, macrophages played a dominant role in scaffold degradation, with fibroblasts playing a contributory role. The significant drops in pH at week 3 appeared to be a good reflection of the exponential increase of scaffold degradation. At a later stage, the magnitude of pH drop detected in the co-culture samples was lower than that of its counterpart cultured with macrophages only, suggesting that fibroblasts and macrophages interacted to buffer the local environment, thereby contributing to the maintenance of pH stability. The modest decline in pH in the presence of cells would less likely induce unfavorable effects on cells. In a parallel study, we also showed that the scaffold was very biocompatible and did not influence cell viabilities [17]. The results here also suggested that the pH drop was not the leading mechanism of scaffold degradation.
Figure 2.

pH value changes under the influence of scaffold as compared with scaffold-free controls. M: macrophages; F: fibroblasts; S: scaffold.
3.3 Assessment of enzyme activities
Turnovers of natural materials by substrate-specific enzymes have been well characterized. On the contrary, since most synthetic polymers are not specific substrates for natural enzymes, the mechanisms of enzyme-mediated polymer degradation are likely to be complex. Limited attention has been devoted to discerning the activities of enzymes on polymer degradation [18, 19] and many other related issues have yet to be explored. It was previously reported that the activities of various capsule-borne enzymes were elevated in a rat subdermal model two weeks after material implantation [20]. However, abnormal expressions of many oxidative and hydrolytic enzymes are associated with diseases. Therefore, understanding of enzyme-mediated degradation of materials has important implications in their accurate applications. Production of H2O2 was elevated when macrophages were exposed to the scaffold [17], suggesting the involvement of reactive oxygen species. In the current investigation, we set out to identify some of the hydrolytic enzymes including non-specific esterase (NSE), lysozyme, collagenases, hyaluronidase, α-glucosidase and β-glucosidase, in scaffold degradation.
It was previously reported that cholesterol esterase and carboxyl esterase activities increased during long term culture of macrophages with polycarbonate urethane [18, 19]. As the structure of PLGA is also abundant in ester bonds, it could be postulated that macrophages are capable of mediating degradation of Dextran/PLGA scaffolds through a comparable mechanism. Therefore, NSE was selected as a hydrolytic enzyme to investigate scaffold degradation (Fig. 3). In the presence of scaffold, NSE activity was elevated in the co-culture samples as well as in the macrophage samples (Fig. 3A and C), with the latter exhibited the highest activity (Fig. 3C and H). The interface of the macrophage-laden scaffold and the culture dish revealed higher NSE activity in the scaffold (Fig. 3G), which was a contrast to the weak NSE activity of on the culture dish. Although no NSE activity was detected from the fibroblasts due to the lack of expression, fibroblasts played a role in modulating macrophage NSE activities.
Figure 3.

Activities of NSE in the presence of Dextran/PLGA scaffold at week 1. (A, C, E) cells in the scaffolds; (B, D, F) cells on the culture dish; (A–B) macrophages/fibroblasts co-culture; (C–D) macrophages; (E–F) fibroblasts; (G) the interface of scaffold and the culture dish; S: scaffold; P: plastic culture dish; arrow: macrophages with higher NSE activities; (H) Scores for NSE activities; M: macrophages; F: fibroblasts; S: scaffold.
Bacterial derived dextran is naturally susceptible to lysozyme degradation [21]. In addition, lysozyme could degrade many other natural and synthetic polymers, including chitosan [22], poly-(HEMA) [23], polyesters [24], etc. Dextran/PLGA scaffold-prompted macrophage production of lysozyme was examined (Fig. 4). Three day after cell seeding, there was an increase in lysozyme activity in both the co-culture sample and macrophage sample incubated with the scaffolds. Lysozyme activities increased noticeably at day 7 and further elevations were detected by day 21. At day 3 and 7, samples with macrophages showed higher lysozyme activities than their co-culture counterparts, whereas, the lysozyme activities of the co-culture samples were higher than those incubated with the macrophages alone at day 21. The results suggested that even though fibroblasts did not produce lysozyme, they played a role in regulating its activity produced by macrophages. It is known that lysozyme production is up-regulated when macrophages are activated. The results here were in good agreement with the finding in a parallel study indicating that more macrophages were activated in the co-culture samples than in the samples cultured with macrophages only at a later stage of culture [17].
