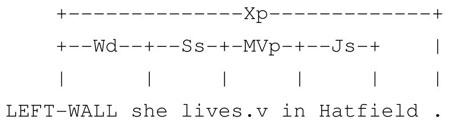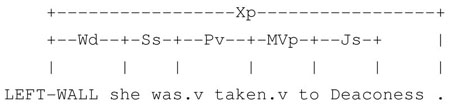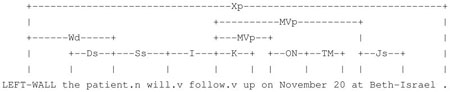Abstract
Objective
Clinical records contain significant medical information that can be useful to researchers in various disciplines. However, these records also contain personal health information (PHI) whose presence limits the use of the records outside of hospitals.
The goal of de-identification is to remove all PHI from clinical records. This is a challenging task because many records contain foreign and misspelled PHI; they also contain PHI that are ambiguous with non-PHI. These complications are compounded by the linguistic characteristics of clinical records. For example, medical discharge summaries, which are studied in this paper, are characterized by fragmented, incomplete utterances and domain-specific language; they cannot be fully processed by tools designed for lay language.
Methods and Results
In this paper, we show that we can de-identify medical discharge summaries using a de-identifier, Stat De-id, based on support vector machines and local context (F-measure = 97% on PHI). Our representation of local context aids de-identification even when PHI include out-of-vocabulary words and even when PHI are ambiguous with non-PHI within the same corpus. Comparison of Stat De-id with a rule-based approach shows that local context contributes more to de-identification than dictionaries combined with hand-tailored heuristics (F-measure = 85%). Comparison with two well-known named entity recognition (NER) systems, SNoW (F-measure = 94%) and IdentiFinder (F-measure = 36%), on five representative corpora show that when the language of documents is fragmented, a system with a relatively thorough representation of local context can be a more effective de-identifier than systems that combine (relatively simpler) local context with global context. Comparison with a Conditional Random Field De-identifier (CRFD), which utilizes global context in addition to the local context of Stat De-id, confirms this finding (F-measure = 88%) and establishes that strengthening the representation of local context may be more beneficial for de-identification than complementing local with global context.
Keywords: automatic de-identification of narrative patient records, local lexical context, local syntactic context, dictionaries, sentential global context, syntactic information for de-identification
1 Introduction
Medical discharge summaries can be a major source of information for many studies. However, like all other clinical records, discharge summaries contain explicit personal health information (PHI) which, if released, would jeopardize patient privacy. In the United States, the Health Information Portability and Accountability Act (HIPAA) provides guidelines for protecting the confidentiality of patient records. Paragraph 164.514 of the Administrative Simplification Regulations promulgated under the HIPAA states that for data to be treated as de-identified, it must clear one of two hurdles.
An expert must determine and document “that the risk is very small that the information could be used, alone or in combination with other reasonably available information, by an anticipated recipient to identify an individual who is a subject of the information.”
Or, the data must be purged of a specified list of seventeen categories of possible identifiers, i.e., PHI, relating to the patient or relatives, household members and employers, and any other information that may make it possible to identify the individual [1]. Many institutions consider the clinicians caring for a patient and the names of hospitals, clinics, and wards to fall under this final category because of the heightened risk of identifying patients from such information [2, 3].
Of the seventeen categories of PHI listed by HIPAA, the following appear in medical discharge summaries: first and last names of patients, of their health proxies, and of their family members; identification numbers; telephone, fax, and pager numbers; geographic locations; and dates. In addition, names of doctors and hospitals are frequently mentioned in discharge summaries; for this study, we add them to the list of PHI. Given discharge summaries, our goal is to find the above listed PHI and to replace them with either anonymous tags or realistic surrogates.
Medical discharge summaries are characterized by fragmented, incomplete utterances and domain-specific language. As such, they cannot be effectively processed by tools designed for lay language text such as news articles [4]. In addition, discharge summaries contain some words that can appear both as PHI and non-PHI within the same corpus, e.g., the word Huntington can be both the name of a person, “Dr. Huntington”, and the name of a disease, “Huntington’s disease”. They also contain foreign and misspelled words as PHI, e.g., John misspelled as Jhn and foreign variants such as Ioannes. These complexities pose challenges to de-identification.
An ideal de-identification system needs to identify PHI perfectly. However, while anonymizing the PHI, such a system needs to also protect the integrity of the data by maintaining all of the non-PHI, so that medical records can later be processed and retrieved based on their inclusion of these terms. Almost all methods that determine whether a target word2, i.e., the word to be classified as PHI or non-PHI, is PHI base their decision on a combination of features related to the target itself, to words that surround the target, and to discourse segments containing the target. We call the features extracted from the words surrounding the target and from the discourse segment containing the target the context of the target. In this paper, we are particularly interested in comparing methods that rely on what we call local context, by which we mean the words that immediately surround the target (local lexical context) or that are linked to it by some immediate syntactic relationship (local syntactic context), and global context, which refers to the relationships of the target with the contents of the discourse segment containing the target. For example, the surrounding k-tuples of words to the left and right of a target are common components of local context, whereas a model that selects the highest probability interpretation of an entire sentence by a Markov model employs sentential global context (where the discourse segment is a sentence).
In this paper, we present a de-identifier, Stat De-id, which uses local context to de-identify medical discharge summaries. We treat de-identification as a multi-class classification task; the goal is to consider each word in isolation and to decide whether it represents a patient, doctor, hospital, location, date, telephone, ID, or non-PHI. We use Support Vector Machines (SVMs), as implemented by LibSVM [5], trained on human-annotated data as a means to this end.
Our representation of local context benefits from orthographic, syntactic, and semantic characteristics of each target word and the words within a ±2 context window of the target. Other models of local context have used the features of words immediately adjacent to the target word; our representation is more thorough as it includes (for a ±2 context) local syntactic context, i.e., the features of words that are linked to the target by syntactic relations identified by a parse of the sentence. This novel representation of local syntactic context uses the Link Grammar Parser [6], which can provide at least a partial syntactic parse even for incomplete and fragmented sentences [7]. Note that syntactic parses can be generally regarded as sentential features. However, in our corpora, more than 40% of the sentences only partially parse. The features extracted from such partial parses represent phrases rather than sentences and contribute to local context. For sentences that completely parse, our representation benefits from syntactic parses only to the extent that they help us relate the target to its immediate neighbors (within 2 links), again extracting local context.
On five separate corpora obtained from Partners Healthcare and Beth Israel Deaconess Medical Center, we show that despite the fragmented and incomplete utterances and the domain-specific language that dominate the text of discharge summaries, we can capture the patterns in the language of these documents by focusing on local context; we can use these patterns for de-identification. Stat De-id, presented in this paper, is built on this hypothesis. It finds more than 90% of the PHI even in the face of ambiguity between PHI and non-PHI, and even in the presence of foreign words and spelling errors in PHI.
We compare Stat De-id with a rule-based heuristic+dictionary approach [8] two named entity recognizers, SNoW [9] and IdentiFinder [10], and a Conditional Random Field De-identifier (CRFD). SNoW and IdentiFinder also use local context; however, their representation of local context is relatively simple and, for named entity recognition (NER), is complemented with information from sentential global context, i.e., the dependencies of entities with each other and with non-entity tokens in a single sentence. CRFD, developed by us for the studies presented in this paper, employs the exact same local context used by Stat De-id and reinforces this local context with sentential global context. In this manuscript, we refer to sentential global context simply as global context. Because medical discharge summaries contain many short, fragmented sentences, we hypothesize that global context will add limited value to local context for de-identification, and that strengthening the representation of local context will be more effective for improving de-identification. We present experimental results to support this hypothesis: On our corpora, Stat De-id significantly outperforms all of SNoW, IdentiFinder, CRFD, and the heuristic+dictionary approach.
The performance of Stat De-id is encouraging and can guide research in identification of entities in corpora with fragmented, incomplete utterances and even domain-specific language. Our results show that even on such corpora, it is possible to create a useful representation of local context and to identify the entities indicated by this context.
2 Background and Related Work
A number of investigators have developed methods for de-identifying medical corpora or for recognizing named entities in non-clinical text (which can be directly applied to at least part of the de-identification problem). The two main approaches taken have been either (a) use of dictionaries, pattern matching, and local rules or (b) statistical methods trained on features of the word(s) in question and their local or global context. Our work on Stat De-id falls into the second of these traditions and differs from others mainly in its use of novel local context features determined from a (perhaps partial) syntactic parse of the text.
2.1 De-identification
Most de-identification systems use dictionaries and simple contextual rules to recognize PHI [8, 11]. Gupta et al. [11], for example, describe the DeID system which uses the U.S. Census dictionaries to find proper names, employs patterns to detect phone numbers and zip codes, and takes advantage of contextual clues (such as section headings) to mark doctor and patient names. Gupta et al. report that, after scrubbing with DeID, of the 300 reports scrubbed, two reports still contained accession numbers, two reports contained clinical trial names, three reports retained doctors’ names, and three reports contained hospital or lab names.
Beckwith et al. [12] present a rule-based de-identifier for pathology reports. Unlike our discharge summaries, pathology reports contain significant header information. Beckwith et al. identify PHI that appear in the headers (e.g., medical record number and patient name) and remove the instances of these PHI from the narratives. They use pattern-matchers to find dates, IDs, and addresses; they utilize well-known markers such as Mr., MD, and PhD to find patient, institution, and physician names. They conclude their scrubbing by comparing the narrative text with a database of proper names. Beckwith et al. report that they remove 98.3% of unique identifiers in pathology reports from three institutions. They also report that on average 2.6 non-PHI phrases per record are removed.
The de-identifier of Berman [13] takes advantage of standard nomenclature available in UMLS. This system assumes that words that do not correspond to nomenclature and that are not in a standard list of stop words are PHI and need to be removed. As a result, this system produces a large number of false positives.
Sweeney’s Scrub system [3] employs numerous experts each of which specializes in recognizing a single class of personally-identifying information, e.g., person names. Each expert uses lexicons and morphological patterns to compute the probability that a given word belongs to the personally-identifying information class it specializes in. The expert with the highest probability determines the class of the word. On a test corpus of patient records and letters, Scrub identified 99–100% of personally-identifying information. Unfortunately, Scrub is a proprietary system and is not readily available for use.
To identify patient names, Taira et al. [14] use a lexical analyzer that collects name candidates from a database and filters out the candidates that match medical concepts. They refine the list of name candidates by applying a maximum entropy model based on semantic selectional restrictions—the hypothesis that certain word classes impose semantic constraints on their arguments, e.g., the verb vomited implies that its subject is a patient. They achieve a precision of 99.2% and recall of 93.9% on identification of patient names in a clinical corpus.
