Abstract
Several mechanisms are expected to rapidly rid mutualisms of genetic variation in partner quality. Variation for mutualist quality, however, appears to be widespread. We used a model legume–rhizobium mutualism to test for evidence that context-dependent selection may maintain variation in partner quality. In a greenhouse experiment using 10 natural populations of Medicago truncatula and two strains of Sinorhizobium medicae, we detected significant genotype×genotype (G×G) interactions for plant fitness, indicating that the most beneficial rhizobium strain depends on the host genotype. In a second experiment using a subset of the plant populations used in the first experiment, we detected significant G×G interactions for both plant and rhizobium fitness. Moreover, the plant population with which rhizobium strains gained the greatest benefit depended on the nitrogen environment. Finally, we found that in a high nitrogen environment, all plant populations had lower fitness when inoculated with a 1 : 1 mixture of strains than with the worse single strain alone, suggesting that nitrogen shifts the exchange of benefits in favour of rhizobia. Our data suggest that genotype, nitrogen and biotic dependency might contribute to the maintenance of genetic variation in mutualist quality when coupled with spatial or temporal heterogeneity in the environment.
Keywords: genotype by environment, coevolution, cheating, symbiosis, cooperation, partner choice
1. Introduction
Many mutualistic relationships, including those between plants and pollinators, legumes and rhizobia, and coral animals and zooxanthellae, appear to have been stable for many millions of years (Bronstein 1994a; Knowlton & Rohwer 2003). Nevertheless, models predict that these relationships should be highly susceptible to invasion by exploiter (also known as cheater) genotypes that take from their symbiotic partner more than they give (Trivers 1970; Denison 2000; Sachs et al. 2004). The invasion of exploiters can be prevented, and thus mutualism stabilized, if individuals are able to exclude or later punish (sanction) less beneficial partners (Bull & Rice 1991; West et al. 2002a). Empirical work has provided evidence for the operation of such mechanisms; yucca plants selectively abort fruits with heavy yucca moth egg loads (Pellmyr & Huth 1994), and legumes provide fewer resources to root nodules containing less beneficial rhizobia (Kiers et al. 2006; Simms et al. 2006). If these mechanisms are highly effective at removing all but the best partner genotypes, then we may expect to find little intraspecific genetic diversity in the quality of mutualists (Bull & Rice 1991; West et al. 2002a). Nevertheless, genetic diversity for partner quality has been found in many mutualisms, including the mutualism between leguminous plants (Fabaceae) and rhizobial bacteria (Smith & Goodman 1999; Simms & Taylor 2002).
In the legume–rhizobium mutualism, plants provide rhizobia with carbon and shelter inside of root nodules, while rhizobia provide plant available nitrogen (N) that they have converted (fixed) from atmospheric N2 (Vance 2002). Data from both agricultural systems (e.g. Miller & Sirois 1982; reviewed in Smith & Goodman 1999; Robinson et al. 2000), as well as a few non-domesticated systems (Parker 1995; Burdon et al. 1999; Mhadhbi et al. 2005), reveal that plant biomass and N content vary when plants are grown in symbiosis with different rhizobium strains. In other words, there exists genetic variation for the benefits plants receive from symbiosis with rhizobia. Moreover, variation exists among rhizobium strains for the capacity to form nodules with different plant genotypes or species (e.g. Robinson et al. 2000; Garau et al. 2005; Mhadhbi et al. 2005).
Genetic variation in partner quality may persist because selection caused by plant sanctions is not effective at removing less optimal genotypes that drift through plant and rhizobium populations. Alternatively, diversity in the quality of mutualists may be maintained if quality is dependent upon the genotype with which a mutualist associates. Such genotype dependency, or genotype×genotype (G×G) interactions, can generate frequency-dependent selection, which may maintain genetic variation within mutualist populations (Bever 1999). Variation in partner benefits may also be maintained, or the loss of diversity slowed, if the optimal mutualistic partner is dependent upon the abiotic environment. For example, from the plant perspective, the benefits of forming symbiosis with rhizobium strains will depend upon the availability of soil N (West et al. 2002b), which is less costly for plants to acquire than biologically fixed N. In fact, the abiotic environment may even cause an interaction to shift from mutualism to parasitism (Neuhauser & Fargione 2004). Finally, variation may be maintained if partner quality depends on the biotic environment. For example, less beneficial rhizobia may be maintained if partner choice or sanctions are less effective when plants are grown with more diverse rhizobium communities, either because of changes in the competitive interactions among rhizobia, or because low-quality rhizobia hide from sanctions in mixed-strain nodules (Denison 2000).
Here, we report on a series of greenhouse experiments with Medicago truncatula Gaertn. (barrel medic) and Sinorhizobium medicae, a model system for investigating genetic mechanisms and evolution in legume–rhizobium symbioses (Cook et al. 1997; Young et al. 2005; Bailly et al. 2006), designed to test for evidence of three types of context dependence that may maintain genetic variation in mutualist quality. We tested for genotype dependency by assessing genetic variation among natural plant populations for the fitness benefits of symbiosis with different rhizobium strains, as well as assessing genetic variation among rhizobium strains for the fitness benefits of symbiosis with plant populations. Significant plant population×rhizobium strain (G×G) interactions for fitness would mean that the relative benefits that partners confer in symbiosis depend on the genotypic context of the interaction. We tested for abiotic environmental dependency by assessing genetic variation in the benefits plants and rhizobia obtain when growing in different nitrogen environments. A significant genotype×nitrogen (genotype×environment, G×E) interaction for plant or rhizobium fitness would mean that the value of interacting with a partner genotype depends on the N environment. Finally, we tested for biotic environmental dependency by evaluating the effects of a mixed community of rhizobia on plant fitness. If plants are effective at either choosing or sanctioning, we expect that plants will be associated most often with the strains which confer high fitness benefits in a mixed population of rhizobia. Alternatively, plants may be unable to choose the most cooperative partners from a mixture of strains. We compared the size and fruit production of plants grown with mixed- with single-genotype rhizobium populations in order to evaluate the ability of plant populations to discriminate among rhizobium strains of unequal quality.
