Abstract
The loss of thymic function with age may be due to diminished numbers of T-cell progenitors and the loss of critical mediators within the thymic microenvironment. To assess the molecular changes associated with this loss, we examined transcriptomes of progressively aging mouse thymi, of different sexes and on caloric-restricted (CR) vs. ad libitum (AL) diets. Genes involved in various biological and molecular processes including transcriptional regulators, stress response, inflammation and immune function significantly changed during thymic aging. These differences depended on variables such as sex and diet. Interestingly, many changes associated with thymic aging are either muted or almost completely reversed in mice on caloric-restricted diets. These studies provide valuable insight into the molecular mechanisms associated with thymic aging and emphasize the need to account for biological variables such as sex and diet when elucidating the genomic correlates that influence the molecular pathways responsible for thymic involution.
Keywords: Thymus, Involution, Aging, Microarray, AGEMAP, Thymocytes, Caloric restriction
1. Introduction
Alterations in immune function with age in both humans and animals are important to the health of aging individuals. Older humans are much more susceptible to microbial infections of the soft tissues, skin, the abdomen and both the urinary and respiratory tracts [1,2]. These older subjects also demonstrate an increased incidence of infectious endocarditis, tuberculosis, meningitis and herpes zoster, and the mortality rates for these diseases in older patients are often two to three times higher than in younger people with the same disease [3-6]. In addition, an increased prevalence of specific cancers and certain autoimmune diseases have been observed with advancing age [6-9]. The increased prevalence of these conditions and the higher morbidity and mortality from infections strongly suggest functional defects in a deteriorating immune system with advancing age [10-16]. Many studies have centered on possible loss or alterations in the number of circulating T lymphocytes and T-cell subsets [17,18]. This focus on T lymphocytes seems reasonable given that the thymus is known to atrophy with progressive aging and correlates with a significant loss in its capability to generate de novo T cells for export into the peripheral T-cell pool [19]. Interestingly, this loss in thymic output with age does not result in any significant change in the total number of peripheral T cells as the maintenance of peripheral T-cell numbers appears to be regulated via a thymus-independent homeostatic process involving expansion of mature peripheral T cells. This peripheral homeostatic expansion of mature T cells results in a much more limited T-cell receptor (TCR) repertoire with age [19,20] and is believed to contribute to the diminished capacity of older T cells to proliferate upon mitogenic and other stimulation when compared with their younger counterparts [20-23]. However, given that the loss in thymic function is one of the earliest and most consistent steps in the progression to immune dysfunction, the involuting thymus seems to be a promising target to which to direct therapy with the specific goal of reversing thymic atrophy and restoring thymopoiesis and T-cell export [19,24-26].
For a number of years in gerontological research, efforts have been made to identify new and unique biomarkers that will define the biological, physiological and chronological changes that occur with age [27]. Numerous studies have focused on the comparison of calorically restricted (CR) primates and rodents to their ad libitum (AL)-fed animal controls, where significant differences in a variety of physiological processes have been observed as well as a prolongation in the animals’ lifespan [28,29]. This delay of aging by CR appears to depend upon a delay, or sometimes ablation, of a broad spectrum of age-associated pathophysiological changes and a 30–50% increase in maximum life span. These studies are quite complicated as CR induces a plethora of biological changes including decreases in oxidative stress [30], glycation or glycoxydation [31,32], body temperature and circulating thyroid hormone levels associated with a hypometabolic state [33,34] as well as alterations in gene expression and protein degradation [29,32] and a number of neuroendocrine and inflammatory changes [35-37]. Despite the identification of many notable physiological differences between such dietary-restricted animals, no specific biomarkers have ever been reproducibly identified that could accurately identify the age of the organism or predict lifespan.