De-identification resembles NER. NER is the task of identifying entities such as people, places, and organizations in narrative text. Most NER tasks are performed on news and journal articles. However, given the similar kinds of entities targeted by de-identification and NER, NER approaches can be relevant to de-identification.
2.2 Named Entity Recognition
Much NER work has been inspired by the Message Understanding Conference (MUC) and by the Entity Detection and Tracking task of Automatic Content Extraction (ACE) conference organized by the National Institute of Standards and Technology. Technologies developed for ACE-2007, for example, have been designed for and evaluated on several individual corpora: a 65000-word Broadcast News corpus, a 47500-word Broadcast Conversations corpus, a 60000-word Newswire corpus, a 47500-word Weblog corpus, a 47500-word Usenet corpus, and a 47500-word Conversational Telephone Speech corpus [15].
One of the most successful named entity recognizers, among the NER systems developed for and outside of MUC and ACE, is IdentiFinder [10]. IdentiFinder uses a Hidden Markov Model (HMM) to learn the characteristics of names that represent entities such as people, locations, geographic jurisdictions, organizations, and dates. For each entity class, IdentiFinder learns a bigram language model, where a word is defined as a combination of the actual lexical unit and various orthographic features. To find the names and classes of all entities, IdentiFinder computes the most likely sequence of entity classes in a sentence given the observed words and their features. The information obtained from the entire sentence constitutes IdentiFinder’s global context.
Isozaki and Kazawa [16] use SVMs to recognize named entities in Japanese text. They determine the entity type of each target word by employing features of the words within two words of the target (a ±2 word window). The features they use include the part of speech and the structure of the word, as well as the word itself.
Roth and Yih’s SNoW system [9] labels the entities and their relationships in a sentence. The relationships expressed in the sentence constitute SNoW’s global context and aid it in creating a final hypothesis about the entity type of each word. SNoW recognizes names of people, locations, and organizations.
Our de-identification solution combines the strengths of some of the abovementioned systems. Like Isozaki et al., we use SVMs to identify the class of individual words (where the class is one of seven categories of PHI or the class non-PHI); we use orthographic information as well as part of speech and local context as features. Like Taira et al., we hypothesize that PHI categories are characterized by their local lexical and syntactic context. However, our approach to de-identification differs from prior NER and de-identification approaches in its use of deep syntactic information obtained from the output of the Link Grammar Parser [6]. We benefit from this information to capture local syntactic context even when parses are partial, i.e., input text contains fragmented and incomplete utterances. We enrich local lexical context with local syntactic context and thus create a more thorough representation of local context. We use our newly defined representation of local context to identify PHI in clinical text.
3 Definitions
We define the PHI found in medical discharge summaries as follows:
Patients: include the first and last names of patients, their health proxies, and family members. Titles, such as Mr., are excluded, e.g., “Mrs. [Lunia Smith]patient was …”.
Doctors: include medical doctors and other practitioners. Again titles, such as Dr., are not considered part of PHI, e.g., “He met with Dr. [John Doe]doctor”.
Hospitals: include names of medical organizations. We categorize the entire institution name as PHI including common words such as hospital, e.g., “She was admitted to [Brigham and Women’s Hospital]hospital”.
IDs: refer to any combination of numbers and letters identifying medical records, patients, doctors, or hospitals, e.g., “Provider Number: [12344]ID”.
Dates: HIPAA specifies that years are not considered PHI, but all other elements of a date are. We label a year appearing in a date as PHI if the date appears as a single lexical unit, e.g., 12/02/99, and as non-PHI if the year exists as a separate token, e.g., 23 March, 2006. This decision was motivated by the fact that many solutions to de-identification and NER classify entire tokens as opposed to segments of a token. Also, once identified, dates such as 12/02/99 can be easily post-processed to separate the year from the rest.
Locations: include geographic locations such as cities, states, street names, zip codes, and building names and numbers, e.g., “He lives in [Newton]location”.
Phone numbers: include telephone, pager, and fax numbers.
4 Hypotheses
We hypothesize that we can de-identify medical discharge summaries even when the documents contain many fragmented and incomplete utterances, even when many words are ambiguous between PHI and non-PHI, and even in the presence of foreign words and spelling errors in PHI. Given the nature of the domain-specific language of discharge summaries, we hypothesize that a thorough representation of local context will be more effective for de-identification than (relatively simpler) local context enhanced with global context; in this manuscript, local context refers to the characteristics of the target and of the words within a ±2 context window of the target whereas global context refers to the dependencies of entities with each other and with non-entity tokens in a sentence.
5 Corpora
We tested our methods on five different corpora, three of which were developed from a corpus of 48 discharge summaries from various medical departments at the Beth Israel Deaconess Medical Center (BIDMC), the fourth of which consisted of authentic data including actual PHI from 90 discharge summaries of deceased patients from Partners HealthCare, and the fifth of which came from a corpus of 889 de-identified discharge summaries, also from Partners. The sizes of these corpora and the distribution of PHI within them are shown in Table 1. The collection and use of these data were approved by the Institutional Review Boards of Partners, BIDMC, State University of New York at Albany, and Massachusetts Institute of Technology.
Table 1.
Number of words in each PHI category in the corpora. Word counts depend on the number and format of inserted surrogates.
| Category | Number of tokens | ||||
|---|---|---|---|---|---|
| Random corpus | Ambiguous corpus | Out-of-vocabulary corpus | Authentic corpus | Challenge corpus | |
| Non-PHI | 17,874 | 19,275 | 17,875 | 112,669 | 444,127 |
| Patient | 1,048 | 1,047 | 1,037 | 294 | 1,737 |
| Doctor | 311 | 311 | 302 | 738 | 7,697 |
| Location | 24 | 24 | 24 | 88 | 518 |
| Hospital | 600 | 600 | 404 | 656 | 5,204 |
| Date | 735 | 736 | 735 | 1,953 | 7,651 |
| ID | 36 | 36 | 36 | 482 | 5,110 |
| Phone | 39 | 39 | 39 | 32 | 271 |
A successful de-identification scheme must achieve two competing objectives: it must anonymize all PHI in the text; however, it must leave intact the non-PHI. Two of the major challenges to achieving these objectives in medical discharge summaries are the existence of ambiguous PHI and the existence of out-of-vocabulary PHI.
Four of our corpora were specifically created to test our system in the presence of these challenges. Three of these artificial corpora were based on the corpus of 48 already de-identified discharge summaries from BIDMC. In this corpus, the PHI had been replaced by [REMOVED] tags (see excerpt below). This replacement had been performed semi-automatically. In other words, the PHI had been removed by an automatic system [8] and the output had been manually scrubbed. Before studying this corpus, our team confirmed its correctness.
HISTORY OF PRESENT ILLNESS: The patient is a 77-year-old woman with long standing hypertension who presented as a Walk-in to me at the [REMOVED] Health Center on [REMOVED]. Recently had been started q.o.d. on Clonidine since [REMOVED] to taper off of the drug. Was told to start Zestril 20 mg. q.d. again. The patient was sent to the [REMOVED] Unit for direct admission for cardioversion and anticoagulation, with the Cardiologist, Dr. [REMOVED] to follow.
SOCIAL HISTORY: Lives alone, has one daughter living in [REMOVED]. Is a non-smoker, and does not drink alcohol.
HOSPITAL COURSE AND TREATMENT: During admission, the patient was seen by Cardiology, Dr. [REMOVED], was started on IV Heparin, Sotalol 40 mg PO b.i.d. increased to 80 mg b.i.d., and had an echocardiogram. By [REMOVED] the patient had better rate control and blood pressure control but remained in atrial fibrillation. On [REMOVED], the patient was felt to be medically stable.
…
We used the definitions of PHI classes in conjunction with local contextual clues to identify the PHI category corresponding to each of the [REMOVED] phrases in this corpus. We used dictionaries of common names from the U.S. Census Bureau, dictionaries of hospitals and locations from online sources, and lists of diseases, treatments, and diagnostic tests from the UMLS Metathesaurus to generate surrogate PHI for three corpora: a corpus populated with random surrogate PHI [8], a corpus populated with ambiguous surrogate PHI, and a corpus populated with out-of-vocabulary surrogate PHI. The surrogate PHI inserted into each of the corpora represent the common patterns associated with each PHI class. The name John K. Smith, for example, can appear as John K. Smith, J. K. Smith, J. Smith, Smith, John, etc. The date July 5th 1982 can be expressed as July 5, 5th of July, 07/05/82, 07-05-82, etc.
5.1 Corpus Populated with Random PHI
We randomly selected names of people from a dictionary of common names from the U.S. Census Bureau, and names of hospitals and locations from online dictionaries in order to generate surrogate PHI for the corpus with random PHI (Details of these dictionaries can be found in Dictionary Information section). After manually tagging the PHI category of each [REMOVED] phrase, we replaced each [REMOVED] with a random surrogate from the correct PHI category and dictionary. In the rest of this paper, we refer to this corpus as the random corpus. The first column of Table 1 shows the breakdown of PHI in the random corpus.
5.2 Corpus Populated with Ambiguous PHI
To generate a corpus containing ambiguous PHI, two graduate students marked medical concepts corresponding to diseases, tests, and treatments in the de-identified corpus. Agreement, as measured by Kappa, on marking these concepts was 93%. The annotators discussed and resolved their differences, generating a single gold standard for medical concepts.
We used the marked medical concepts to generate ambiguous surrogate PHI with which to populate the de-identified corpus. In addition to the people, hospital, and location dictionaries employed in generating the random corpus, we also used lists of diseases, treatments, and diagnostic tests from the UMLS Metathesaurus in order to locate examples of medical terms that occur in the narratives of our records and to deliberately inject these terms into the surrogate patients, doctors, hospitals, and locations (with appropriate formatting). This artificially enhanced the occurrence of challenging examples such as “Mr. Huntington suffers from Huntington’s disease” where the first occurrence of “Huntington” is a PHI and the second is not. The ambiguous terms we have injected into the corpus were guaranteed to appear both as PHI and as non-PHI in this corpus.
In addition to the ambiguities resulting from injection of medical terms into patients, doctors, hospitals, and locations, this corpus also already contained ambiguities between dates and non-PHI. Many dates appear in the format 11/17, a common format for reporting medical measurements. In our corpus, 14% of dates are ambiguous with non-PHI. After injection of ambiguous medical terms, 49% of patients, 79% of doctors, 100% of locations, and 14% of hospitals are ambiguous with non-PHI. In return, 20% of non-PHI are ambiguous with PHI. In the rest of this paper, we refer to this corpus as the ambiguous corpus. The second column of Table 1 shows the distribution of PHI in the ambiguous corpus; Table 2 shows the distribution of tokens that are ambiguous between PHI and non-PHI.
Table 2.