2. Material and methods
(a) Plant×rhizobium experiment
In order to test for genetic variation among plant populations for the fitness benefits they receive from rhizobium strains, we grew M. truncatula plants from 10 geographically distinct populations in symbiosis with one of two rhizobium strains. The 10 M. truncatula populations, seeds from which were obtained from USDA, ARS, National Genetic Resources Program (see figure 1 in the electronic supplementary material), represent a large portion of the natural geographical range of M. truncatula (Ronfort et al. 2006). As the genetic relationship among seeds from each population is unknown, we tested for genetic variation among populations of M. truncatula. Plants were inoculated with either S. medicae strain ABS7 (A) or S. medicae strain WSM540 (W). Strain ABS7 (obtained from K. A. VandenBosch, University of MN) is commonly used for dissecting the genetic mechanisms underlying signalling between plants and rhizobia, initiation of nodulation and N fixation (e.g. Cook et al. 1997). Strain WSM540 (obtained from P. H. Graham, University of MN) is a Sardinian isolate in the Western Australia Soil Microbiology Collection (Garau et al. 2005).
Prior to planting, seeds were manually scarified, sterilized (1 min in 70% ethanol, followed by 5 min in commercial bleach) and sown in autoclave-sterilized peat pellets (Jiffy, Norway). After planting, seeds were placed in the dark for 24 h and then moved to the laboratory bench to germinate. After one week, 20 seedlings (with peat pellets) from each plant×rhizobium combination (600 plants in total) were transplanted to individual pots containing steam-sterilized field soil and randomized in a common greenhouse environment, where they were given adequate water until senescence. Low levels of contamination among pots may have occurred; therefore, our treatments should be conservatively interpreted as different rhizobium environments rather than rhizobium monocultures per se. Contamination, if any did occur, should result in smaller differences among treatments, making our results conservative.
Sinorhizobium medicae cultures were grown in yeast mannitol broth with 3.7 μM FeCl3 (Vincent 1970), at 30°C for 48 h. Immediately before inoculating plants, inoculum cell density was adjusted to approximately 106 cells ml−1 (based on OD670) by diluting liquid cultures with sterile water. Each plant received 1 ml of inoculum within 1 day of germination and a second inoculation (using the same procedure) after being moved to the greenhouse.
We estimated the quality of rhizobium strains for plant populations by measuring the number of leaves on each plant six weeks after planting and counting the total number of mature fruits produced. Midway through the experiment, 226 plants on one greenhouse bench died due to exposure to intense artificial light during a greenhouse malfunction. These plants were excluded from the analysis of fruit number, leaving 374 plants.
(b) N-addition experiment
In a separate experiment designed to test for N-dependent variation in mutualist quality, 10 replicates of each of the four populations from the plant×rhizobium experiment were inoculated with either A or W (as in the plant×rhizobium experiment) or an equal mixture of A and W. The mixed inoculum was prepared by combining A and W cells in a 1 : 1 ratio immediately before inoculation (all plants received the same total number of cells). Half of the plants in this experiment (120 plants) were fertilized with 50 ml of 1.0 mM KNO3 three times per week starting approximately one week after germination. The other 120 plants were not fertilized. The four plant populations used in this experiment were chosen based on their divergent fitness responses to symbiosis with A and W strains in the plant×rhizobium experiment, as described above (figure 1 in the electronic supplementary material).
To estimate plant growth and fitness, we measured leaf number eight weeks after planting and counted the number of mature fruits and seeds produced by each plant. Early growth in the greenhouse may be an important component of fitness in natural environments, where seedlings may experience intense competition. The competition-free greenhouse environment may ameliorate these differences during later reproductive stages—potentially leading to more conservative estimates of G×G interactions during fruit production.
We also estimated rhizobium fitness by counting the total number of nodules produced on each plant as well as the mean length and branch number of 10 randomly selected nodules collected from each plant. Nodules were stored atop silica gel and cotton wool at 4°C in individual 1.5 ml tubes until measurement. After drying, the length of the longest branch and the number of branches (one, two to three or more than three) on each nodule were scored. Preliminary analyses revealed that nodule length and branches were positively correlated (rd.f.=145=0.53, p<0.0001), nodule number and the number of branches per nodule were negatively correlated (rd.f.=144=−0.26, p=0.002) and nodule length and number were not significantly correlated (rd.f.=144=−0.02, p=0.98). Therefore, we also estimated nodule volume per plant as the product of nodule length×nodule number.
In a preliminary experiment, we found that the number of reproductive offspring inside a nodule, estimated as the number of colonies produced from crushing and plating a nodule, was positively correlated with nodule length (rd.f.=33=0.59, p<0.001). Data on nodule number and size, therefore, appear to be appropriate for estimating the effect of symbiosis on rhizobium fitness.
(c) Data analysis
(i) Single inoculum treatments
For the plant×rhizobium experiment, we used mixed model ANOVA (PROC MIXED, SAS Institute) to test whether genetic variation among plant populations, rhizobium strains (A and W) and their interaction affected the fitness benefits obtained by plants. For this experiment, rhizobium strain was included as a fixed effect, and plant population and the plant population×rhizobium strain interaction were included as random effects. The significance of random effects was determined by comparing the difference in −2×ln L between models differing by the inclusion of each random effect with a Χ2-distribution with one degree of freedom.