Microarray analysis has been an efficient way to profile the changes associated with disease onset, especially in the arena of both cancer and infectious disease [27]. By identifying the global gene expression profiles between disease and non-disease states, researchers are often able to isolate individual genes that may be used either as markers for disease, or as molecular targets as well as whole pathways that may be involved in the studied disease. Similar studies have now been initiated to examine age-related changes in a number of tissues and cellular subsets by microarray analysis in an effort to identify genes or patterns of gene expression associated with physiological aging, lifespan, and age-related disease states [27,38-42]. Only a few studies to date have focused on immunologic aging with the majority of studies focusing on an examination of bulk T-cell populations [43]. Limited replication of analysis, small sample sizes and poor array validation have restricted the interpretation of such data with regard to age-related immunological changes. To date, no studies have focused on gene expression changes within the aging thymus using microarray analysis with an additional focus on additional biological variables such as gender and dietary restrictions.
In an effort to understand the potential forces driving age-associated thymic involution and the cellular pathways involved in these phenomena, we have performed microarray analyses on thymi derived from young and old mice to identify differences in gene expression patterns which may be attributed to aging. We have included mice of both sexes in this study to determine sex-specific differences in aging, as well as mice that have been placed on various dietary regimens. Our results demonstrate that many of the putative thymic aging-responsive genes are in fact dependent upon variables like sex and diet, some of which are independent of age. Only a small fraction of the total gene products examined demonstrated thymic aging-dependent gene expression changes that were also independent of sex or diet. Complex biological interactions are therefore responsible for the breadth of alterations in gene expression generally observed in the expression analyses of progressively aging thymi.
2. Materials and methods
2.1. Mice
Specific pathogen-free C57BL/6 mice were purchased through the Office of Biological Resources and Resource Development of the National Institute on Aging (Bethesda, MD). All mice were maintained in an AAALAC-certified barrier facility and were acclimated for 2 weeks before use. All mice were fed autoclaved food. Water was ingested ad libitum. All mice with evidence of disease (e.g., enlarged spleen, gross tumors) were excluded from these studies. The distribution and characteristics of the animals utilized in this study are described in Table 1. In this study, mice were fed either diets consisting of 100% regular feed (ad libitum) or caloric-restricted (CR) mice as follows: Up to 13 weeks of age, 100% regular feed, followed by 90% fortified feed for 1 week, 75% for 1 week, then 60% fortified feed after that until the age at which the mice were sacrificed. For the verification studies, flash-frozen thymi for RNA and protein analyses from both C57BL/6 and Balb/C mice were obtained from the NIA aged rodent tissue bank (Bethesda, MD).
Table 1.
Mouse age, gender and diet distribution for cDNA array experiments
| Mice at age of sacrifice
| |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 month | 6 months
|
16 months
|
24 months
|
||||||||||
|
Ad libitum |
Ad libitum |
Restricted
|
Ad libitum |
Restricted
|
Ad libitum |
Restricted
|
|||||||
| M | F | M | F | M | F | M | F | M | F | M | F | M | F |
| 5 | 5 | 5 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 3 | 5 |
| Total = 67 | |||||||||||||
The mice were sacrificed at 1, 6, 16 or 24 months of age. Each age group except the youngest included mice placed on either an ad libitum (AL) or a caloric-restricted (CR) diet. These groups were further divided to include both male and female mice in the numbers indicated along the bottom row of the above table.
2.2. RNA extraction and microarray analysis
All data from microarray experiments have been deposited into the GEO Accession Database (number in progress). For each array sample, the RNA was prepared using the entire thymus. The tissue was processed using a Bead Beater (Bio-Spec, Bartlesville, OK) followed by RNA purification using the RNEasy Mini Kit (Qiagen, Valencia, CA). The RNA was examined for quantity and quality using a Bioanalyzer (Agilent Technologies, Palo Alto, CA). Five micrograms of each RNA sample was used in a PCR reaction with [33P]dCTP (Valeant, Costa Mesa, CA). Radiolabelled cDNA was allowed to hybridize overnight at 43 °C to the mouse 17K DNA filters developed by the Laboratory of Genetics, NIA [44]. The hybridized filters were washed and placed under imaging screens for 3 days to allow for sufficient exposure. The images were developed and scanned in a Storm Phosphoimager (Molecular Dynamics, Piscataway, NJ) and the data was extracted using ArrayPro Software (Media Cybernetics, San Diego, CA).