Distribution of words, i.e., tokens, that are ambiguous between PHI and non-PHI.
| Category | Number of ambiguous tokens in the ambiguous corpus | Number of ambiguous tokens in the challenge corpus |
|---|---|---|
| Non-PHI | 3,787 | 39,374 |
| Patient | 514 | 158 |
| Doctor | 247 | 1,083 |
| Location | 24 | 44 |
| Hospital | 86 | 1,910 |
| Date | 201 | 81 |
| ID | 0 | 4 |
| Phone | 0 | 1 |
5.3 Corpus Populated with Out-of-Vocabulary PHI
The corpus containing out-of-vocabulary PHI was created by the same process used to generate the random corpus. However, instead of using dictionaries, we generated surrogates by randomly selecting word lengths and letters, e.g., “O. Ymfgi was admitted …”. Almost all generated patient, doctor, location, and hospital names were consequently absent from common dictionaries. In the rest of this paper, we refer to this corpus as the out-of-vocabulary (OoV) corpus. The third column of Table 1 shows the distribution of PHI in the OoV corpus.
5.4 Authentic Discharge Summary Corpus
In addition to the artificial corpora, we obtained and used a corpus of authentic discharge summaries with genuine PHI about deceased patients. In the rest of this paper, we refer to this corpus as the authentic corpus.
The authentic corpus contained approximately 90 discharge summaries of various lengths from various medical departments from Partners HealthCare. This corpus differed from the artificial corpora obtained from BIDMC in both the writing style and in the distribution and frequency of use of PHI. However, it did contain the same basic categories of PHI. Three annotators manually marked the PHI in this corpus so that each record was marked three times. Agreement among the annotators, as measured by Kappa, was 100%. As with artificial corpora, we automatically de-identified the narrative portions of these records. The fourth column of Table 1 shows the break-down of PHI in the authentic discharge summary corpus.
5.5 Challenge Corpus
Finally, we obtained and used a separate, larger corpus of 889 discharge summaries, again from Partners HealthCare. This corpus, which had formed the basis for a workshop and shared-task on de-identification organized at the 2006 AMIA Fall Symposium, had been manually de-identified and all authentic PHI in it had been replaced with realistic out-of-vocabulary or ambiguous surrogates [17]. Of the surrogate PHI tokens in this corpus, 73% of patients, 67% of doctors, 56% of locations, and 49% of hospitals were out-of-vocabulary. 10% of patients, 15% of doctors, 10% of locations, and 37% of hospitals were ambiguous with non-PHI. This corpus thus combined the challenges of out-of-vocabulary and ambiguous PHI de-identification. In the rest of this paper, we refer to this corpus as the challenge corpus. The fifth column of Table 1 shows the breakdown of PHI in the challenge discharge summary corpus.
In general, our corpora include non-uniform representation of various PHI categories. What is more, in terms of overall number of tokens, although our authentic and challenge corpora are larger than the standard corpora used for NER shared-tasks organized by NIST, our random, ambiguous, and out-of-vocabulary corpora contain very few examples of some of the PHI categories, e.g., 24 examples of locations. Therefore, in this manuscript, while we maintain the distinction among the PHI categories for classification, we report results on the aggregate set of PHI consisting of patients, doctors, locations, hospitals, dates, IDs, and phone numbers. We measure the performance of systems in differentiating this aggregate set of PHI from non-PHI. We report significance test results on the aggregate set of PHI and on non-PHI separately. Finally, we analyze the performance of our system on individual PHI categories only to understand its strengths and weaknesses on our data so as to identify potential courses of action for future work.
While access to more and larger corpora is desirable, freely-available corpora for training de-identifiers are not common, and until de-identification research becomes more successful and accepted, it will require large investments in human reviewers to create them. Even then, multiple rounds of human review of the records may not be satisfactory for the Institutional Review Boards to allow widespread and unhindered use of clinical records for de-identification research. Even for those who can obtain the data, the use of the records may be limited to a particular task [17].
6 Methods: Stat De-id
Categories of PHI are often characterized by local context. For example, the word Dr. before a name invariably suggests that the name is that of a doctor. While titles such as Dr. provide easy context markers, other clues may not be as straightforward, especially when the language of documents is dominated by fragmented and incomplete utterances. We created a representation of local context that is useful for recognizing PHI even in fragmented, incomplete utterances.
We devised Stat De-id, a de-identifier that uses SVMs, to classify each word in the sentence as belonging to one of eight categories: doctor, location, phone, address, patient, ID, hospital, or non-PHI. Stat De-id uses features of the target, as well as features of the words surrounding the target in order to capture the contextual clues human annotators found useful in de-identification. We refer to the features of the target and its close neighbors as local context.
Stat De-id is distinguished from similar approaches in its use of syntactic information extracted from the Link Grammar Parser [6]. Despite the fragmented nature of the language of discharge summaries, we can obtain (partial) syntactic parses from the Link Grammar Parser [7] and we can use this information in creating a representation of local context. We augment the syntactic information with semantic information from medical dictionaries, such as the Medical Subject Headings (MeSH) of the Unified Medical Language System (UMLS) [18]. Stat De-id will be freely available through the i2b2 Hive, https://www.i2b2.org/resrcs/hive.html, a common tools distribution mechanism of the National Centers for Biological Computing project on Informatics for Integrating Biology and the Bedside (i2b2) that partially funded its development.
6.1 Support Vector Machines
Given a collection of data points represented by multi-dimensional vectors and class labels, (xi, yi), i = 1, …, l where xi ∈ Rn and yi ∈ {1, −1}l, SVMs [5, 19] optimize:
| (1) |
| (2) |
where C>0 is the penalty parameter, ξi is a measure of misclassification error, and w is the normal to the plane. This optimization maps input training vectors, xi, to a higher dimensional space given by the function, φ. SVMs find a hyperplane that in this space best separates the data points according to their class. To prevent over-fitting, the hyperplane is chosen so as to maximize the distance between the hyperplane and the closest data point in each class. The data points that are closest to the discovered hyperplane are called the support vectors. Given a data point whose class is unknown, an SVM determines on which side of the hyperplane the point lies and labels it with the corresponding class [19]. The kernel function:
| (3) |
plays a role in determining the optimal hyperplane and encodes a “similarity measure between two data points” [20]. In this paper, we explore a high dimensional feature space which can be prone to over-fitting. To minimize this risk, we employ the linear kernel
| (4) |
and investigate the impact of various features on de-identification.
The choice of SVMs over other classifiers is motivated by their capability to robustly handle large feature sets; in our case, the number of features in the set is on the order of thousands. SVMs tend to be robust to the noise that is frequently present in such high dimensional feature sets [19]. In addition, while “classical learning systems like neural networks suffer from their theoretical weakness, e.g., back-propagation usually converges only to locally optimal solutions.” [21], in comparison to other neural classifiers, SVMs are often more successful in finding globally optimum solutions [22].
Conditional Random Fields (CRF) [23] provide a viable alternative to SVMs. Just like SVMs, CRFs can “handle many dependent features”; however, unlike SVMs, they also can “make joint inference over entire sequences” [24]. In our case, predictions over entire sequences correspond to global context. Given our interest in using local context, for the purposes of this manuscript, we focus primarily on SVMs. We use CRFs as a basis for comparison, in order to gauge the contribution of sentential global context to local context. We employ the multi-class SVM implementation of LIBSVM [5]. This implementation builds a multi-class classifier from several binary classifiers using one-against-one voting.
6.2 Knowledge Representation
We use a vector to represent our features for use with an SVM. In this vector, each row corresponds to a single target and each column represents the possible values of all features of all targets in the training corpus. For example, suppose the first feature under consideration is dictionary information, i.e., the dictionaries that the target appears in, and the second is the part of speech of the target. Let w be the number of unique dictionaries relevant to the training corpus, and p be the number of unique parts of speech in the training corpus. Then, the first w columns of the feature vector represent the possible dictionaries. We mark the dictionaries that contain the target by setting the value of their entry(ies) (where an entry is the intersection of a row and a column) to one; all other dictionary entries for that target will be zero. Similarly, let’s assume that the next p columns of the vector represent the possible values of parts of speech extracted from the training corpus; we mark the part of speech of the target by setting that entry to one and leaving the other part-of-speech entries at zero.
The vector that is fed into the SVM is concatenation of individual feature vectors that capture the target itself, the lexical bigrams of the target, use of capitalization, punctuation, or numbers in the target, and the length of the target, part-of-speech of the target, as well as syntactic bigrams, MeSH IDs, dictionary information, and section headings of the target.
We evaluate our system using cross-validation. At each round of cross-validation, we recreate the feature vector based on the training corpus used for that round, i.e., the feature vector does not overfit to the validation set.
6.3 Lexical and Orthographic Features
6.3.1 The target itself
Some words consistently occur as non-PHI. Taking the word itself into consideration allows the classifier to learn that certain words, such as and and they, are never PHI.
We incorporate the target word feature into our knowledge representation using a vector of all unique words in the training corpus. We mark unique words after normalization using UMLS’s Norm [18]. We mark each target word feature by setting the value of the entry corresponding to the target to one and leaving all other entries at zero.
6.3.2 Lexical Bigrams
The context of the target can reveal its identity. For example, in a majority of the cases, the bigram admitted to is followed by the name of a hospital. Similarly, the bigram was admitted is preceded by the patient. To capture such indicators of PHI, we consider uninterrupted strings of two words occurring before and after the target. We refer to these strings as lexical bigrams.
We keep track of lexical bigrams using a vector that contains entries for both left and right lexical bigrams of the target. The columns of the vector correspond to all lexical bigrams that are observed in the training corpus. We mark the left and right lexical bigrams of a target by setting their entries to one and leaving the rest of the lexical bigram entries at zero.
6.3.3 Capitalization
Orthographic features such as capitalization can aid identification of PHI. Most names, i.e., names of locations as well as people, usually begin with a capital letter. We represent capitalization information in the form of a single column vector which for each target (row) contains an entry of one if the target is capitalized and zero if it is not.
6.3.4 Punctuation
Dates, phone numbers, and IDs tend to contain punctuation. Including information about the presence or absence of “−” or “/” in the target helps us recognize these categories of PHI. Punctuation information is incorporated into the knowledge representation in a similar manner to capitalization.
6.3.5 Numbers
Dates, phone numbers, and IDs consist of numbers. Information about the presence or absence of numbers in the target can help us assess the probability that the target belongs to one of these PHI categories. Presence of numbers in a target is incorporated into the knowledge representation in a similar manner to capitalization.
6.3.6 Word Length
Certain entities are characterized by their length, e.g., phone numbers. For each target, we mark its length in terms of characters by setting the vector entry corresponding to its length to one.
6.4 Syntactic Features
6.4.1 Part of Speech
Most PHI instances are more likely to be nouns than adjectives or verbs. We obtain information about the part of speech of words using the Brill tagger [25]. Brill first uses lexical lookup to assign to each word its most likely part-of-speech tag; it then refines each tag, as necessary, based on the tags immediately surrounding it.