For the N-addition experiment, we used ANOVA (PROC GLM, SAS Institute) to test for variation among plant populations, rhizobium strains, N treatments and their interactions. Since we have three estimates of rhizobium fitness (nodule number, length and branches), we tested for significant effects of plant population and N environment using MANOVA, followed by univariate ANOVAs for each individual fitness measure (as well as nodule volume). For these analyses, plant population was treated as a fixed variable because the four populations were chosen based on results from the plant×rhizobium experiment.
(ii) Mixed inoculum treatment
Our primary interest in the mixed inoculum treatment was to determine whether plant populations are able to preferentially associate with the more beneficial rhizobium strain, and whether this ‘choosiness’ might vary among plant populations. To test this, we compared the fitness of plants inoculated with a mixture of rhizobium strains with those inoculated with a single strain. We calculated the relative performance (RP) of each plant grown with mixed inoculum compared with performance when grown with a single rhizobium strain as
where frAW is a plant's fruit production with the mixed inoculum treatment and fr(A+W)/2 is the mean of the two single inoculum treatments (calculated separately for each population in plant×rhizobium experiment, and for each population×nitrogen treatment combination in the N-addition experiment). We also estimated RP based on leaf number (RPleaves), as well as seed production (RPseeds; N-addition experiment only). An RP=0 is expected if plants randomly sample rhizobia from their environment and there are no interactions between rhizobia after nodule formation. RP>0 is expected if plants preferentially associate with more beneficial rhizobium strains, whereas RP<0 is expected if plant fitness with the mixed inoculum is lower than expected based on random association. For the plant×rhizobium experiment, we analysed RPleaves and RPfruits using PROC MIXED (SAS Institute) for the random effect of plant population. For the N-addition experiment, we analysed RPleaves, RPfruits and RPseeds using PROC GLM (SAS Institute) for the fixed effects of population, nitrogen and their interaction. We note that the estimate of RP should be viewed with some caution because it does not incorporate variance associated with calculating the means of the single inoculum treatments.
3. Results
(a) Plant fitness
In the plant×rhizobium experiment, we detected a significant plant population×rhizobium strain effect on plant growth and fruit production. This interaction, along with several rhizobium-dependent changes in the rank order of plant growth and fitness (table 1; figure 2 in the electronic supplementary material) and the lack of an overall difference between the effects of the two rhizobium strains (strain main effect p>0.1; table 2), indicates that the rhizobium strain conferring greatest benefit depended upon the plant population. In other words, neither strain was universally more beneficial. Plant populations, however, did differ in their overall growth and fitness (population main effect p<0.0001; table 2), although the population with the highest growth differed between the two strain environments (table 1). The rank order of plant populations in each rhizobium environment also differed between leaf number and fruit number (table 1; figure 2 in the electronic supplementary material).
Table 1.
Mean plant growth and fitness of 10 M. truncatula populations in symbiosis with one of the two strains (A or W) of S. medicae.
| leaf number (s.e.) | fruit number (s.e.) | |||
|---|---|---|---|---|
| population | A | W | A | W |
| 1 | 16.5(1.6) | 14.9(1.6) | 6.0(2.2) | 8.8(2.0) |
| 2 | 16.7(1.9) | 21.3(1.6) | 2.3(2.3) | 5.1(2.2) |
| 3 | 17.5(2.0) | 16.7(1.7) | 10.2(2.5) | 15.2(2.4) |
| 4 | 16.0(1.6) | 12.5(1.6) | 23.9(2.5)a | 16.7(2.2) |
| 5 | 17.5(1.6) | 16.5(1.7) | 1.7(2.4) | 1.8(2.2) |
| 6 | 16.6(1.6) | 15.7(1.6) | 13.8(2.2) | 14.4(2.3) |
| 7 | 23.8(3.1)a | 15.8(3.4) | 21.0(4.2) | 14.0(4.8) |
| 8 | 23.9(1.7)a | 19.5(1.6) | 23.0(2.2)a | 24.9(2.2)a |
| 9 | 17.8(1.6) | 24.8(1.7)a | 13.2(2.2) | 4.5(2.6) |
| 10 | 11.3(1.9) | 14.2(1.7) | 2.1(2.4) | 8.0(2.6) |
Population with the highest performance with each strain is given in bold; where two values are highlighted, these means are not significantly different.
Table 2.
Mixed model analysis of plant growth and fitness with single-strain inoculum treatments. (For random effects, Χ2 (ln-likelihood ratio) is shown. *p≤0.05; ****p≤0.0001)
| leaf no. | fruit no. | |
|---|---|---|
| random effects | ||
| population | 19.6**** | 117.4**** |
| pop×strain | 3.9* | 4.1* |
| fixed effects | F1,8 | F1,8 |
| strain | 0.01 | 0.18 |
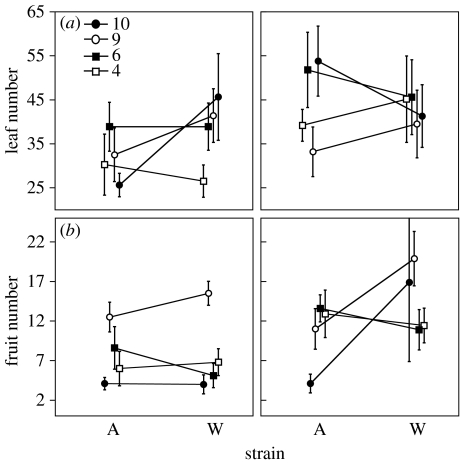
Results from the N-addition experiment were similar to those from the plant×rhizobium experiment. In particular, we found a significant plant population×rhizobium strain interaction, no significant main effect of strain and significant variation among plant populations (table 3). As expected, N-fertilized plants were significantly larger and produced more fruits and seeds than unfertilized plants. The plant population×rhizobium strain interaction, which explained a significant amount of variation in leaf number and fruit production, did not explain a significant amount of variation in seed production (table 3; p=0.20). Nevertheless, seed and fruit numbers were highly correlated (rd.f.=241=0.86, p<0.0001), and these two measures showed similar patterns (see figure 3a in the electronic supplementary material).