2.3. Statistical data analysis
All data were processed in Excel spreadsheets using a Z score statistical analysis method developed at NIA [45]. In order to be selected for the final gene list, the expression value of a particular gene had to be at least 1.5 times different from the Z score of the control. Differences were considered statistically significant only if they had a p value less than 0.01.
2.4. Gene expression profiles
The selected gene lists were uploaded along with their Z scores to the Microarray Data Analysis website of the National Human Genome Research Institute (http://arrayanalysis.nih.gov/). Using distance-based gene selection, gene expression profiles were created in order to visualize differences between age, gender and diet.
2.5. Thymocyte isolation
Freshly extracted thymi from mice of various ages were dissociated in RPMI using a syringe and forceps. Cell clumps were broken up with repeated pipetting and then pouring through 70 μm nylon mesh cell strainers (BD Falcon, Bedford, MA). The cells were washed once after which the red blood cells were lysed with ammonium chloride buffer. The thymocytes were then washed twice in RPMI followed by PBS, counted, and pelleted. Cell pellets were either used directly in the Qiagen RNAEasy mini kits for RNA preparation or lysed in RIPA buffer containing protease and phosphatase inhibitors (Sigma, St. Louis, MO) to use in Western blot analysis.
2.6. Real-time PCR
Array results were verified by semi-quantitative RT-PCR. One microgram of RNA from each array sample or from thymus or thymocyte RNA samples was used to make cDNA with the TaqMan Reverse Transcription kit (Applied Biosystems, Branchburg, NJ) or the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). One microliter of each cDNA sample was then used to measure quantity using the SYBR Green PCR master mix (Applied Biosystems) and reactions were run on the ABI Prism 7700 Sequence Detector (Applied Biosystems). The results were normalized to GAPDH or 18S using the QuantumRNA universal 18S (Ambion, Austin, TX) and were also used to determine relative quantities. The primers used were:
CCT3: forward (F)—GCCATTCTTCGAGAGATTCAA, reverse (R)—TGCATCTGCTGCTCTAGGAA; CTSL: F—AGAAGGGGAGGCTTTTTCAG, R—TGCCGGTCTTAAGGAACATC; DOCK7: F—TTGATGGAAACCTGGCTACTG, R—TGCAGATAGACTGCACTTTGG; EIF3S10: F—CAAAGGAGCGAGAGAAGGAA, R—CCAGTTGTTCAACCTGTTTGG; EIF4G2: F—TGCTTCTCGTTTCAGTGCTT, R—TTCATTTGCGGAGTTGTTTG; ETFB: F—AAAGTGGACCTTTTGTTCCTG, R—TCAATTTCCCGTTCCAC TTT; HSPA8: F—TTGCTTTCACGGACACAGAG, fR—GCATCATTCACCACCATGAA; HSPCA: F—CA AGACCAACCAATGGAGGA, R—TCAGGCTCTCGTAACGGATT; HSPCB: F—TCGGCCTATCTAGTTGCAGA, R—TGAGGTGAAGGATCACTTTGG; IGH6: F—ATGGAATGGACCTGGGTCTT, R—ATGTCCAG GCCTCTGCTTTA; IGHG: F—TGCAGTCTGGACTCTACACTATGA, R—GGGCATTTGTGACACTCCTT; NR1I3: F—TTGCATATCTCACTCAACACTACG, R—TCCTGGAGATGCAGTCCTTT; NR3C1: F—TTCCTTGGGGGCTATGAACT, R—AAGCTTCATCGGAGCACA; SMARCD2: F—ATTCCGAAAAC GCCTGCT, R—GGATTCGCTGAGGTAGAACCTT; VIL2: F—GG ATTTCCTACCTGGCTGAA, R—TT GACTTGCAGGA AGAAGAGC.