In addition to the part of speech of the target, we also consider the parts of speech of the words within a ±2 context window of the target. This information helps us capture some syntactic patterns without fully parsing the text. We include part of speech information in our knowledge representation via a vector that contains entries for all parts of speech present in the training corpus. We mark the part of speech of a target by setting its entry to one and leaving the rest of the part-of-speech entries in the vector at zero.
6.4.2 Syntactic Bigrams
Syntactic bigrams capture the local syntactic dependencies of the target, and we hypothesize that particular types of PHI in discharge summaries occur within similar syntactic structures. For example, patients are often the subject of the passive construction was admitted, e.g., “John was admitted yesterday”. The same syntactic dependency exists in the sentence “John, who had hernia, was admitted yesterday”, despite the differences in the immediate lexical context of John and was admitted.
Link Grammar Parser
To extract syntactic dependencies between words, we use the Link Grammar Parser. This parser’s computational efficiency, robustness, and explicit representation of syntactic dependencies make it appealing for use even on our fragmented text [7, 26, 27].
The Link Grammar Parser models words as blocks with left and right links. This parser imposes local restrictions on the type of links, out of 107 main link types, that a word can have with surrounding words. A successful parse of a sentence satisfies the link requirements of each word in the sentence.
The Link Grammar Parser has several features that increase robustness in the face of ungrammatical, incomplete, fragmented, or complex sentences. In particular, the lexicon contains generic definitions “for each of the major parts of speech: noun, verb, adjective, and adverb.” When the parser encounters a word that does not appear in the lexicon, it replaces the word with each of the generic definitions and attempts to find a valid parse.
This parser can also be set to enter a less scrupulous “panic mode” if a valid parse is not found within a given time limit. The panic mode comes in very handy when text includes fragmented or incomplete utterances; this mode allows the parser to suspend some of the link requirements so that it can output partial parses [7]. As we are concerned with local context, these partial parses are often sufficient.
Using Link Grammar Parser Output as an SVM Input
The Link Grammar Parser produces the following structure for the sentence:3
“John lives with his brother.”
This structure shows that the verb lives has an Ss connection to its singular subject John on the left and an MVp connection to its modifying preposition with on the right. To use such syntactic dependency information obtained from the Link Grammar Parser, we created a novel representation that captures the syntactic context, i.e., the immediate left and right dependencies, of each word. We refer to this novel representation as “syntactic n-grams”; syntactic n-grams capture all the words and links within n connections of the target. For example, for the word lives in the parsed sentence, we extract all of its immediate right connections (where a connection is a pair consisting of the link name and the word linked to)—in this case the set {(with, MVp)}. We represent the right syntactic unigrams of the word with this set of connections. For each element of the right unigram set thus extracted, we find all of its immediate right connections—in this case {(brother, Js)}. The right syntactic bigram of the word lives is then {{(with, MVp)}, {(brother, Js)}}. The left syntactic bigram of lives, obtained through a similar process, is {{(LEFT WALL, Wd)}, {(John, Ss)}}. For words with no left or right links, we create their syntactic bigrams using the two words immediately surrounding them with a link value of NONE. Note that when words have no links, this representation implicitly reverts back to uninterrupted strings of words (which we refer to as lexical n-grams).
To summarize, the syntactic bigram representation consists of: the right-hand links originating from the target; the words linked to the target through single right-hand links (call this set R1); the right-hand links originating from the words in R1; the words connected to the target through two right-hand links (call this set R2); the left-hand links originating from the target; the words linked to the target though single left-hand links (call this set L1); the left-hand links originating from the words in L1; and the words linked to the target through two left-hand links (call this set L2). The vector representation of syntactic bigrams sets the entries corresponding to L1, R1, L2, R2, and their links to target to one; the rest of the entries are set to zero.
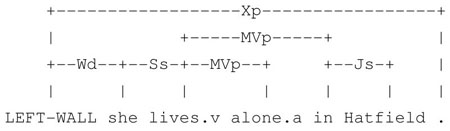
In our corpus, syntactic bigrams provide stable, meaningful local context. We find that they are particularly useful in eliminating the (sometimes irrelevant) local lexical context often introduced by relative clauses, (modifier) prepositional phrases, and adverbials. Even when local lexical context shows much variation, syntactic bigrams remain stable. For example, consider the sentences “She lives in Hatfield”, “She lives by herself in Hatfield”, and “She lives alone in Hatfield”,, which we adopted from actual examples in our data. In these sentences, the lexical bigrams of Hatfield differ; however, in all of them, Hatfield has the left syntactic bigram {{(lives, MVp), {(in, Js)}}.
“She lives in Hatfield.”
“She lives alone in Hatfield.”
“She lives by herself in Hatfield.4”
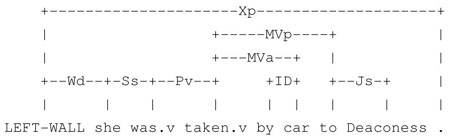
Similarly, in the sentences “She was taken to Deaconess Hospital”, “She was taken by car to Deaconess Hospital”, and “She was taken by his brother to Deaconess Hospital”, lexical local context of Deaconess varies but local syntactic context remains stable.5
“She was taken to Deaconess.”
“She was taken by car to Deaconess.”
“She was taken by her brother to Deaconess.”
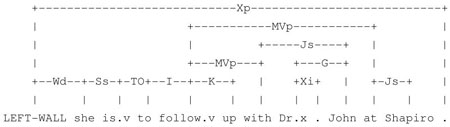
In our corpus, we find that some verbs, e.g., live, admit, discharge, transfer, follow up, etc., have stable local syntactic context which can be relied on even in the presence of much variation in local lexical context. For example, a word that has the left syntactic bigram of {{(follow, MVp), {(on, ON)}} is usually a date; a word that has the left syntactic bigram of {{(follow, MVp), {(with, Js)}} is usually a doctor; and a word that has the left syntactic bigram of {{(follow, MVp)}, {(at, Js)}} is usually a hospital.
“The patient will follow up on November 20 at Beth-Israel.6”
“She is to follow up with Dr. John at Shapiro.7”
6.5 Semantic Features
6.5.1 MeSH ID
We use the MeSH ID of the noun phrase containing the target as a feature representing the word. MeSH maps biological terms to descriptors, which are arranged in a hierarchy. There are 15 high-level categories in MeSH: e.g., A for Anatomy, B for Organism, etc. Each category is divided up to a depth of 11. MeSH descriptors have unique tree numbers which represent their position in this hierarchy. We find the MeSH ID of phrases by shallow parsing the text to identify noun phrases and exhaustively searching each phrase in the UMLS Metathesaurus. We conjecture that this feature will be useful in distinguishing medical non-PHI from PHI: unlike most PHI, medical terms such as diseases, treatments, and tests have MeSH ID’s.
We include the MeSH ID’s in our knowledge representation via a vector that contains entries for all MeSH ID’s in the training corpus. We mark the MeSH ID of a target by setting its entry to one and leaving the rest of the MeSH ID entries at zero.
6.5.2 Dictionary Information
Dictionaries are useful in detecting common PHI. We use information about the presence of the target and of words within a ±2 word window of the target in location, hospital, and name dictionaries. The dictionaries used for this purpose include:
-
A dictionary of names, from U.S. Census Bureau [29], consisting of:
1,353 male first names, including the 100 most common male first names in the U.S., covering approximately 90% of the U.S. population.
4,401 female first names, including the 100 most common female first names in the U.S., covering approximately 90% of the U.S. population.
90,000 last names, including the 100 most common last names in the U.S., covering 90% of the U.S. population.
A dictionary of locations, from U.S. Census [30] and from WorldAtlas [31], consisting of names of 3606 major towns and cities in New England (the location of the hospital from which the corpora were obtained), in the U.S., and around the world.
And, a dictionary of hospitals, from Douglass [8], consisting of names of 369 hospitals in New England.
We added to these a dictionary of dates, consisting of names and abbreviations of months, e.g., January, Jan, and names of the days of the week. The overlap of these dictionaries with each of our corpora is shown in Table 3. The incorporation of dictionary information into the vector representation has been discussed in the Knowledge Representation section.
Table 3.
Percentage of words that appear in name, location, hospital, and month dictionaries used by Stat De-id and by the heuristic+dictionary approach.
| Corpus | Patients in names dict. | Doctors in names dict. | Locations in location dict. | Hospitals in hospital dict. | Dates in month dict. | Non-PHI in names dict. | Non-PHI in location dict. | Non-PHI in hospitals dict. | Non-PHI in month dict. |
|---|---|---|---|---|---|---|---|---|---|
| Random | 86.45% | 86.50% | 87.5% | 87.5% | 12.65% | 15.87% | 9.19% | 14.10% | 0.07% |
| Authentic | 78.57% | 70.33% | 54.55% | 80.18% | 21.97% | 16.12% | 10.19% | 12.74% | 0.02% |
| Ambiguous | 86.53% | 86.50% | 100% | 87.5% | 12.64% | 19.53% | 10.50% | 14.03% | 0.08% |
| OoV | 2.51% | 1.99% | 0% | 19.56% | 12.65% | 15.87% | 9.19% | 14.10% | 0.07% |
| Challenge | 14.10% | 17.20% | 11.40% | 26.59% | 5.15% | 15.36% | 11.32% | 8.61% | 0.06% |
6.5.3 Section Headings
Discharge summaries have a repeating structure that can be exploited by taking into consideration the heading of the section in which the target appears, e.g., HISTORY OF PRESENT ILLNESS. In particular, the headings help determine the types of PHI that appear in the templated parts of the text. For example, dates follow the DISCHARGE DATE heading. The section headings have been incorporated into the feature vector by setting the entry corresponding to the relevant section heading to one and leaving the entries corresponding to the rest of section headings at zero.
7 Baseline Approaches
We compared Stat De-id with a scheme that relies heavily on dictionaries and hand-built heuristics [8], with Roth and Yih’s SNoW [9], with BBN’s IdentiFinder [10], and with our in-house Conditional Random Field De-identifier (CRFD). SNoW, IdentiFinder, and CRFD take into account dependencies of entities with each other and with non-entity tokens in a sentence, i.e., sentential global context, while Stat De-id focuses on each word in the sentence in isolation, using only local context provided by a few surrounding words and the words linked by close syntactic relationships. We chose these baseline schemes to explore the contributions of local and global context to de-identification in clinical narrative text.
While we cross-validated SNoW and Stat De-id on our corpora, we did not have the trainable version of IdentiFinder available for our use. Thus, we were unable to train this system on the training data used for Stat De-id and SNoW, but had to use it as trained on news corpora. Clearly, this puts IdentiFinder at a relative disadvantage, so our analysis intends not so much to draw conclusions about the relative strengths of these systems but to study the contributions of different features. In order to strengthen our conclusions about contributions of global and local context to de-identification, we compare Stat De-id with CRFD, which adds sentential global context to the features employed by Stat De-id and, like Stat De-id and SNoW, is cross-validated on our corpora.