Table 3.
ANOVAs for growth and fitness of plants in the single-strain inoculum treatments grown in either low or high nitrogen. (†p≤0.1; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001. Numerator d.f. shown for each effect; denominator d.f. for leaf number=135, fruit number=134, seed number=134.)
| d.f. | leaf no. | fruit no. | seed no. | |
|---|---|---|---|---|
| population | 3 | 6.89*** | 9.86**** | 6.85*** |
| strain | 1 | 0.93 | 1.04 | 0.44 |
| nitrogen | 1 | 70.17**** | 8.87** | 8.00** |
| pop×strain | 3 | 2.83* | 2.69* | 1.56 |
| pop×nitrogen | 3 | 2.48† | 1.70 | 1.85 |
| strain×nitrogen | 1 | 0.77 | 0.62 | 0.04 |
| pop×strain×nitrogen | 3 | 0.63 | 0.54 | 0.58 |
As in the plant×rhizobium experiment, rhizobium quality depended on plant population, and the rank order of plant populations changed with the rhizobium environment (figure 1). Again, however, the patterns differed between leaf and fruit numbers. Early in development, the favoured plant population differed depending on nitrogen treatment and rhizobium strain, but plant population 9 had the highest fruit number in both rhizobium and N environments (figure 1).
Figure 1.
Mean number of (a) leaves (±s.e.) and (b) fruits (±s.e.) produced by each of the four M. truncatula populations in symbiosis with two strains of S. medicae either (left column) without or (right column) with added nitrogen.
(b) Rhizobium fitness
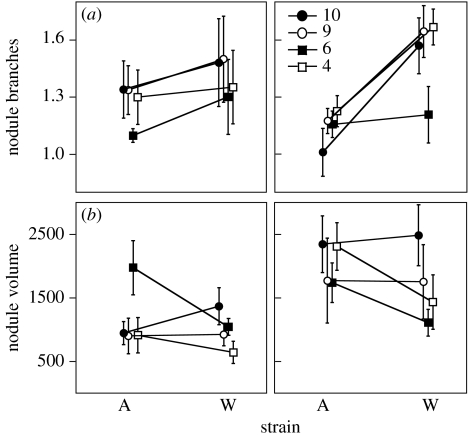
In the N-addition experiment, in which we estimated rhizobium fitness, the MANOVA revealed that rhizobium strains differed in their fitness, and that fitness was affected by N supply, plant population, as well as interactions between these factors (table 4). Several of these interactions are of interest in understanding mutualism evolution. The plant population×rhizobium strain (G×G) effect suggests that the benefits rhizobia obtain from symbiosis with plants depend upon the plant population they infect—an important prerequisite for coevolution. The strain×N effect (significant for nodule branches) indicates that the benefits of symbiosis for rhizobia also depend upon the abiotic N environment. The population×N effect indicates that the benefits of symbiosis with plant populations depended on the N environment, e.g. the effects of plant populations 6 and 10 on strain A were N dependent (figure 2b). The MANOVA also revealed a significant population×strain×N effect, suggesting that the G×G interactions between plant and rhizobium strains depended on the N environment; however, this effect was not significant in the univariate analyses (table 4).
Table 4.
MANOVA (Pillai's test statistic) and separate ANOVAs for rhizobium fitness estimates in symbiosis with 10 plant populations, in either high or low nitrogen. (*p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001. Numerator d.f. shown; NN, nodule number (denominator d.f.=134); NB, nodule branches (129); NL, nodule length of 0.1 mm (130); NV, nodule volume (129).)
| MANOVA | d.f. | NN | NB | NL | NV | |
|---|---|---|---|---|---|---|
| population | 8.58**** | 3 | 4.42** | 10.76**** | 9.89**** | 6.95*** |
| strain | 25.01**** | 1 | 4.42** | 70.28**** | 40.63**** | 1.08 |
| nitrogen | 17.02**** | 1 | 20.88**** | 15.82**** | 14.41*** | 34.09**** |
| pop×strain | 2.32* | 3 | 3.78* | 4.92** | 1.79 | 3.60* |
| pop×nitrogen | 3.33* | 3 | 2.08 | 3.18* | 6.52*** | 2.91* |
| strain×nitrogen | 3.42* | 1 | 0.04 | 5.02* | 0.75 | 0.14 |
| pop×strain×nitrogen | 1.95* | 3 | 0.92 | 1.45 | 1.63 | 1.22 |
Figure 2.
Fitness response of two S. medicae strains in symbiosis with four populations of M. truncatula either (left column) without or (right column) with added nitrogen. Rhizobium fitness was estimated by (a) nodule branches (±s.e.) and (b) nodule volume (±s.e.).
Despite these significant interactions, we found little evidence that either the biotic or abiotic environment reversed the rank order of rhizobium strains. Instead, strain W had the highest fitness in most plant and N environments (figure 2), so our results largely reflect differences in the degree to which plant population and N affect the fitness of strains A and W. Nevertheless, we did detect some evidence of plant population-dependent changes in rank for nodule volume. Specifically, the rank fitness of A and W differed between plant populations 6 and 10 in the low N treatment (figure 2b); however, the difference between A and W with population 10 was not significant after multiple comparisons.