2.7. Western blot analysis
Equal amounts of protein from total thymus or thymocyte pellets were run in a 10% Tris–glycine gel and transferred to nitrocellulose on the Novex gel-blot system (Invitrogen, Carlsbad, CA). The nitrocellulose filters were then probed using specific antibodies to the glucocorticoid receptor (GR), the immunoglobulin heavy chain M (IgM), IgG and GAPDH obtained from Abcam (Cambridge, MA). In addition, the antibody to eIF4G was also utilized (Santa Cruz Biotechnology Inc, Santa Cruz, CA). The HRP-conjugated secondary antibodies were from Amersham (Piscataway, NJ). Bands were visualized using the ECL Plus western blotting detection reagents (Amersham) and CL-X Posure film from Pierce (Rockford, IL).
2.8. Immunohistochemistry (IHC)
Paraffin sections of thymus from young (3.5 months) and old (24 months) C57/BL6 and Balb/C mice were sliced at 5 μm of thickness and mounted onto positively charged slides by Paragon (Baltimore, MD). The slides were then steamed for 30 min in the presence of antigen unmasking solution (Vector Laboratories, Burlingame, CA). These slides were subsequently washed and then blocked with rodent block (Vector Laboratories) for 1 h, followed by an overnight at 4 °C incubation with anti-GR antibody (Abcam, Cambridge, MA). The cells were washed again with PBS for 30 min and then probed with a biotin-conjugated secondary antibody for 1 h (Vector Laboratories). Staining was visualized using a Lab Vision ABC (streptavidin/biotin) detection system (Fremont, CA). After probing, the slides were then washed, mounted in permount and examined on a Zeiss Axiovert 200 microscope using Axio-Vision software (Zeiss, Thornwood, NY).
3. Results
3.1. Microarray analysis reveals distinct transcriptional changes with thymic aging
To examine the molecular events associated with aging in mammals, we utilized cDNA-based microarray analysis to elucidate the transcriptional response to the aging process in the murine thymus. In the current study, a total of 67 mice were utilized and divided into seven separate categories. Four age groups of mice, namely 1-, 6-, 16-, and 24-month-old animals, were placed on ad libitum diets. The three additional groups also included mice at the ages of 6, 16 and 24 months of age fed a 60% caloric-restricted diet beginning at 4 months of age. These conditions are summarized in Table 1. The thymi of these various animals were obtained and utilized in microarray analysis to assess any differences with age. Out of the 17,000 genes on the cDNA array, 1066 genes significantly changed with age in these various groups. A complete list of all genes that changed with age can be accessed at (GEO Accession in progress). Many replicate genes were spotted at several different locations on the filters to verify reproducibility of the results. The robust quality of the array is reflected in the fact that the statistical analysis of the data identifies the repeated spots (data not shown).
Using the in-house Microarray Data Analysis website of the National Human Genome Research Institute (http://arrayanalysis.nih.gov/), a distance-based gene selection analysis was performed on this data set and an expression profile was generated (Fig. 1a). Dramatic changes in gene expression were observed between mice aged 1 month and 6 months (240 genes changed; 130 up and 110 down) and those aged 16 months (123 genes changed; 93 up and 30 down) or 24 months (1066 genes changed; 587 up and 479 down) (Fig. 1b). The early changes in expression may possibly reflect the known peak of thymic decline between the initiation of puberty and the midlife period in mice [46]. The results in Table 2 list some of the genes demonstrating the greatest increases or decreases when comparing the oldest mice to the youngest mice on the ad libitum diet. A complete list of these genes can be accessed at (GEO Accession in progress). Most of the proteins encoded by these genes are fairly equally divided between the nucleus and the cytoplasm as well as a large number of genes associated with the plasma membrane. The most common genes that changed with age include transcription regulators such as various ETS, forkhead P and the SMARC transcription factors. There were also a number of genes coding for enzymes, such as ATP synthases, MAP kinases and ubiquitin-conjugating enzymes. These proteins perform a variety of functions within the cells and are most commonly associated with cellular growth and proliferation, cell death, cancer and protein synthesis. A striking observation was that of the four most significantly changed genes involved in DNA repair and recombination, every one was down-regulated. The relevance of these gene changes to thymic aging remains to be determined.