Figure 4.
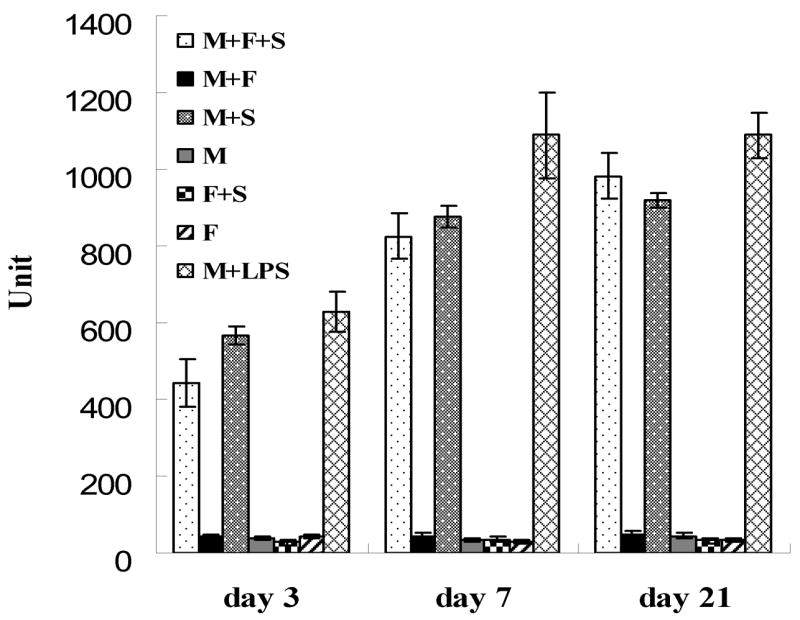
Lysozyme activities under the influence of Dextran/PLGA scaffold. M: macrophages; F: fibroblasts; S: scaffold.
α-glucosidase and β-glucosidase are lysosomal enzymes with important roles in carbohydrate metabolism [25]. Dextran could be metabolized into glucose, and thus is a potential substrate for these two enzymes. Hyaluronidases and collagenases degrade hyaluronan and collagen, respectively. However, they are also known to non-specifically degrade other substrates [26, 27]. Collectively, these four enzymes are produced by both fibroblasts and macrophages. Due to the limited sensitivities of the substrate-specific assays, hyaluronidases, α-glucosidase and β-glucosidase were not detectable in all samples (data not shown). Therefore, real-time PCR, a more sensitive method, was used to detect the differential expressions of α-glucosidase, β-glucosidase, hyaluronidase-1 and gelatinase (MMP9) in the presence of scaffold at the mRNA level (Fig. 5A). With the exception of β-glucosidase, all other enzymes were up-regulated throughout the entire culture span, strongly suggesting their roles in scaffold degradation. In addition, the expressions of α-glucosidase, hyaluronidase-1 and gelatinase were elevated in fibroblasts and macrophages, separately or co-cultured, which further confirmed that both fibroblasts and macrophages were capable of degrading the scaffold. Moreover, the expression levels of these enzymes in the co-culture samples were not simply the arithmetical sum of those observed in the samples cultured with either fibroblasts or macrophages, implicating the interactions of the two cell types. However, the total activities of all collagenases, measured by using fluorescent-labeled gelatin, did not show any significant increase (data not shown). Our previous study [13] showed that MMP-1 was not regulated in the presence of scaffold, which could be inferred as not all collagenases were pertinent to scaffold degradation. Collectively, lysozyme, NSE, α-glucosidase, hyaluronidase-1 as well as gelatinase were up-regulated during scaffold degradation despite both PLGA and dextran were not specific substrates for these enzymes. It is highly probable that there are other cell-secreted hydrolases capable of degrading the scaffold non-specifically.
Figure 5.
Relative quantitative real-time PCR analyses of gene expressions. (A): enzymes; (B): receptors; M: macrophages; F: fibroblasts; S: scaffold.