7.1 Heuristic+dictionary Scheme
Most traditional de-identification approaches use dictionaries and hand-tailored heuristics. We obtained one such system that identifies PHI by checking to see if the target words occur in hospital, location, and name dictionaries, but not in a list of common words [8]. Simple contextual clues, such as titles, e.g., Mr., and manually determined bigrams, e.g., lives in, are also used to identify PHI not occurring in dictionaries. We ran this rule-based system on each of the artificial and authentic corpora. Note that the discharge summaries obtained from the BIDMC had been automatically de-identified by this approach prior to manual scrubbing. The dictionaries used by Stat De-id were identical to the dictionaries of this system.
7.2 SNoW
Roth and Yih’s SNoW system [9] recognizes people, locations, and organizations. This system takes advantage of words in a phrase, surrounding bigrams and trigrams of words, the number of words in the phrase, and information about the presence of the phrase or constituent words in people and location dictionaries to determine the probability distribution of entity types and relationships between the entities in a sentence. This system uses the probability distributions and constraints imposed by relationships on the entity types to compute the most likely assignment of relationships and entities in the sentence. In other words, SNoW uses its beliefs about relationships between entities, i.e., the global context of the sentence, to strengthen or weaken its hypothesis about each entity’s type.
We cross-validated SNoW on each of the artificial and authentic corpora, but only on the entity types it was designed to recognize, i.e., people, locations, and organizations. For each corpus, tenfold cross-validation trained SNoW on 90% of the corpus and validated it on the remaining 10%.
7.3 IdentiFinder
IdentiFinder, described in more detail in the Background and Related Work section, uses HMMs to find the most likely sequence of entity types in a sentence given a sequence of words. Thus, it uses the global context of the entities in a sentence. IdentiFinder is distributed pre-trained on news corpora. We obtained and used this system out-of-the-box.
7.4 Conditional Random Field De-identifier (CRFD)
We built CRFD, a de-identifier based on Conditional Random Fields (CRF) [23], which, like SVMs, can handle a very large number of features, but which makes joint inferences over entire sequences. For our purposes, following the example of IdentiFinder, sequences are set to be sentences. CRFD employs exactly the same local context features used by Stat De-id. However, the use of conditional random fields allows this de-identifier to also take into consideration sentential global context while predicting PHI; CRFD finds the optimal sequence of PHI tags over the complete sentence. We use the CRF implementation provided by IIT Bombay [32] and cross-validate (tenfold) CRFD on each of our corpora.
8 Evaluation Methods
8.1 Precision, Recall, and F-measure
We evaluated the de-identification and NER systems on four artificial and one authentic corpora. We evaluated Stat De-id using tenfold cross-validation; in each round of cross-validation we extracted features only from the training corpus, trained the SVM only on these features, and evaluated performance on a held-out validation set. To compare with the performance of baseline systems, we computed precision, recall, and F-measures for each system. Precision for class x is defined as Where β is the number of correctly classified instances of class x and B is the total number of instances classified as class x. Recall for class x is defined as where v is the number of correctly classified instances of x and V is the total V number of instances of x in the corpus. The metric that is of most interest in de-identification is recall for PHI. Recall measures the percentage of PHI that is correctly identified and should ideally be very high. We are also interested in maintaining the integrity of the data, i.e., avoiding the classification of non-PHI as PHI. This is captured by precision. In this paper, we also compute F-measure, which is the harmonic mean of precision and recall, given by:
| (5) |
and provides a single number that can be used to compare systems. In the biomedical informatics literature, precision is referred to as positive predictive value and recall is referred to as sensitivity.
In this paper, the purpose of de-identification is to find all PHI and not to distinguish between types of PHI. Therefore, we group the seven PHI classes into a single PHI category, and compute precision and recall for PHI versus non-PHI. In order to study the performance on each PHI type, in the section on Multi-Class SVM Results and Implications for Future Research, we present the precision, recall and F-measure for each individual PHI class. More details can be found in Sibanda [33].
8.2 Statistical Significance
Precision, recall, and F-measure represent proportions of populations. In trying to determine the difference in performance of two systems, we therefore employ the z-test on two proportions. We test the significance of the differences in F-measures on PHI and the differences in F-measures on non-PHI [34–36].
Given two system outputs, the null hypothesis is that there is no difference between the two proportions, i.e., H0: p1 = p2. The alternate hypothesis states that there is a difference between the two proportions, i.e., H1: p1 ≠ p2. At the significance level α, the z-statistic is given by:
| (6) |
where
| (7) |
n1 and n2 refer to sample sizes. A z-statistic of ±1.96 means that the difference between the two proportions is significant at α = 0.05. All significance tests in this paper are run at this α.
9 Results and Discussion
9.1 De-identifying Random and Authentic Corpora
We first de-identified the random and authentic corpora. On the random corpus, Stat De-id significantly outperformed all of IdentiFinder, CRFD, and the heuristic+dictionary baseline. Its F-measure on PHI was 97.63% compared to IdentiFinder’s 68.35%, CRFD’s 81.55%, and the heuristic+dictionary scheme’s 77.82% (see Table 4).8 We evaluated SNoW only on the three kinds of entities it is designed to recognize. We found that it recognized PHI with an F-measure of 96.39% (see Table 5). In comparison, when evaluated only on the entity types SNoW could recognize, Stat De-id achieved a comparable F-measure of 97.46%. On the authentic corpus, Stat De-id significantly outperformed all other systems (see Table 6 and Table 7). F-measure differences from SNoW were also significant.
Table 4.
Precision, recall, and F-measure on random corpus. IFinder refers to IdentiFinder and H+D refers to heuristic+dictionary approach. Highest F-measures are in bold. The F-measure differences from Stat De-id, in PHI and in non-PHI, that are significant at α = 0.05 are marked with an * in all of the tables in this paper.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.34% | 96.92% | 97.63% |
| IFinder | PHI | 62.21% | 75.83% | 68.35%* |
| H+D | PHI | 93.67% | 66.56% | 77.82%* |
| CRFD | PHI | 81.94% | 81.17% | 81.55%* |
| Stat De-id | Non-PHI | 99.53% | 99.75% | 99.64% |
| IFinder | Non-PHI | 96.15% | 92.92% | 94.51%* |
| H+D | Non-PHI | 95.07% | 99.31% | 97.14%* |
| CRFD | Non-PHI | 98.91% | 99.05% | 98.98%* |
Table 5.
Evaluation of SNoW and Stat De-id on recognizing people, locations, and organizations found in the random corpus. Note that these are the only entity types SNoW was built to recognize. Highest F-measures are in bold. The difference in PHI F-measures between SNoW and Stat De-id is not significant at α = 0.05. The difference in non-PHI F-measures is significant at the same α and marked as such with an *.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.31% | 96.62% | 97.46% |
| SNoW | PHI | 95.18% | 97.63% | 96.39%* |
| Stat De-id | Non-PHI | 99.64% | 99.82% | 99.73% |
| SNoW | Non-PHI | 99.75% | 99.48% | 99.61%* |
Table 6.
Evaluation on authentic discharge summaries. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.46% | 95.24% | 96.82% |
| IFinder | PHI | 26.17% | 61.98% | 36.80%* |
| H+D | PHI | 82.67% | 87.30% | 84.92%* |
| CRFD | PHI | 91.16% | 84.75% | 87.83%* |
| Stat De-id | Non-PHI | 99.84% | 99.95% | 99.90% |
| IFinder | Non-PHI | 98.68% | 94.19% | 96.38%* |
| H+D | Non-PHI | 99.58% | 99.39% | 99.48%* |
| CRFD | Non-PHI | 99.62% | 99.86% | 99.74%* |
Table 7.
Evaluation of SNoW and Stat De-id on authentic discharge summaries. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.40% | 93.75% | 96.02% |
| SNoW | PHI | 96.36% | 91.03% | 93.62%* |
| Stat De-id | Non-PHI | 99.90% | 99.98% | 99.94% |
| SNoW | Non-PHI | 99.86% | 99.95% | 99.90%* |
The superiority of Stat De-id over the heuristic+dictionary approach suggests that using dictionaries with only simple, incomplete contextual clues is not as effective for recognizing PHI. The superiority of Stat De-id over IdentiFinder, CRFD, and SNoW suggest that, on our corpora, a system using (a more complete representation of) local context performs as well as (and sometimes better than) systems using (weaker representations of) local context combined with global context.
9.2 De-identifying the Ambiguous Corpus
Ambiguity of PHI with non-PHI complicates the de-identification process. In particular, a greedy de-identifier that removes all keyword matches to possible PHI would remove Huntington from both the doctor’s name, e.g., “Dr. Huntington”, and the disease name, e.g., “Huntington’s disease”. Conversely, use of common words as PHI, e.g., “Consult Dr. Test”, may result in inadequate anonymization of some PHI.
When evaluated on such a challenging data set where some PHI were ambiguous with non-PHI, Stat De-id accurately recognized 94.27% of all PHI: its performance measured in terms of F-measure was significantly better than that of IdentiFinder, SNoW, CRFD, and the heuristic+dictionary scheme on both the complete corpus (see Table 8 and Table 9) and on only the ambiguous entries in the corpus (see Table 10) for both PHI and non-PHI at α = 0.05. For example, the patient name Camera in “Camera underwent relaxation to remove mucous plugs.” is missed by all baseline schemes but is recognized correctly by Stat De-id.
Table 8.
Evaluation on the corpus containing ambiguous data. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 96.37% | 94.27% | 95.31% |
| IFinder | PHI | 45.52% | 69.04% | 54.87%* |
| H+D | PHI | 79.69% | 44.25% | 56.90%* |
| CRFD | PHI | 81.84% | 78.08% | 79.92%* |
| Stat De-id | Non-PHI | 99.18% | 99.49% | 99.34% |
| IFinder | Non-PHI | 95.23% | 88.22% | 91.59%* |
| H+D | Non-PHI | 92.52% | 98.39% | 95.36%* |
| CRFD | Non-PHI | 98.12% | 98.78% | 98.45%* |
Table 9.
Evaluation of SNoW and Stat De-id on ambiguous data. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 95.75% | 93.24% | 94.48% |
| SNoW | PHI | 92.93% | 91.57% | 92.24%* |
| Stat De-id | Non-PHI | 99.33% | 99.59% | 99.46% |
| SNoW | Non-PHI | 99.17% | 99.31% | 99.24%* |
Table 10.