Fig. 1.
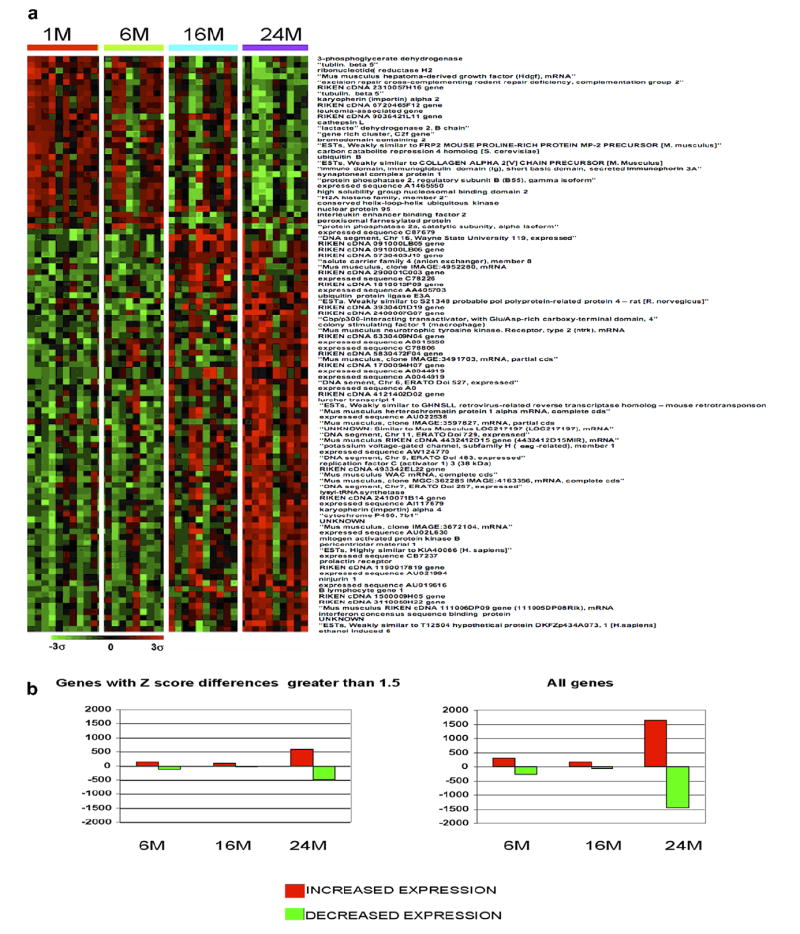
Gene expression profiles of genes which change with age. (a) This profile was created using the Ingenuity program for the genes that exhibited the greatest degree of change when comparing the oldest thymic group to the youngest. The brighter the red, the higher the expression level above the mean for all genes, and the brighter the green the lower the expression level. Expression levels are statistically significant in the 24-month age group. The other age groups are included to exhibit expression trends and may or may not be statistically significant. (b) Comparison of the total numbers of genes affected at each age group.
Table 2.