3.4 Receptors involved in scaffold degradation
Cell surface receptors are specific to their diverse natural ligands [28]. Macrophages and fibroblasts can interact with various biomaterials [29, 30], but the mechanisms of recognizing and internalizing degraded synthetic biomaterials are incompletely understood. Understanding this process is important for designing future biomaterials for applications such as drug delivery systems targeting specific receptors [31]. Hitherto, most related studies have been focused on inhalable natural particles [32, 33] and metallic bone substitutes [34, 35]. In our investigation, we attempted to screen for some of the receptors that were previously shown to associate with biomaterial degradation and might be involved in scaffold degradation. The expressions of putative receptors: TLR4, macrophage receptor with collagenous structure (MARCO), CD204, CD44 and uPARAP/Endo180 were detected with real-time PCR. TLR4 is mainly a receptor for LPS but also associates with biomaterial activation of macrophages [36]. MARCO is a major receptor for unopsonized particles and is known to mediate silica uptake [37, 38]. CD204 is a variant of the class A scavenger receptor capable of uptaking titanium dioxide, silica, diesel particles and latex beads [37, 38]. CD44 is a major receptor that binds to degraded hyaluronan, collagen as well as fibronectin [39]. uPARAP/Endo180 is an essential receptor for collagen uptake and degradation [40].
Figure 5B showed that none of the receptors of interest were regulated 3 days after cell seeding. However, the expressions of CD204 and TLR4 were obviously higher after 1 and 3 weeks. Moreover, there was no change in the expression of MARCO, CD44, uPARAP/Endo180 throughout the entire culture span. The regulated receptors were mainly macrophage receptors, indicating the dominant role of macrophages in scaffold degradation. In contrast, none of the tested fibroblasts receptors were regulated. Therefore, the scaffold degradation/clearance by fibroblast could be mediated by other mechanisms. The magnitudes of CD204 and TLR4 expression in macrophages could not be fully accounted for their activities in the co-culture samples, which implied interaction of the two cell types during the scaffold degradation. Up-regulation of TLR4 was consistent with the observation that the scaffold could activate macrophages, with or without fibroblasts [17]. In addition, macrophage activations are generally associated with phagocytosis of biomaterials [41–43]. Increased expression of TLR4 during scaffold degradation suggested its role in removing the debris of degraded scaffold through receptor-mediated phagocytosis. As we have alluded to previously, CD204 plays a role in the uptake of many materials. However, to our knowledge, its role in interacting with either PLGA or dextran has not yet been reported. Its elevated expression suggests that CD204 may be involved in recognition/internalization of the degraded debris. Since TLR4 and CD204 are multi-functional that could influence macrophage profoundly, the roles and mechanisms of these two regulated receptors in scaffold degradation have to be further investigated. The lack of regulations of CD44 and uPARAP/Endo 180 expressions suggested that the turnover of ECM deposited by cells was not altered, and thus, the increased expression of gelatinase and hyaluronidase-1 were responsible for scaffold degradation rather than the altered ECM turnover. It is highly possible that other receptors are involved in scaffold degradation and the elimination of degraded scaffold could also be mediated by non-receptor mediated phagocytosis.
3.5 In vivo degradation
The biodegradability of the scaffold was evaluated in mice subdermal implant models. Three days after implantation, the sizes of scaffolds decreased to approximately half of their pre-implant sizes (Fig. 6A). One week later, the sizes of the scaffolds further decreased to approximately one-fourth of their pre-implant sizes and the scaffolds were encapsulated by very thin fibrous tissues (Fig. 6B). After three weeks, three out of four implants were completely resorbed with the adjoining tissues fully restored. The remnant of the scaffold further decreased to approximately one-tenth of its original size (Fig 6C). The morphologies of these retrieved scaffolds (Fig. 7A and B) showed noticeable degradation one week post-implantation. The in vivo degradation of the scaffold was considerably faster than that of their in vitro counterpart. The extent of in vivo erosion in one week (Fig. 7) was comparable to that of observed after one month of exposure in the co-culture model (Fig. 1E), this further underscored the extended time span needed for any credible in vitro model system intended to emulate in vivo degradations. Nonetheless, the pattern of erosion for the implanted scaffold fibers bore remarkable resemblance to their counterparts subjected to cell-mediated erosion. These result signified that the fibroblast/macrophage co-culture model could be utilized as an in vitro tool to evaluate and study the mechanisms of biodegradation of materials under the influence of biological systems. The information obtained could guide the design of biomaterials and their selection for different applications.