(c) Mixed inoculum
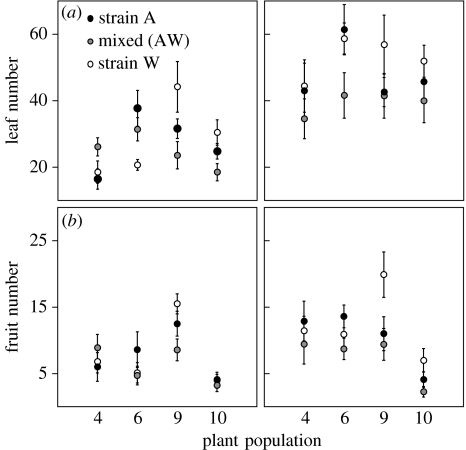
In the N-addition experiment, analyses of variance revealed that the effects of mixed- versus single-strain inoculum environments on plant growth and seed production (RPleaves and RPseeds) differed among the plant populations (table 5). Moreover, plant response to the mixed- versus single-strain inocula depended upon the N environment (significant population×N for RPleaves; table 5). This interaction was not significant for RPfruits or RPseeds, but the response of plant populations to the mixed inoculum was unchanged (table 5, figure 3, and figure 3b in the electronic supplementary material). Separate analyses revealed that populations differed in their response to the mixed-strain relative to single-strain inocula only in the ambient N treatment (results not shown). In other words, without N addition, some plant populations produced more leaves and fruits than expected if plants are unable to differentiate between strains, while others produced fewer leaves and fruits (figure 3). Perhaps more interesting, however, we found that all plant populations in the high N treatment were smaller and produced fewer fruits when grown with the mixed inoculum than would be expected if the effect of a strain mixture were simply an additive function of each strain alone (RPleaves, RPfruits and RPseeds all<0). In fact, for all plant populations in the high N environment (as well as for two populations in the low N environment), growth and fruit production were even lower when grown with the mixed inoculum than with the worse single strain alone (figure 3).
Table 5.
ANOVAs for the effect of plant population and nitrogen on the performance of plants in the mixed inoculum treatment, relative to the mean of the two single inoculum treatments, in either high or low nitrogen. (*p≤0.05; **p≤0.01. F-values shown; denominator d.f.=63.)
| d.f. | RPleaves | RPfruits | RPseeds | |
|---|---|---|---|---|
| population | 3 | 4.21** | 2.02 | 3.37* |
| nitrogen | 1 | 4.00* | 2.08 | 0.32 |
| pop×nitrogen | 3 | 5.71** | 1.01 | 0.68 |
Figure 3.
Mean number of (a) leaves (±s.e.) and (b) fruits (±s.e.) produced by each of the four M. truncatula populations in three rhizobium inoculation treatments (strain A alone, strain W alone or a 1 : 1 mixture of A and W), either (left column) without or (right column) with added nitrogen.
4. Discussion
If either partner choice (Bull & Rice 1991) or sanctions (West et al. 2002a) allow mutualists to associate with only those partners that provide the greatest net fitness benefit, then we may expect that populations will harbour little genetic variation for mutualist quality. Nevertheless, genetic variation in mutualist quality appears to be widespread (Smith & Goodman 1999; Simms & Taylor 2002). Here, we detected evidence for three types of context dependency that may help to explain the presence of that genetic variation. In the Medicago–Sinorhizobium system we studied, the fitness benefits that plants and rhizobia obtained from their mutualistic partner depended upon the genotype of both the plant and rhizobium, the abiotic environment and the rhizobium community in which plants were grown.
Although the ecology and evolution of mutualistic interactions have received much attention (reviewed by Bronstein 1994a), relatively few studies have investigated the extent of genetic variation in mutualist quality in natural systems. We found significant among-population variation in the benefits plants obtained from associating with different rhizobium strains. A handful of other studies have documented plant genetic variation in natural populations (Parker 1995; Wilkinson et al. 1996; Burdon et al. 1999; Simms et al. 2006), suggesting that such variation may be widespread. Fewer studies, however, have found genetic variation in the benefits rhizobia derive from forming symbiosis with different plant genotypes (Miller & Sirois 1982; Spoerke et al. 1996). Using three nodule characteristics as measures of rhizobium fitness, we found that the fitness benefits rhizobium strains obtained from associating with plants also depended upon the rhizobium strain–plant population combination. In other words, from neither the plant nor the rhizobium perspective is there a universally most beneficial partner. Such genetic variation indicates that legume–rhizobium mutualisms may be evolutionarily dynamic, with the benefits of symbiosis depending on the genotypes with which partners interact (Thompson 1988; Bronstein 1994b; Parker 1995).
(a) Frequency dependence and the maintenance of VG for partner quality
The significant plant genotype×rhizobium genotype interactions raise the possibility that genetic variation (VG) in mutualist quality is maintained by frequency-dependent selection. Bever (1999) showed that in a simple two-host (e.g. HA and HB), two-symbiont (e.g. SA and SB) population, negative frequency dependence will maintain genetic variation in mutualist quality if HA derives more benefit from symbiosis with symbiont SA than SB, while symbiont SB derives more benefit than SA from HA, and vice versa for HB. Under this scenario, as host HA increases in frequency, symbiont SB will also increase in frequency. In turn, an environment where symbiont SB is more common than SA results in a shift in the relative fitness of host genotypes and an increase in the relative frequency of HB (i.e. the frequency of HA feeds back negatively on its own fitness via its symbiont). In positive frequency dependence, by contrast, host HA receives and confers the greatest benefit when forming a symbiosis with SA (an increase in the frequency of host HA would feed back positively on its own fitness). While the potential for negative frequency dependence to maintain genetic variation is well known (Kimura & Ohta 1971; Bever 1999), positive frequency dependence can also maintain genetic variation in partner populations if populations are spatially structured (i.e. when populations are not well mixed; Molofsky et al. 2001).