Genes with the greatest differences in expression when comparing the oldest age group to the youngest age group
| Gene function | No. of genes | Examples | Entrez ID | Fold change |
|---|---|---|---|---|
| Cellular growth and proliferation | 110 | Exportin 4 | 57258 | +3.9 |
| Tnfrsf12a (tumor necrosis factor receptor superfamily) | 27279 | +3.19 | ||
| Cell death | 106 | Dock7 | 67299 | +3.57 |
| Cancer | 104 | RioK1 (rio kinase) | 71340 | +3.43 |
| Gnb2-rs1 (guanine nucleotide binding protein) | 14694 | −4.05 | ||
| Protein synthesis/maintenance | 100 | ZKscan1 (Zinc finger with KRAB and scan domains) | 74570 | +3.53 |
| Zfp294 (Zinc finger) | 78913 | +3.33 | ||
| Eef1a1 (elongation factor) | 13627 | −4.00 | ||
| PPIA (peptidyl isomerase) | 19034 | −4.14 | ||
| Cell cycle | 86 | Cullin 1 | 26965 | +4 |
| Chek1 (checkpoint kinase 1) | 12649 | +3.63 | ||
| Cyclin H | 66671 | −3.84 | ||
| Gene expression and transcription | 72 | TCF7 (T-cell-specific transcription factor) | 21414 | −3.58 |
| Preb (prolactin regulatory element) | 50907 | +3.46 | ||
| Ribosomal S9 | 76846 | −3.53 | ||
| ARbp (ribosomal phosphoprotein) | 11837 | −3.69 | ||
| NR1i3 (nuclear receptor subfamily) | 12355 | +3.36 | ||
| HAT1(histone aminotransferase) | 107435 | +3.26 | ||
| DNA replication, recombination, and repair | 63 | Hmgb2p (high-mobility group, pseudogene) | 15352 | −3.66 |
| Hmgn2 (high-mobility group, binding) | 15331 | −4.95 | ||
| H2A, z (histone 2A) | 51788 | −3.86 | ||
| Hnrpa2b1 (heteronuclear riboprotein) | 53379 | −3.54 | ||
| Cellular assembly and organization | 46 | Tubulin A1 | 22142 | −4.32 |
| Cell morphology | 44 | Actin, β | 11461 | −4.58 |
| Post-translational modification | 41 | Ubiquitin B | 22187 | −3.61 |
| Cathepsin L | 13039 | −5.70 | ||
| Other genes of highly significant weight | SLC16A2 (solute carrier) | 20502 | +3.40 | |
| SLC25 (solute carrier) | 11740 | −3.57 | ||
| IgH6 (immunoglobulin) | 16019 | +3.70 | ||
| IgHg | 380794 | +3.60 | ||
| LDH2 (lactase dehydrogenase) | 16832 | −3.55 |
This table includes the grouping of gene products into their functional classes as determined by the Ingenuity gene ontology program. The examples shown here encompass the 16 most significantly upregulated (bolded) and 16 most significantly down regulated genes (italicized) in the data set. All data presented is statistically significant at the 24-month age group.
Given that the thymus undergoes significant involution with progressive aging, one would expect to see changes in genes that are associated with T-cell development and thymic function (Table 3). Examples of such genes include thymosin beta 10 (Tmsb10), whose expression decreased with age (−2.72-fold) but was restored in older mice on a caloric-restricted diet (+2.90-fold). Tmsb10 is a member of a family of proteins originally isolated from calf thymus and it is critical for proper maintenance of the actin cytoskeleton within cells [47]. Tmsb4, which is in the same family, has previously been shown to decline with age [48]. Several other immune-related genes were also downregulated with aging including cathepsin L (which ranged in value from −2.53-fold at 6 months to −5.70-fold at 24 months), a protease that has been shown to be involved in MHC class II processing in cortical thymic epithelial cells [49]. Interestingly, expression of β-actin RNA also decreased (−4.58-fold) with age in the array analysis and this may correspond to reports demonstrating that actin polymerization is impaired in aged mouse T lymphocytes [50]. All of these above changes were consistent with the literature regarding age-associated changes to the thymus, lending credence to our observations. Whether these changes in expression are due to the global changes in the thymic structure and make-up or due simply to the loss of thymocyte numbers needs to be addressed. For example, the data in Fig. 2a confirms our microarray results demonstrating an age-associated increase in IgM and IgG heavy chain levels using real time RT-PCR on mRNA derived from total thymus and purified thymocytes. This increase is also evident in the protein expression of these immunoglobulin genes in aged thymocytes Fig. 2b. These data suggest that the changes in the expression of certain genes such as immunoglobulins may be due to changes in the cellular make-up of the thymocyte subpopulations rather than global changes in the thymic structure or thymic cellularity.