Figure 6.
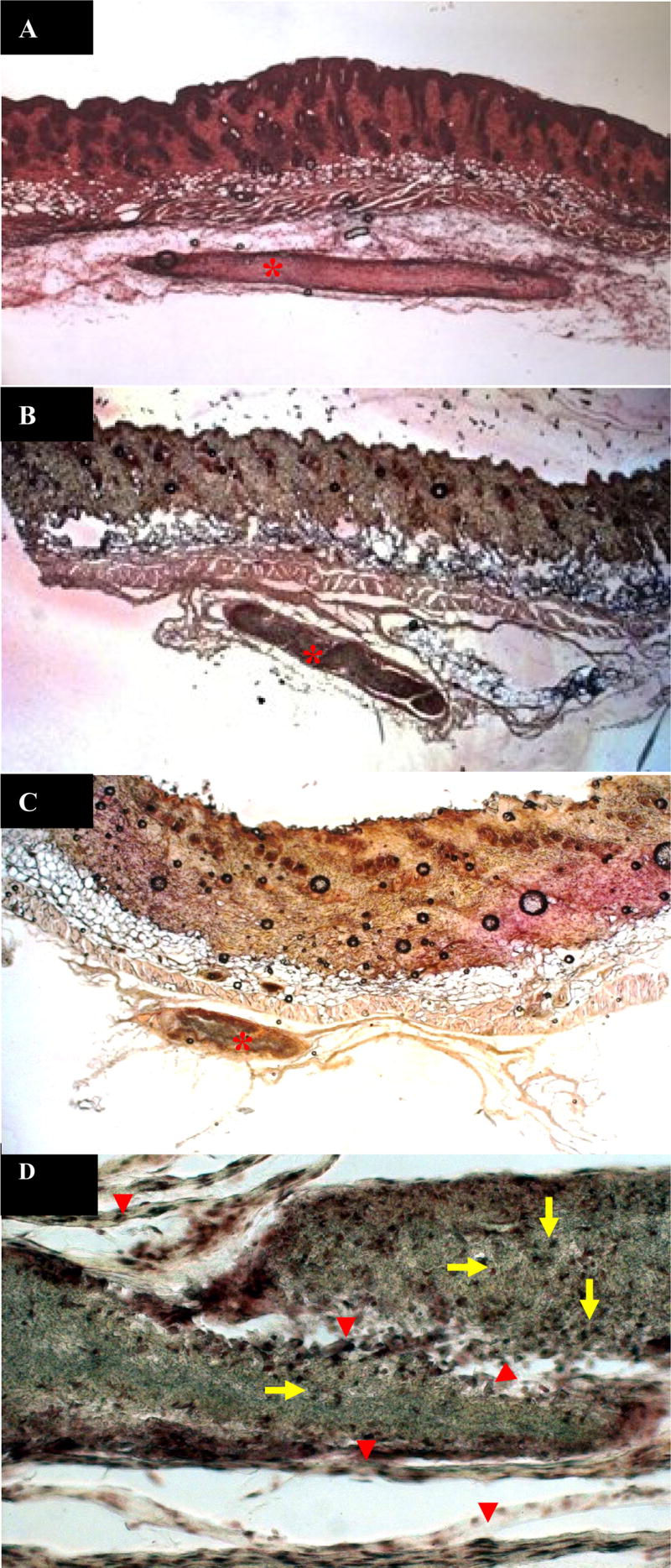
H&E staining of the implanted Dextran/PLGA scaffolds in the mice subdermal implantation model. (A) 3 days; (B) 1 week; and (C–D) 3 weeks after implantation. Star: the implants; Arrow: macrophages; Arrow head: fibroblasts; A–C: 40 X; D: 200 X.
Figure 7.
SEM of degraded Dextran/PLGA scaffolds retrieved from in vivo implantation 1 week post-surgery.
4. Conclusion
The Dextran/PLGA scaffold could be degraded by fibroblasts and macrophages, cultured separately or together. The activities of lysozyme, gelatinase, hyaluronidase-1, NSE, and α-glucosidase as well as the expressions of cell surface receptors CD204 and TLR4 were upregulated, suggesting their involvements in the cell-mediated scaffold degradation. The results of in vivo subdermal implantation of the Dextran/PLGA scaffold further confirmed its good biodegradability.