Evaluation only on ambiguous people, locations, and organizations found in ambiguous data. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 94.02% | 92.08% | 93.04% |
| IFinder | PHI | 50.26% | 67.16% | 57.49%* |
| H+D | PHI | 58.35% | 30.08% | 39.70%* |
| SNoW | PHI | 91.80% | 87.83% | 89.77%* |
| CRFD | PHI | 74.15% | 71.15% | 72.62%* |
| Stat De-id | Non-PHI | 98.28% | 98.72% | 98.50% |
| IFinder | Non-PHI | 92.26% | 85.48% | 88.74%* |
| H+D | Non-PHI | 86.19% | 95.31% | 90.52%* |
| SNoW | Non-PHI | 97.34% | 98.27% | 97.80%* |
| CRFD | Non-PHI | 95.84% | 96.89% | 96.37%* |
9.3 De-identifying the Out-of-Vocabulary Corpus
In many cases, discharge summaries contain foreign or misspelled words, i.e., out-of-vocabulary words, as PHI. An approach that simply looks up words in a dictionary of proper nouns may fail to anonymize such PHI. We hypothesized that on the data set containing out-of-vocabulary PHI, context would be the key contributor to de-identification. As expected, the heuristic+dictionary method recognized PHI with the lowest F-measures on this data set (see Table 11 and Table 12). Again, Stat De-id outperformed all other approaches significantly (α = 0.05), obtaining an F-measure of 97.44% for recognizing out-of-vocabulary PHI in our corpus, while IdentiFinder, CRFD, and the heuristic+dictionary scheme had F-measures of 53.51%, 80.32%, and 38.71% respectively (see Table 11).
Table 11.
Evaluation on the out-of-vocabulary corpus. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.12% | 96.77% | 97.44% |
| IFinder | PHI | 52.44% | 54.62% | 53.51%* |
| H+D | PHI | 88.24% | 24.79% | 38.71%* |
| CRFD | PHI | 82.01% | 78.71% | 80.32%* |
| Stat De-id | Non-PHI | 99.54% | 99.74% | 99.64% |
| IFinder | Non-PHI | 93.52% | 92.97% | 93.25%* |
| H+D | Non-PHI | 90.32% | 99.53% | 94.70%* |
| CRFD | Non-PHI | 98.43% | 99.01% | 98.72%* |
Table 12.
Evaluation of SNoW and Stat De-id on the people, locations, and organizations found in the out-of-vocabulary corpus. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.04% | 96.49% | 97.26% |
| SNoW | PHI | 96.50% | 95.08% | 95.78%* |
| Stat De-id | Non-PHI | 99.67% | 99.82% | 99.74% |
| SNoW | Non-PHI | 99.53% | 99.67% | 99.60%* |
Of only the out-of-vocabulary PHI, 96.49% were accurately identified by Stat De-id. In comparison, the heuristic+dictionary approach accurately identified those PHI that could not be found in dictionaries 11.15% of the time, IdentiFinder recognized these PHI 57.33% of the time, CRFD recognized them 84.75% of the time, and SNoW gave an accuracy of 95.08% (see Table 13). For example, the fictitious doctor name Znw was recognized by Stat De-id but missed by all other systems in the sentence “Labs showed hyperkalemia (increased potassium), …, discussed with primary physicians (Znw) and cardiologist (P. Nwnrgo).”
Table 13.
Recall on only the out-of-vocabulary PHI. Highest recall is in bold. The differences from Stat De-id are significant at α = 0.05.
| Method | Stat De-id | IFinder | SNoW | H+D | CRFD |
|---|---|---|---|---|---|
| Recall | 96.49% | 57.33%* | 95.08%* | 11.15%* | 84.75%* |
9.4 De-identifying the Challenge Corpus
The challenge corpus combines the difficulties posed by out-of-vocabulary and ambiguous PHI. Being the largest of our corpora, we expect the results on this corpus to be most reliable. Consistent with our observations on the rest of the corpora, Stat De-id outperformed all other systems significantly (α = 0.05) on the challenge corpus, obtaining an F-measure of 98.03% (see Table 14 and Table 15). The performance of IdentiFinder on this corpus is the worst (F-measure = 33.20%), followed by the heuristic+dictionary approach (F-measure = 43.95%) and CRFD (F-measure = 85.57%). When evaluated only on the entity types SNoW could recognize, Stat De-id achieved an F-measure of 97.96% in recognizing PHI, significantly outperforming SNoW with F-measure = 96.21%.
Table 14.
Evaluation on the challenge corpus. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.69% | 97.37% | 98.03% |
| IFinder | PHI | 25.10% | 49.10% | 33.20%* |
| H+D | PHI | 36.24% | 55.84% | 43.95%* |
| CRFD | PHI | 86.37% | 84.79% | 85.57%* |
| Stat De-id | Non-PHI | 99.83% | 99.92% | 99.86% |
| IFinder | Non-PHI | 97.25% | 92.47% | 94.80%* |
| H+D | Non-PHI | 97.67% | 94.95% | 96.29%* |
| CRFD | Non-PHI | 99.55% | 99.65% | 99.60%* |
Table 15.
Evaluation of SNoW and Stat De-id on the people, locations, and organizations found in the challenge corpus. Highest F-measures are in bold. The F-measure differences from Stat De-id in PHI and in non-PHI are significant at α = 0.05.
| Method | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Stat De-id | PHI | 98.98% | 96.96% | 97.96% |
| SNoW | PHI | 98.73% | 93.81% | 96.21%* |
| Stat De-id | Non-PHI | 99.90% | 99.97% | 99.93% |
| SNoW | Non-PHI | 99.80% | 99.96% | 99.88%* |
9.5 Feature Importance
To understand the gains of Stat De-id, we determined the relative importance of each feature by running Stat De-id with the following restricted feature sets on the random, authentic, and challenge corpora:
The target words alone.
The syntactic bigrams alone.
The lexical bigrams alone.
The part of speech (POS) information alone.
The dictionary-based features alone.
The MeSH features alone.
The orthographic features alone.
Table 16 shows that running Stat De-id only with the target word, i.e., a linear SVM with keywords as feature, would give an F-measure of 57% on the random corpus. In comparison, Table 4 shows that Stat De-id with the complete feature set gives an F-measure of 97%. Similarly, Stat De-id with only the target word gives an F-measure of 80% on the authentic corpus (Table 17). When employed with all of the features, the F-measure rises to 97% (Table 6). Finally, the target word by itself can recognize 65% of PHI in the challenge corpus whereas Stat De-id with the complete feature set gives an F-measure of 98%. The observed improvements on each of the corpora suggest that the features that contribute to a more thorough representation of local context also contribute to more accurate de-identification. Note that keywords are much more useful on the authentic corpus than on the random and challenge corpora. This is because there are more and varied PHI in the random and challenge corpora. In contrast, in the authentic corpus, many person and hospital names repeat, making keywords informative. Regardless of this difference, on both corpora, as the overall feature set improves, so does the performance of Stat De-id.
Table 16.
Comparison of features for random corpus. For all pairs of features, the differences between F-measures for PHI and the differences between F-measures for non-PHI are significant at α = 0.05. The only exceptions are the difference of F-measures in non-PHI of lexical bigrams and POS information (marked by †), the difference in F-measures in PHI of MeSH and orthographic features (marked by ‡), and the difference in F-measures in non-PHI of MeSH and orthographic features (marked by •). Best F-measures are in bold.
| Feature | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Target words | Non-PHI | 91.61% | 98.95% | 95.14% |
| PHI | 86.26% | 42.03% | 56.52% | |
| Lexical bigrams | Non-PHI | 95.61% | 98.10% | 96.84%† |
| PHI | 85.43% | 71.14% | 77.63% | |
| Syntactic bigrams | Non-PHI | 96.96% | 98.72% | 97.83% |
| PHI | 90.76% | 80.20% | 85.15% | |
| POS information | Non-PHI | 94.85% | 98.38% | 96.58%† |
| PHI | 86.38% | 65.84% | 74.73% | |
| Dictionary | Non-PHI | 88.99% | 99.26% | 93.85% |
| PHI | 81.92% | 21.41% | 33.95% | |
| MeSH | Non-PHI | 86.49% | 100% | 92.75%• |
| PHI | 0% | 0% | 0%‡ | |
| Orthographic | Non-PHI | 86.49% | 100% | 92.75%• |
| PHI | 0% | 0% | 0%‡ |
Table 17.
Comparison of features for authentic corpus. For all pairs of features, the differences between F-measures for PHI and the differences between F-measures for non-PHI are significant at α = 0.05. The only exceptions are the difference of F-measures in non-PHI of lexical bigrams and syntactic bigrams (marked by †) and the difference of F-measures in non-PHI of MeSH and orthographic features (marked by ‡). Best F-measures are in bold.
| Feature | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Target words | Non-PHI | 98.79% | 99.94% | 99.36% |
| PHI | 97.64% | 67.38% | 79.74% | |
| Lexical bigrams | Non-PHI | 98.46% | 99.83% | 99.14%† |
| PHI | 92.75% | 58.47% | 71.73% | |
| Syntactic bigrams | Non-PHI | 98.55% | 99.87% | 99.21%† |
| PHI | 94.66% | 60.97% | 74.17% | |
| POS information | Non-PHI | 97.95% | 99.63% | 98.78% |
| PHI | 81.99% | 44.64% | 57.81% | |
| Dictionary | Non-PHI | 97.11% | 99.89% | 98.48% |
| PHI | 88.11% | 21.14% | 34.10% | |
| MeSH | Non-PHI | 96.37% | 100% | 98.15%‡ |
| PHI | 0% | 0% | 0% | |
| Orthographic | Non-PHI | 96.39% | 99.92% | 98.12%‡ |
| PHI | 22.03% | 0.61% | 1.19% |
The results in Table 16 also show that, when used alone, lexical and syntactic bigrams are two of the most useful features for de-identification of the random corpus. The same two features constitute the most useful features for de-identification of the challenge corpus (Table 18). In the authentic corpus (Table 17), target word and syntactic bigrams are the most useful features. All of random, authentic, and challenge corpora highlight the relative importance of local context features; in all three corpora, context is more useful than dictionaries, reflecting the repetitive structure and language of discharge summaries.
Table 18.
Comparison of features for challenge corpus. For all pairs of features, the differences between F-measures for PHI and the differences between F-measures for non-PHI are significant at α = 0.05. Best F-measures are in bold.
| Feature | Class | Precision | Recall | F-measure |
|---|---|---|---|---|
| Target words | Non-PHI | 96.90% | 99.87% | 98.36% |
| PHI | 96.05% | 49.56% | 65.38% | |
| Lexical bigrams | Non-PHI | 97.34% | 99.69% | 98.50% |
| PHI | 91.99% | 56.87% | 70.29% | |
| Syntactic bigrams | Non-PHI | 97.50% | 99.74% | 98.61% |
| PHI | 93.44% | 59.61% | 72.79% | |
| POS information | Non-PHI | 96.04% | 99.42% | 97.70% |
| PHI | 79.33% | 35.24% | 48.80% | |
| Dictionary | Non-PHI | 94.26% | 99.90% | 96.99% |
| PHI | 69.70% | 3.79% | 7.19% | |
| MeSH | Non-PHI | 94.05% | 100% | 96.93% |
| PHI | 0% | 0% | 0% | |
| Orthographic | Non-PHI | 96.05% | 99.60% | 97.79% |
| PHI | 84.67% | 35.30% | 49.83% |
On all three corpora, syntactic bigrams outperform lexical bigrams in recognizing PHI. The F-measure difference between the syntactic and lexical bigrams is significant for PHI on all of the random, challenge, and authentic corpora. Most prior approaches to de-identification/NER have used only lexical bigrams, ignoring syntactic dependencies. Our experiments suggest that syntactic context is more informative than lexical context for the identification of PHI in discharge summaries, even though these records contain fragmented and incomplete utterances.