Although significant G×G interactions suggest that frequency dependence may contribute to the maintenance of genetic variation in mutualist quality, visual inspection of plant and rhizobium fitness rankings (figures 1 and 2) suggests that frequency-dependent selection alone will not maintain variation. In particular, there exist few cases in which a plant population experiences the highest fitness with one rhizobium strain, yet confers higher fitness to the other rhizobium strain. A formal set of criteria for establishing frequency dependence set forth in Bever (1999) confirms these visual observations; we detected evidence for frequency dependence in only 10 out of 96 pairwise interactions, all of which were indicative of positive frequency dependence (table 1 in the electronic supplementary material). Therefore, our data suggest that negative frequency dependence may play little role in the maintenance of genetic variation in mutualist quality that we detected. In contrast, our data suggest that positive frequency dependence, although not widespread, may contribute to the maintenance of genetic variation in this mutualism. However, more data on a broader range of naturally co-occurring plant and rhizobium partners are necessary to fully assess the potential for frequency dependence to maintain genetic variation in this mutualism. Moreover, little is known about the spatial structure in plant and rhizobium interactions, which is necessary for positive frequency dependence to maintain genetic variation (Simms & Taylor 2002).
Results from our N-addition experiment also suggest a role for spatial heterogeneity in the maintenance of genetic variation in mutualist quality. In particular, we found that fertilizing with small amounts of inorganic N altered the genotype dependency of fitness benefits conferred to rhizobia by plants (figure 2). Since N availability is likely to vary in space and time (Boerner et al. 1998; Wilson & Thompson 2005), environmentally dependent selection may facilitate the maintenance of genetic variation in legume–rhizobium communities (Gomulkiewicz et al. 2003; Nuismer et al. 2003).
(b) Effects of a biological market on plant fitness
We also detected evidence for biotic environmental dependence, a third form of context dependency in mutualistic interactions. In natural systems, an individual plant is likely to encounter multiple rhizobium genotypes that may vary in quality, i.e. a ‘biological market’ (Noë & Hammerstein 1995; Simms & Taylor 2002). Therefore, understanding plant response to a mixed community of rhizobia is necessary for understanding plant–rhizobium coevolution. Mutualism theory predicts that the value of a traded resource decreases as its external availability increases (West et al. 2002b; Neuhauser & Fargione 2004). Consequently, the range of rhizobium genotypes that are mutualists will decrease as soil N increases, so selection should favour increased plant choosiness of rhizobium partners when N levels are high. However, we found that added N resulted in all plant populations performing worse with a mixture of rhizobium strains, compared with their mean performance with each strain singly.
Surprisingly, for all populations in high N, and for two of the four populations in low N, plants were smaller and produced fewer fruits when inoculated with a mixture of the two rhizobium genotypes than with the worse strain alone. This effect is difficult to explain; while plants may sample rhizobia at different rates (Dowling & Broughton 1986), differential sampling is expected to result in plant fitness intermediate between the fitness with each strain individually. Sanctioning rhizobia (particularly in high N environments) might be costly because plants form nodules before sanctioning (Kiers et al. 2006). This cost could account for lower plant performance in the mixed inoculum if plants sanction only when infected with multiple rhizobium genotypes (otherwise, plants in the worse single strain treatment should be spending equal resources sanctioning rhizobia). This seems unlikely, however, as micro-environmental and developmental factors, not just genetic, should affect the benefits gained from a given nodule. In fact, plants do appear to sanction the same genotype differently depending on the environmental conditions (Kiers et al. 2006). A second potential explanation for the poor performance of plants in mixed inoculum is strain–strain antagonism. Such negative interactions have been documented in the soil (Schwinghamer & Brockwell 1978; Hafeez et al. 2005) and between bacterial parasites in other host systems (Massey et al. 2004); however, we acknowledge that there is no evidence for such competition between rhizobia in planta.
(c) Understanding the fitness of rhizobia
Owing to a lack of data on rhizobial reproduction and fitness, the best method for estimating the benefits rhizobia receive from symbiosis with plants is not known. Some of our results differed among rhizobium fitness estimates; for example, the strain×N interaction affected the number of nodule branches, but not their length or total number. As each nodule branch has its own meristem, greater branching probably results in higher reproductive potential on a per nodule basis because the reproductive cells in indeterminate M. truncatula nodules probably remain near the actively growing meristem (Denison 2000). Branching, however, was negatively correlated with the total number of nodules on a plant, potentially indicating a tradeoff between these two components of rhizobium fitness. Nodule length is positively and significantly correlated with the number of viable cells inside a nodule in our system, in the alfalfa–Sinorhizobium meliloti symbiosis (W. C. Ratcliff 2005, personal communication), as well as in the determinate Lupinus–Bradyrhizobium symbiosis (Simms et al. 2006). Unlike branching, nodule length was uncorrelated with nodule number. Therefore, it appears that nodule volume may be a good estimate of the reproductive benefits rhizobia obtain from forming nodules with plants.
An important caveat, however, is that any nodule-based estimate of fitness ignores survival and growth outside of the host plant. Moreover, traits that affect soil survival and growth may be unrelated to nodule size; for example, bacterial storage compounds such as PHB may be important determinants of rhizobium fitness in the soil, yet PHB concentration may not be correlated with nodule number or size. Unfortunately, few, if any, data are available for evaluating the relationships between rhizobium fitness inside of plants and rhizobium fitness in the soil (Denison 2000). Multigenerational experiments, in which the relationship between nodule traits (such as nodule size) and rhizobium genotype frequencies in the rhizosphere can be specifically investigated, are needed to understand the fitness impacts of symbiosis for rhizobia, as well as how precisely nodule measures predict evolutionary changes in rhizobium populations.