Acknowledgments
This study was supported by the NIH (DK068401, WC). We would also thank Dr. Michael Hadjiargyrou, Dr. Anil Dhundale and Dr. Howard Fleit for their insights.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mano JF, Silva GA, Azevedo HS, Malafaya PB, Sousa RA, Silva SS, Boesel LF, Oliveira JM, Santos TC, Marques AP, Neves NM, Reis RL. Natural origin biodegradable systems in tissue engineering and regenerative medicine: present status and some moving trends. J R Soc Interface. 2007 Dec;4(17):999–1030. doi: 10.1098/rsif.2007.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L, Ding J. In vitro degradation of three-dimensional porous poly(D,L-lactide-co-glycolide) scaffolds for tissue engineering. Biomaterials. 2004 Dec;25(27):5821–30. doi: 10.1016/j.biomaterials.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Onyeri S, Siewe M, Moshfeghian A, Madihally SV. In vitro characterization of chitosan-gelatin scaffolds for tissue engineering. Biomaterials. 2005 Dec;26(36):7616–27. doi: 10.1016/j.biomaterials.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 4.Santerre JP, Woodhouse K, Laroche G, Labow RS. Understanding the biodegradation of polyurethanes: from classical implants to tissue engineering materials. Biomaterials. 2005 Dec;26(35):7457–70. doi: 10.1016/j.biomaterials.2005.05.079. [DOI] [PubMed] [Google Scholar]
- 5.Dadsetan M, Christenson EM, Unger F, Ausborn M, Kissel T, Hiltner A, Anderson JM. In vivo biocompatibility and biodegradation of poly(ethylene carbonate) J Control Release. 2003 Dec;93(3):259–70. doi: 10.1016/j.jconrel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Ratner BD, Hoffman AS, Schoen FJ, Lemons JE. Biomaterials science: an introduction to materials in medicine. 2. Elsevier; Amsterdam: 2004. [Google Scholar]
- 7.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005 Jan-Feb;11(1–2):1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 8.Adams DO, Hamilton TA. The cell biology of macrophage activation. Annu Rev Immunol. 1984;2:283–318. doi: 10.1146/annurev.iy.02.040184.001435. [DOI] [PubMed] [Google Scholar]
- 9.Arlein WJ, Shearer JD, Caldwell MD. Continuity between wound macrophage and fibroblast phenotype: analysis of wound fibroblast phagocytosis. Am J Physiol. 1998 Oct;275(4 Pt 2):1041–8. doi: 10.1152/ajpregu.1998.275.4.R1041. [DOI] [PubMed] [Google Scholar]
- 10.Rabinovitch M. Professional and non-professional phagocytes: an introduction. Trends Cell Biol. 1995 Mar;5(3):85–7. doi: 10.1016/s0962-8924(00)88955-2. [DOI] [PubMed] [Google Scholar]
- 11.Meier B, Radeke HH, Selle S, Habermehl GG, Resch K, Sies H. Human fibroblasts release low amounts of reactive oxygen species in response to the potent phagocyte stimulants, serum-treated zymosan, N-formyl-methionyl-leucyl-phenylalanine, leukotriene B4 or 12-O-tetradecanoylphorbol 13-acetate. Biol Chem Hoppe Seyler. 1990 Oct;371(10):1021–5. doi: 10.1515/bchm3.1990.371.2.1021. [DOI] [PubMed] [Google Scholar]
- 12.Christenson EM, Dadsetan M, Wiggins M, Anderson JM, Hiltner A. Poly(carbonate urethane) and poly(ether urethane) biodegradation: in vivo studies. J Biomed Mater Res A. 2004 Jun;69(3):407–16. doi: 10.1002/jbm.a.30002. [DOI] [PubMed] [Google Scholar]
- 13.Pan H, Jiang H, Chen W. Interaction of dermal fibroblasts with electrospun composite polymer scaffolds prepared from dextran and poly lactide-co-glycolide. Biomaterials. 2006 Jun;27(17):3209–20. doi: 10.1016/j.biomaterials.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Hasilik A, Neufeld EF. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J Biol Chem. 1980 May;255(10):4937–45. [PubMed] [Google Scholar]