We conjecture that the lexical context of PHI is more variable than their syntactic context because many English sentences are filled with clauses, adverbs, etc., that separate the subject from its main verb. The Link Grammar Parser can recognize these interjections so that the words break up lexical context but not syntactic context. For example, the word supposedly gets misclassified by lexical bigrams as PHI when encountered in the sentence “Trantham, Faye supposedly lives at home with home health aide and uses a motorized wheelchair”. This is because the verb lives which appears on the right-hand side of supposedly is a strong lexical indicator for PHI. If we parse this sentence with the Link Grammar Parser, we find that the right-hand link for the word supposedly is (lives, E) where E is the link for “verb-modifying adverbs which precede the verb” [6]. This link is not an indicator of patient names and helps mark supposedly as non-PHI.
9.6 Local vs. Global Context
Table 19 shows that the local context features of SNoW and IdentiFinder are also quite powerful. When employed with the same learning algorithm utilized by Stat De-id, the individual local feature sets of SNoW and IdentiFinder give performance F-measures above 86%. Note that Stat De-id’s local context outperforms the local context of SNoW and IdentiFinder on all corpora. This result supports the hypothesis that developing a more thorough representation of local context can benefit de-identification. Our experiments with CRFD were designed specifically to address the relative value of improved local vs. global context features. Our results show that a CRF-based system using exactly the same local context feature set as Stat De-id performs significantly worse than Stat De-id on all corpora (Tables 4, 6, 8, 10, 11, 13, and 14). Thus, on our corpora and for CRFD, attending to global consistency in addition to a rich set of local features actually hurts performance. This strongly supports our hypothesis that improved local features are dominant for de-identification.
Table 19.
Comparison of local context feature sets of Stat-Deid, SNoW, and IdentiFinder, evaluated individually, with SVMs, on each of the corpora. Highest F-measures are in bold. All differences from the corresponding All Stat De-id F-measures are significant at α = 0.05 and marked as such with an *.
| Corpus | Feature | Class | Precision | Recall | F-measure |
|---|---|---|---|---|---|
| Random | All Stat De-id/All CRFD | Non-PHI | 99.53% | 99.75% | 99.64% |
| PHI | 98.34% | 96.92% | 97.63% | ||
| All local context features of SNoW | Non-PHI | 98.79% | 99.36% | 99.08%* | |
| PHI | 95.76% | 92.23% | 93.96%* | ||
| All local context features of IdentiFinder | Non-PHI | 99.35% | 99.18% | 99.27%* | |
| PHI | 94.33% | 95.85% | 95.33%* | ||
| Authentic | All Stat De-id/All CRFD | Non-PHI | 99.84% | 99.95% | 99.90% |
| PHI | 98.46% | 95.24% | 96.82% | ||
| All local context features of SNoW | Non-PHI | 99.72% | 99.94% | 99.83%* | |
| PHI | 98.42% | 92.67% | 95.46%* | ||
| All local context features of IdentiFinder | Non-PHI | 99.66% | 99.92% | 99.79%* | |
| PHI | 97.75% | 91.04% | 94.28%* | ||
| Ambiguous | All Stat De-id/All CRFD | Non-PHI | 99.18% | 99.49% | 99.34% |
| PHI | 96.37% | 94.27% | 95.31% | ||
| All local context features of SNoW | Non-PHI | 98.15% | 98.98% | 98.56%* | |
| PHI | 92.51% | 87.11% | 89.73%* | ||
| All local context features of IdentiFinder | Non-PHI | 97.62% | 98.49% | 98.05%* | |
| PHI | 88.86% | 83.42% | 86.06%* | ||
| OoV | All Stat De-id/All CRFD | Non-PHI | 99.54% | 99.74% | 99.64% |
| PHI | 98.12% | 96.77% | 97.44% | ||
| All local context features of SNoW | Non-PHI | 98.95% | 99.45% | 99.20%* | |
| PHI | 96.10% | 92.67% | 94.33%* | ||
| All local context features of IdentiFinder | Non-PHI | 99.12% | 99.17% | 99.14%* | |
| PHI | 94.23% | 93.87% | 94.05%* | ||
| Challenge | All Stat De-id/All CRFD | Non-PHI | 99.83% | 99.92% | 99.86% |
| PHI | 98.69% | 97.37% | 98.03% | ||
| All local context features of SNoW | Non-PHI | 99.50% | 99.86% | 99.68%* | |
| PHI | 97.72% | 92.14% | 94.85%* | ||
| All local context features of IdentiFinder | Non-PHI | 99.50% | 99.89% | 99.70%* | |
| PHI | 98.21% | 92.15% | 95.08%* |
10 Multi-Class SVM Results and Implications for Future Research
The goal of this paper is to separate PHI from non-PHI for de-identification purposes. We have so far shown that Stat De-id recognizes 94–97% of the PHI and outperforms all other systems. However, from a policy perspective, the adequacy of the performance of Stat De-id depends on the PHI that are missed. Not all PHI are equally strong identifiers of individuals. For example, failing to remove 6% of the names would have different policy implications than failing to remove 6% of dates. Therefore, in order to put the performance figures into perspective, we evaluate Stat De-id on each type of PHI on the authentic and challenge corpora.
The results in Table 20 and Table 21 show that Stat De-id performs relatively poorly in recognizing phone and location PHI classes. Of the 88 location words in the authentic corpus, 22% are classified as non-PHI and 16% are classified as hospital names. For example, the location Hollist in the sentence “The patient lives at home in Hollist with his parents.” is missed. The errors in the location class arise because there are too few positive examples in the training set to learn the context distinguishing locations. Furthermore, the context for locations often overlaps with the context for hospitals.
Table 20.
Multi-class classification results for Stat De-id on authentic and challenge corpora.
| Class | Precision | Recall | F-measure | |||
|---|---|---|---|---|---|---|
| Authentic | Challenge | Authentic | Challenge | Authentic | Challenge | |
| Non-PHI | 99.84% | 99.83% | 99.94% | 99.92% | 99.89% | 99.88% |
| Patient | 98.94% | 97.17% | 95.24% | 96.72% | 97.05% | 96.94% |
| Doctor | 98.48% | 98.64% | 96.34% | 97.37% | 97.40% | 98.00% |
| Location | 92.73% | 91.98% | 57.95% | 75.29% | 71.33% | 82.80% |
| Hospital | 94.15% | 98.63% | 90.70% | 96.58% | 92.39% | 97.59% |
| Date | 98.23% | 97.23% | 96.83% | 96.86% | 97.52% | 97.04% |
| ID | 98.16% | 98.51% | 99.38% | 98.53% | 98.76% | 98.52% |
| Phone | 90.48% | 97.84% | 59.38% | 83.76% | 71.70% | 90.26% |
Table 21.
Multi-class confusion matrix for Stat De-id on authentic corpus.
| Predicted | Non-PHI | Patient | Doctor | Location | Hospital | Date | ID | Phone |
|---|---|---|---|---|---|---|---|---|
| Actual | ||||||||
| Non-PHI | 112,605 | 2 | 4 | 0 | 17 | 33 | 8 | 0 |
| Patient | 12 | 280 | 1 | 0 | 0 | 0 | 0 | 1 |
| Doctor | 24 | 1 | 711 | 0 | 2 | 0 | 0 | 0 |
| Location | 19 | 0 | 3 | 51 | 14 | 0 | 0 | 1 |
| Hospital | 54 | 0 | 3 | 4 | 595 | 0 | 0 | 0 |
| Date | 58 | 0 | 0 | 0 | 4 | 1,891 | 0 | 0 |
| ID | 3 | 0 | 0 | 0 | 0 | 0 | 479 | 0 |
| Phone | 11 | 0 | 0 | 0 | 0 | 1 | 1 | 19 |
Of the 32 phone numbers in the authentic corpus, 34% are misclassified as non-PHI. For example, the number 234-907-1924 is missed in the sentence “DR. JANE DOE (234-907-1924)”. Again, these errors arise because there are too few phone numbers in the training set.
Despite being person names, patients and doctors are rarely misclassified as each other, although they sometimes do get misclassified as non-PHI. This is because the honorifics used with the patients’ and doctors’ names help differentiate between the two. However, the honorifics are absent from some of the names. When these names also lack local context, this leaves Stat-Deid to its best guess, i.e., non-PHI. For example, the name “Jn Smth” which consists of rare tokens and appears on a line all by itself, with no context, gets misclassified as non-PHI. 4% of patients and 3% of doctors in the authentic corpus are misclassified as non-PHI. These observations generalize to the challenge corpus also.
Misclassifying patient and doctor names as non-PHI is detrimental for de-identification. Similarly, misclassifying phone numbers as non-PHI can cause serious privacy concerns as today’s technology allows us to cross-reference a single phone number with a search engine, e.g., Google, and link it directly with individuals. To minimize the risk of revealing PHI and of easy re-identification, we plan to improve performance on PHI with stereotypical formats, such as names, phone numbers, social security numbers, medical record numbers, dates, and addresses, by enhancing Stat De-id with the patterns employed by rule-based systems, not to make a final determination of whether something matching the pattern is PHI, but as an additional input feature. Such features can be drawn from available dictionaries of names, places, etc., to augment what can be learned automatically from labeled corpora.
Note that, in general, even after de-identification, it may be possible to deduce the identity of individuals mentioned in the records, for example, by combining the de-identified data with other publicly available information or by studying some indirect identifiers that may be mentioned in the records but that do not fall into one of the PHI categories defined by HIPAA [37]. To further reduce the risk of re-identification, such indirect identifiers may also need to be removed or generalized. Unfortunately, the problem of minimizing data loss by generalizing or removing data is computationally intractable [38], so only heuristic methods are typically employed [39]. The category of doctors is one indirect identifier that is not included in PHI defined by HIPAA but is included in the PHI marked by Stat De-id.
11 Conclusions
In this paper, we have shown that we can de-identify clinical text, characterized by fragmented and incomplete utterances, using local context 94–97% of the time. Our representation of local context is novel; it includes novel syntactic features which provide us with useful linguistic information even when the language of documents is fragmented. The results presented imply that de-identification can be performed even when corpora are dominated by fragmented and incomplete utterances, even when many words in the corpora are ambiguous between PHI and non-PHI, and even when many PHI include out-of-vocabulary terms. Structure and repetitions in the language of documents can be exploited for this purpose.