5. Conclusions
Mechanisms that limit cheaters, if highly effective, may be expected to rid populations of variation in mutualist quality, yet mutualisms in nature appear to harbour extensive intraspecific genetic variation for the benefits conferred to, and received from, the interacting partners. Our experiments suggest that frequency-dependent G×G interactions themselves are unlikely to maintain genetic variation. Instead, our data point to spatial or temporal heterogeneity as necessary for maintaining genetic variation in partner quality in this mutualism. Genetic structure in and among natural populations, patchiness in the availability of nitrogen and variability in the biotic environment (such as the herbivore or soil microbial communities) are all probable candidates for generating heterogeneous selection on this mutualism in nature. More empirical data on the effects of these factors on partners from natural communities will further elucidate the relative importance and ecological reality of these hypotheses.
Acknowledgments
We thank S. Short, S. Chowdhury and C. Kelly for their assistance in the greenhouse and laboratory as well as J. Lau, D. Moeller and two anonymous reviewers for their comments that much improved this manuscript. We also thank NSF (DEB-0508305) and the Center for Community Genetics at the University of Minnesota for their financial support.
Supplementary Material
References
- Bailly X, Olivieri I, De Mita S, Cleyet-Marel J.C, Bena G. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Mol. Ecol. 2006;15:2719–2734. doi: 10.1111/j.1365-294X.2006.02969.x. [DOI] [PubMed] [Google Scholar]
- Bever J.D. Dynamics within mutualism and the maintenance of diversity: inference from a model of interguild frequency dependence. Ecol. Lett. 1999;2:52–61. doi:10.1046/j.1461-0248.1999.21050.x [Google Scholar]
- Boerner R.E.J, Scherzer A.J, Brinkman J.A. Spatial patterns of inorganic N, P availability, and organic C in relation to soil disturbance: a chronosequence analysis. Appl. Soil Ecol. 1998;7:159–177. doi:10.1016/S0929-1393(97)00037-1 [Google Scholar]
- Bronstein J.L. Our current understanding of mutualism. Q. Rev. Biol. 1994a;69:31–51. doi:10.1086/418432 [Google Scholar]
- Bronstein J.L. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. 1994b;9:214–217. doi: 10.1016/0169-5347(94)90246-1. doi:10.1016/0169-5347(94)90246-1 [DOI] [PubMed] [Google Scholar]
- Bull J.J, Rice W.R. Distinguishing mechanisms for the evolution of co-operation. J. Theor. Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- Burdon J.J, Gibson A.H, Searle S.D, Woods M.J, Brockwell J. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. J. Appl. Ecol. 1999;36:398–408. doi:10.1046/j.1365-2664.1999.00409.x [Google Scholar]
- Cook D.R, VandenBosch K, deBruijn F.J, Huguet T. Legume models get the nod. Plant Cell. 1997;9:275–281. doi:10.1105/tpc.9.3.275 [Google Scholar]
- Denison R.F. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 2000;156:567–576. doi: 10.1086/316994. doi:10.1086/316994 [DOI] [PubMed] [Google Scholar]
- Dowling D.N, Broughton W.J. Competition for nodulation of legumes. Annu. Rev. Microbiol. 1986;40:131–157. doi: 10.1146/annurev.mi.40.100186.001023. doi:10.1146/annurev.mi.40.100186.001023 [DOI] [PubMed] [Google Scholar]
- Garau G, Reeve W.G, Brau L, Deiana P, Yates R.J, James D, Tiwari R, O'Hara G.W, Howieson J.G. The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil. 2005;276:263–277. doi:10.1007/s11104-005-0374-0 [Google Scholar]
- Gomulkiewicz R, Nuismer S.L, Thompson J.N. Coevolution in variable mutualisms. Am. Nat. 2003;162:S80–S93. doi: 10.1086/378705. doi:10.1086/378705 [DOI] [PubMed] [Google Scholar]
- Hafeez F.Y, Naeem F.I, Naeem R, Zaidi A.H, Malik K.A. Symbiotic effectiveness and bacteriocin production by Rhizobium leguminosarum bv. viciae isolated from agriculture soils in Faisalabad. Environ. Exp. Bot. 2005;54:142–147. doi:10.1016/j.envexpbot.2004.06.008 [Google Scholar]
- Kiers E.T, Rousseau R.A, Denison R.F. Measured sanctions: legume hosts detect quantitative variation in rhizobium cooperation and punish accordingly. Evol. Ecol. Res. 2006;8:1077–1086. [Google Scholar]
- Kimura M, Ohta T. Princeton University Press; Princeton, NJ: 1971. Theoretical aspects of population genetics. [PubMed] [Google Scholar]
- Knowlton N, Rohwer F. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 2003;162:S51–S62. doi: 10.1086/378684. doi:10.1086/378684 [DOI] [PubMed] [Google Scholar]
- Massey R.C, Buckling A, Ffrench-Constant R. Interference competition and parasite virulence. Proc. R. Soc. B. 2004;271:785–788. doi: 10.1098/rspb.2004.2676. doi:10.1098/rspb.2004.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhadhbi H, Jebara M, Limam F, Huguet T, Aouani M.E. Interaction between Medicago truncatula lines and Sinorhizobium meliloti strains for symbiotic efficiency and nodule antioxidant activities. Physiol. Plant. 2005;124:4–11. doi:10.1111/j.1399-3054.2005.00489.x [Google Scholar]