- 15.Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Chemical chaperones increase the cellular activity of N370S beta -glucosidase: a therapeutic strategy for Gaucher disease. Proc Natl Acad Sci U S A. 2002 Nov;99(24):15428–33. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.www.faizyme.com/assahyal.htm
- 17.Pan H, Jiang H, Chen W. A Fibroblast/Macrophage Co-culture Model for Evaluation of the Biocompatibility and Immunocompatibility of an Electrospun Dextran/PLGA scaffold. Submitted. [Google Scholar]
- 18.Labow RS, Meek E, Matheson LA, Santerre JP. Human macrophage-mediated biodegradation of polyurethanes: assessment of candidate enzyme activities. Biomaterials. 2002 Oct;23(19):3969–75. doi: 10.1016/s0142-9612(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 19.Santerre JP, Labow RS, Adams GA. Enzyme-biomaterial interactions: effect of biosystems on degradation of polyurethanes. J Biomed Mater Res. 1993 Jan;27(1):97–109. doi: 10.1002/jbm.820270113. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Basalyga DM, Simionescu A, Isenburg JC, Simionescu DT, Vyavahare NR. Elastin calcification in the rat subdermal model is accompanied by up-regulation of degradative and osteogenic cellular responses. Am J Pathol. 2006 Feb;168(2):490–8. doi: 10.2353/ajpath.2006.050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shockman GD, Daneo-Moore L, Kariyama R, Massidda O. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microb Drug Resist. 1996 Spring;2(1):95–8. doi: 10.1089/mdr.1996.2.95. [DOI] [PubMed] [Google Scholar]
- 22.Yomota C, Komuro T, Kimura T. Studies on the degradation of chitosan films by lysozyme and release of loaded chemicals. Yakugaku Zasshi. 1990 Jun;110(6):442–8. doi: 10.1248/yakushi1947.110.6_442. [DOI] [PubMed] [Google Scholar]
- 23.Lord MS, Stenzel MH, Simmons A, Milthorpe BK. Lysozyme interaction with poly(HEMA)-based hydrogel. Biomaterials. 2006 Mar;27(8):1341–5. doi: 10.1016/j.biomaterials.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Calvo P, Vila-Jato JL, Alonso MJ. Effect of lysozyme on the stability of polyester nanocapsules and nanoparticles: stabilization approaches. Biomaterials. 1997 Oct;18(19):1305–10. doi: 10.1016/s0142-9612(97)00061-6. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Quinn B, Witte DP, Grabowski GA. Gaucher disease mouse models: point mutations at the acid beta-glucosidase locus combined with low-level prosaposin expression lead to disease variants. J Lipid Res. 2005 Oct;46(10):2102–13. doi: 10.1194/jlr.M500202-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Girard N, Maingonnat C, Bertrand P, Tilly H, Vannier JP, Delpech B. Human monocytes synthesize hyaluronidase. Br J Haematol. 2002 Oct;119(1):199–203. doi: 10.1046/j.1365-2141.2002.03733.x. [DOI] [PubMed] [Google Scholar]
- 27.Ala-aho R, Kähäri VM. Collagenases in cancer. Biochimie. 2005 Mar–Apr;87(3–4):273–86. doi: 10.1016/j.biochi.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 29.Ziats NP, Miller KM, Anderson JM. In vitro and in vivo interactions of cells with biomaterials. Biomaterials. 1988 Jan;9(1):5–13. doi: 10.1016/0142-9612(88)90063-4. [DOI] [PubMed] [Google Scholar]
- 30.Anderson JM, Miller KM. Biomaterial biocompatibility and the macrophage. Biomaterials. 1984 Jan;5(1):5–10. doi: 10.1016/0142-9612(84)90060-7. [DOI] [PubMed] [Google Scholar]