Experiments on our corpora suggest that local context plays an important role in de-identification of narratives characterized by fragmented and incomplete utterances. This fact remains true even when the narratives contain uncommon PHI instances that are not present in easily obtainable dictionaries and even when PHI are ambiguous with non-PHI. The more thorough the local context, the better the performance; and strengthening the representation of local context may be more beneficial for de-identification than complementing local with global context.
Acknowledgments
This work was supported in part by the National Institutes of Health through research grants 1 RO1 EB001659 from the National Institute of Biomedical Imaging and Bioengineering and through the NIH Roadmap for Medical Research, Grant U54LM008748. IRB approval has been granted for the studies presented in this manuscript. We thank the anonymous reviewers for their insightful comments and constructive feedback.
Footnotes
This is a thoroughly revised and extended version of the preliminary draft “Role of Local Context in De-identification of Ungrammatical, Fragmented Text” which was presented at the conference of the North American Chapter of Association for Computational Linguistics/Human Language Technology (NAACL-HLT 2006) in June 2006.
Or a target phrase, though we will refer here only to target words.
Wd links the main clause back to the LEFT-WALL; Ss links singular nouns to singular verb forms; MVp connects verbs to their (prepositional) modifying phrases; Js links prepositions to their objects; Ds links determiners to nouns; and Xp links periods to words [28].
“J connects prepositions to their objects” [28].
Pv links verb be to the following passive participle; MVa connects verbs to adverbs; “ID marks the idiomatic strings found in the link grammar dictionary” [28].
“I connects verbs with infinitives”; “K connects verbs with particles like in, out,” etc.,; “ON connects the preposition on to time expressions”; “TM connects month names to day numbers” [28].
“TO connects verbs and adjectives which take infinitival complements to the word to”; “G connects proper nouns together in series”; Xi connects punctuation symbols to abbreviations [28].
Throughout this paper, the baseline F-measures that are significantly different from the corresponding F-measure of Stat De-id at α = 0.05 are marked with * in the tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Özlem Uzuner, University at Albany, State University of New York, Draper 114, 135 Western Ave., Albany, NY 12222
Tawanda C. Sibanda, Massachusetts Institute of Technolology, Computer Science and Artificial Intelligence Laboratory, 32 Vassar Street, Cambridge, MA 02139
Yuan Luo, University at Albany, State University of New York, Draper 114, 135 Western Ave., Albany, NY 12222
Peter Szolovits, Massachusetts Institute of Technolology, Computer Science and Artificial Intelligence Laboratory, 32 Vassar Street, Cambridge, MA 02139
References
- 1. [Accessed May 9, 2007];Health information portability and accountability act. § 164.514, URL: http://www.hhs.gov/ocr/AdminSimpRegText.pdf.
- 2.Malin B, Airoldi E. The effects of location access behavior on re-identification risk in a distributed environment. In: Danezis G, Golle P, editors. 6th International Workshop on Privacy Enhancing Technologies, Privacy Enhancing Technologies, Lecture Notes in Computer Science. Vol. 4258. Springer; Berlin/Heidelberg, Germany: 2006. pp. 413–429. [Google Scholar]
- 3.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. In: Cimino JJ, editor. Proceedings of the American Medical Informatics Association. Hanley & Belfus, Inc; Philadelphia, PA, USA: 1996. pp. 333–337. [PMC free article] [PubMed] [Google Scholar]
- 4.Lovis C, Baud RH. Fast exact string pattern matching algorithms adapted to the characteristics of the medical language. Journal of the American Medical Informatics Association. 2000;7(4):378–391. doi: 10.1136/jamia.2000.0070378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C, Lin C. LIBSVM: a library for support vector machines, Manual. Department of Computer Science and Information Engineering, National Taiwan University; Taipei, Taiwan: 2001. [Accessed October 6, 2007]. URL: http://www.csie.ntu.edu.tw/~cjlin/libsvm/ [Google Scholar]
- 6.Sleator D, Temperley D. Parsing English with a link grammar, Technical Report CMU-CS-91-196. Computer Science Department, Carnegie Mellon University; Pittsburgh, PA, USA: 1991. [Google Scholar]
- 7.Grinberg D, Lafferty J, Sleator D. A robust parsing algorithm for link grammars, Technical Report, CMU-CS-95-125. Computer Science Department, Carnegie Mellon University; Pittsburgh, PA, USA: 1995. [Google Scholar]
- 8.Douglass M. Master’s thesis. Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology; Cambridge, MA, USA: Feb, 2005. Computer assisted de-identification of free text nursing notes. [Google Scholar]
- 9.Roth D, Yih W. Probabilistic reasoning for entity and relation recognition. In: Lenders W, editor. Proceedings of the 19th International Conference on Computational Linguistics; COLING, Association for Computational Linguistics, Morristown, NJ, USA. 2002. pp. 1–7. [Google Scholar]
- 10.Bikel D, Schwartz R, Weischedel R. An algorithm that learns what’s in a name. Machine Learning Journal Special Issue on Natural Language Learning. 1999 February;34(13):211–231. [Google Scholar]
- 11.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. American Journal of Clinical Pathology. 2004;121(2):176–186. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 12.Beckwith B, Mahaadevan R, Balis U, Kuo F. Development and evaluation of an open source software tool for de-identification of pathology reports. BMC Medical Informatics and Decision Making. 2006;6:12. doi: 10.1186/1472-6947-6-12. (Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman J. Concept-match medical data scrubbing: how pathology text can be used in research. Archives of Pathology and Laboratory Medicine. 2003;127(6):680–686. doi: 10.5858/2003-127-680-CMDS. [DOI] [PubMed] [Google Scholar]
- 14.Taira R, Bui A, Kangarloo H. Identification of patient name references within medical documents using semantic selectional restrictions. In: Kohane I, editor. Proceedings of the American Medical Informatics Association. Hanley & Belfus, Inc; Philadelphia, PA, USA: 2002. pp. 757–761. [PMC free article] [PubMed] [Google Scholar]
- 15.The ACE 2007 evaluation plan. [Accessed August 28, 2007]; URL: http://www.nist.gov/speech/tests/ace/ace07/doc/ace07-evalplan.v1.3a.pdf.
- 16.Isozaki H, Kazawa H. Efficient support vector classifiers for named entity recognition. In: Lenders W, editor. Proceedings of the 19th International Conference on Computational Linguistics; COLING, Association for Computational Linguistics, Morristown, NJ, USA. 2002. pp. 390–396. [Google Scholar]
- 17.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. Journal of the American Medical Informatics Association. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unified Medical Language System. [Accessed October 2, 2007]; [web page] 2006, URL: http://www.nlm.nih.gov/pubs/factsheets/umls.html.
- 19.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20(3):273–297. [Google Scholar]
- 20.Sewell M. Support vector machines. [Accessed July 9, 2007]; URL: http://www.svm.org.
- 21.Rychetsky M. Algorithms and Architectures for Machine Learning based on Regularized Neural Networks and Support Vector Approaches. Shaker Verlag GmBH; Germany: 2001. [Google Scholar]
- 22.Burges C. A tutorial on support vector machines for pattern recognition. Knowledge Discovery and Data Mining. 1998;2(2):121–167. [Google Scholar]
- 23.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and sequence data. In: Brodley CE, Danyluk AP, editors. Proceedings of International Conference on Machine Learning. Morgan Kaufmann; San Francisco: 2001. pp. 282–289. [Google Scholar]
- 24.Peng F, McCallum A. Accurate information extraction from research papers using conditional random fields. In: Hirschberg J, editor. Proceedings of Human Language Technology Conference/North American Chapter of the Association for Computational Linguistics Annual Meeting; Association for Computational Linguistics, Morristown, NJ, USA. 2004. pp. 329–336. [Google Scholar]
- 25.Brill E. A simple rule-based part of speech tagger. In: Bates M, Stock O, editors. 3rd Conference on Applied Natural Language Processing; Association for Computational Linguistics, Morristown, NJ, USA. 1992. pp. 152–155. [Google Scholar]
- 26.Sutcliffe R, Brehony T, McElligott A. The grammatical analysis of technical texts using a link parser, Technical Note UL-CSIS-94-13. Department of Computer Science and Information Systems; University of Limerick, Ireland: 1994. [Google Scholar]
- 27.Pyysalo S, Ginter F, Pahikkala T, Boberg J, Jarvinen J, Salakoski T. Evaluation of two dependency parsers on biomedical corpus targeted at protein-protein interactions. International Journal of Medical Informatics. 2006;75(6):430–442. doi: 10.1016/j.ijmedinf.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Index to link grammar documentation. [Accessed July 12, 2007]; URL: http://www.link.cs.cmu.edu/link/dict/index.html.
- 29.U.S. Census Bureau. 1990 census name files. [Accessed July 9, 2007];1999 URL: http://www.census.gov/genealogy/names/
- 30.U.S. Census Bureau. Census 2000 urbanized area and urban cluster information. [Accessed July 9, 2007];2004 URL: http://www.census.gov/geo/www/ua/uaucinfo.html#lists.
- 31.Worldatlas.com. [Accessed July 9, 2007]; URL: http://worldatlas.com.
- 32.Sarawagi S. Conditional random fields implementation. [Accessed September 21, 2007]; URL: http://crf.sourceforge.net/introduction/
- 33.Sibanda T. Master’s Thesis. Department of Electrical Engineering and Computer Science; Massachusetts Institute of Technology: Jun, 2006. Was the patient cured? Understanding semantic categories and their relationships in patient records. [Google Scholar]
- 34.Wong K, Phua K. Statistics Made Simple for Healthcare and Social Science Professionals and Students. University Putra Malaysia Press; Serdang, Selangor, Malaysia: 2006. [Google Scholar]
- 35.Osborn C. Statistical Applications for Health Information Management. 2. Jones & Bartlett Publishers; Boston: 2006. [Google Scholar]
- 36. [Accessed July 12, 2007];Z-test for two proportions. URL: http://www.dimensionresearch.com/resources/calculators/ztest.html.
- 37.Sweeney L. Computational disclosure control: a primer on data privacy protection. [Accessed September 27, 2007];2001 URL: http://www.swiss.ai.mit.edu/classes/6.805/articles/privacy/sweeney-thesis-draft.pdf.
- 38.Vinterbo S. Privacy: a machine learning view. IEEE Transactions on Knowledge and Data Engineering. 2004 August;16(8):939–948. [Google Scholar]
- 39.Lasko T. PhD Thesis. Department of Electrical Engineering and Computer Science; Massachusetts Institute of Technology: Aug, 2007. Spectral anonymization of data. [Google Scholar]