- Miller R.W, Sirois J.C. Relative efficiency of different alfalfa cultivar–Rhizobium meliloti strain combinations for symbiotic nitrogen fixation. Appl. Environ. Microbiol. 1982;43:764–768. doi: 10.1128/aem.43.4.764-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky J, Bever J.D, Antonovics J. Coexistence under positive frequency dependence. Proc. R. Soc. B. 2001;268:273–277. doi: 10.1098/rspb.2000.1355. doi:10.1098/rspb.2000.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser C, Fargione J.E. A mutualism–parasitism continuum model and its application to plant–mycorrhizal interactions. Ecol. Model. 2004;177:337–352. doi:10.1016/j.ecolmodel.2004.02.010 [Google Scholar]
- Noë R, Hammerstein P. Biological markets. Trends Ecol. Evol. 1995;10:336–339. doi: 10.1016/s0169-5347(00)89123-5. [DOI] [PubMed] [Google Scholar]
- Nuismer S.L, Gomulkiewicz R, Morgan M.T. Coevolution in temporally variable environments. Am. Nat. 2003;162:195–204. doi: 10.1086/376582. doi:10.1086/376582 [DOI] [PubMed] [Google Scholar]
- Parker M.A. Plant fitness variation caused by different mutualist genotypes. Ecology. 1995;76:1525–1535. doi:10.2307/1938154 [Google Scholar]
- Pellmyr O, Huth C.J. Evolutionary stability of mutualism between yuccas and yucca moths. Nature. 1994;372:257–260. doi:10.1038/372257a0 [Google Scholar]
- Robinson K.O, Beyene D.A, van Berkum P, Knight-Mason R, Bhardwaj H.L. Variability in plant–microbe interaction between Lupinus lines and Bradyrhizobium strains. Plant Sci. 2000;159:257–264. doi: 10.1016/s0168-9452(00)00345-9. doi:10.1016/S0168-9452(00)00345-9 [DOI] [PubMed] [Google Scholar]
- Ronfort, J., Bataillon, T., Santoni, S., Delalande, M., David, J. L. & Prosperi, J. 2006 Microsatellite diversity and broad scale geographic strcutre in a model legume: building a set of nested core collections for studying naturally occurring variation in Medicago truncatula BMC Plant Biol 6, 28. [DOI] [PMC free article] [PubMed]
- Sachs J.L, Mueller U.G, Wilcox T.P, Bull J.J. The evolution of cooperation. Q. Rev. Biol. 2004;79:135–160. doi: 10.1086/383541. doi:10.1086/383541 [DOI] [PubMed] [Google Scholar]
- Schwinghamer E.A, Brockwell J. Competitive advantage of bacteriocin and phage producing strains of Rhizobium trifolii in mixed culture. Soil Biol. Biochem. 1978;10:383–387. doi:10.1016/0038-0717(78)90062-7 [Google Scholar]
- Simms E.L, Taylor D.L. Partner choice in nitrogen-fixation mutualisms of legumes and rhizobium. Int. Comp. Biol. 2002;42:369–380. doi: 10.1093/icb/42.2.369. doi:10.1093/icb/42.2.369 [DOI] [PubMed] [Google Scholar]
- Simms E.L, Taylor D.L, Povich J, Shefferson R.P, Sachs J.L, Urbina M, Tausczik Y. An empirical test of partner choice mechanisms in a wild legume–rhizobium interaction. Proc. R. Soc. B. 2006;273:77–81. doi: 10.1098/rspb.2005.3292. doi:10.1098/rspb.2005.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.P, Goodman R.M. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 1999;37:473–491. doi: 10.1146/annurev.phyto.37.1.473. doi:10.1146/annurev.phyto.37.1.473 [DOI] [PubMed] [Google Scholar]
- Spoerke J.M, Wilkinson H.H, Parker M.A. Nonrandom genotypic associations in a legume–Bradyrhizobium mutualism. Evolution. 1996;50:146–154. doi: 10.1111/j.1558-5646.1996.tb04481.x. doi:10.2307/2410789 [DOI] [PubMed] [Google Scholar]
- Thompson J.N. Variation in interspecific interactions. Annu. Rev. Ecol. Syst. 1988;19:65–87. doi:10.1146/annurev.es.19.110188.000433 [Google Scholar]
- Trivers R.L. The evolution of reciprocal altruism. Q. Rev. Biol. 1970;46:35–57. [Google Scholar]
- Vance C.P. Root–bacteria interactions: symbiotic N2 fixation. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots: the hidden half. Marcel Dekker, Inc; New York, NY: 2002. pp. 839–868. [Google Scholar]
- Vincent J.M. Blackwell Scientific Publications; Oxford and Edinburgh, UK: 1970. A manual for the practical study of root-nodule bacteria. [Google Scholar]
- West S.A, Kiers E.T, Simms E.L, Denison R.F. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. B. 2002a;269:685–694. doi: 10.1098/rspb.2001.1878. doi:10.1098/rspb.2001.1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.A, Kiers E.T, Pen I, Denison R.F. Sanctions and mutualism stability: when should less beneficial mutualists be tolerated? J. Evol. Biol. 2002b;15:830–837. doi:10.1046/j.1420-9101.2002.00441.x [Google Scholar]
- Wilkinson H.H, Spoerke J.M, Parker M.A. Divergence in symbiotic compatibility in a legume–Bradyrhizobium mutualism. Evolution. 1996;50:1470–1477. doi: 10.1111/j.1558-5646.1996.tb03920.x. doi:10.2307/2410884 [DOI] [PubMed] [Google Scholar]
- Wilson T.B, Thompson T.L. Soil nutrient distributions of mesquite-dominated desert grasslands: changes in time and space. Geoderma. 2005;126:301–315. doi:10.1016/j.geoderma.2004.10.002 [Google Scholar]
- Young N.D, Cannon S.B, Sato S, Kim D, Cook D.R, Town C.D, Roe B.A, Tabata S. Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Phys. 2005;137:1174–1181. doi: 10.1104/pp.104.057034. doi:10.1104/pp.104.057034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.