- 31.Chellat F, Merhi Y, Moreau A, Yahia L. Therapeutic potential of nanoparticulate systems for macrophage targeting. Biomaterials. 2005 Dec;26(35):7260–75. doi: 10.1016/j.biomaterials.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 32.Arredouani MS, Palecanda A, Koziel H, Huang YC, Imrich A, Sulahian TH, Ning YY, Yang Z, Pikkarainen T, Sankala M, Vargas SO, Takeya M, Tryggvason K, Kobzik L. MARCO is the major binding receptor for unopsonized particles and bacteria on human alveolar macrophages. J Immunol. 2005 Nov;175(9):6058–64. doi: 10.4049/jimmunol.175.9.6058. [DOI] [PubMed] [Google Scholar]
- 33.Palecanda A, Kobzik L. Receptors for unopsonized particles: the role of alveolar macrophage scavenger receptors. Curr Mol Med. 2001 Nov;1(5):589–95. doi: 10.2174/1566524013363384. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Ferguson DJ, Quinn JM, Simpson AH, Athanasou NA. Biomaterial particle phagocytosis by bone-resorbing osteoclasts. J Bone Joint Surg Br. 1997 Sep;79(5):849–56. doi: 10.1302/0301-620x.79b5.7780. [DOI] [PubMed] [Google Scholar]
- 35.Fujikawa Y, Itonaga I, Kudo O, Hirayama T, Taira H. Macrophages that have phagocytosed particles are capable of differentiating into functional osteoclasts. Mod Rheumatol. 2005;15(5):346–51. doi: 10.1007/s10165-005-0424-8. [DOI] [PubMed] [Google Scholar]
- 36.Xia ZD, Triffitt JT. A review on macrophage responses to biomaterials. Biomed Mater. 2006;1:1–9. doi: 10.1088/1748-6041/1/1/R01. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton RF, Jr, Thakur SA, Mayfair JK, Holian A. MARCO mediates silica uptake and toxicity in alveolar macrophages from C57BL/6 mice. J Biol Chem. 2006 Nov;281(45):34218–26. doi: 10.1074/jbc.M605229200. [DOI] [PubMed] [Google Scholar]
- 38.Palecanda A, Paulauskis J, Al-Mutairi E, Imrich A, Qin G, Suzuki H, Kodama T, Tryggvason K, Koziel H, Kobzik L. Role of the scavenger receptor MARCO in alveolar macrophage binding of unopsonized environmental particles. J Exp Med. 1999 May;189(9):1497–506. doi: 10.1084/jem.189.9.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu RK, Droll A, Cooper DL, Dougherty ST, Dirks JF, Dougherty GJ. Molecular mechanisms regulating the hyaluronan binding activity of the adhesion protein CD44. J Neurooncol. 1995 Dec;26(3):231–9. doi: 10.1007/BF01052626. [DOI] [PubMed] [Google Scholar]
- 40.Engelholm LH, List K, Netzel-Arnett S, Cukierman E, Mitola DJ, Aaronson H, Kjoller L, Larsen JK, Yamada KM, Strickland DK, Holmbeck K, Dano K, Birkedal-Hansen H, Behrendt N, Bugge TH. uPARAP/Endo180 is essential for cellular uptake of collagen and promotes fibroblast collagen adhesion. J Cell Biol. 2003 Mar;160(7):1009–15. doi: 10.1083/jcb.200211091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branchaud RM, Garant LJ, Kane AB. Pathogenesis of mesothelial reactions to asbestos fibers. Monocyte recruitment and macrophage activation. Pathobiology. 1993;61(3–4):154–63. doi: 10.1159/000163784. [DOI] [PubMed] [Google Scholar]
- 42.Saad B, Matter S, Ciardelli G, Uhlschmid GK, Welti M, Neuenschwander P, Suter UW. Interactions of osteoblasts and macrophages with biodegradable and highly porous polyesterurethane foam and its degradation products. J Biomed Mater Res. 1996 Nov;32(3):355–66. doi: 10.1002/(SICI)1097-4636(199611)32:3<355::AID-JBM8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 43.Grandjean-Laquerriere A, Tabary O, Jacquot J, Richard D, Frayssinet P, Guenounou M, Laurent-Maquin D, Laquerriere P, Gangloff S. Involvement of toll-like receptor 4 in the inflammatory reaction induced by hydroxyapatite particles. Biomaterials. 2007 Jan;28(3):400–4. doi: 10.1016/j.biomaterials.2006.09.015. [DOI] [PubMed] [Google Scholar]