Table 3.
Changes in immune-related genes with age
| Gene | Gene name | Entrez Gene ID | Change at 24 months | Function |
|---|---|---|---|---|
| Actb | Actin, β, cytoplasmic | 11461 | −4.58 | Impaired in older T cells |
| Actr3 | ARP3 actin-related protein 3 homolog (yeast) | 74117 | −2.01 | Involved in actin polymerization for migration of leukocytes |
| Arf1 | ADP-ribosylation factor 1 | 11840 | −1.75 | Cytoskeletal remodeling of lymphoid cells |
| Biklk | Bcl2-interacting killer-like | 12124 | −1.61 | B cell-specific tyrosine kinase |
| Calm1 | Calmodulin 1 | 12313 | −2.38 | Upregulated in activated T cell lines |
| Capns1 | Calpain, small subunit 1 | 12336 | −2.88 | Plays important role in CD4+ T-cell selection in the thymus |
| Ctsl | Cathespin L | 13039 | −5.70 | CD4+ T-cell selection in the thymus |
| Cullin1 | Cullin 1 | 26965 | 4.00 | T cell production |
| Diablo | Diablo homolog (Drosophilia) | 66593 | −2.51 | T cell apoptosis |
| Ets2 | E26 avian leukemia oncogene 2, 3’ domain | 23872 | −2.03 | Proliferation and survival of thymocytes |
| Hmgn2 | High mobility group nucleosomal binding domain 2 | 15331 | −2.29 | Associated with immune diseases |
| Hspa8 | Heat-shock 70 kDa protein 8 | 15481 | −2.79 | Induce CTL; carries antigenic peptides |
| Hspca | Heat-shock protein 1, α | 15519 | −2.82 | High levels in rabbit thymus |
| Hspcb | Heat-shock protein 1, β | 15516 | −3.28 | High levels in rabbit thymus |
| Lsm2 | LSM2 homolog, U6 small nuclear RNA associated (Saccharomyces cerevisiae) | 27756 | −1.97 | Variant predominantly expressed in thymus |
| Mapk8 | Mitogen-activated protein kinase 8 | 26419 | 2.10 | B cell survival |
| Mcm8 | Minichromosome maintenance deficient 3 (Saccharomyces cerevisiae) | 17215 | −1.74 | B cell response to T-dependent antigens |
| Ncl | Nucleolin | 17975 | −2.92 | Thymocyte apoptosis and differentiation |
| Neu1 | Neuraminidase 1 | 18010 | −1.57 | IL-4 synthesis in T cells |
| Nr3c1 | Nuclear receptor subfamily 3, group C, member 1 | 14815 | −2.01 | Necessary for thymocyte survival |
| Ntrk2 | Mus musculus neurotrophic tyrosine kinase, receptor, type 2 (Ntrk2) | 18212 | 2.93 | Thymocyte differentiation |
| Pcna | Proliferating cell nuclear antigen | 18538 | −1.75 | Decreased thymocyte proliferation |
| Ptk2 | PTK2 protein tyrosine kinase 2 | 14083 | −2.09 | Chemokine response |
| Rpl7 | Ribosomal protein L7 | 19989 | −2.98 | High levels of anti-RPL7 IgG in SLE |
| Socs3 | Suppressor of cytokine signaling 3 | 12702 | −1.65 | Immune regulation |
| Tia1 | Cytotoxic granule-associated RNA binding protein 1 | 21841 | −1.51 | Expressed mainly in T cells and NK cells |
| Tmsb10 | Thymosin, β 10 | 19240 | −2.72 | Thymosin, β 10 |
| Tra1 | Tumor rejection antigen gp96 | 22027 | −2.38 | Antitumor immunity; CD8+ T-cell responses |
| Tubb5 | Tubulin, β 5 | 22154 | −2.40 | Abundance in spleen, thymus, and brain |
| Ywhah | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, η polypeptide | 22629 | −2.15 | Control of T cell apoptosis |
| Ywhaz | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ polypeptide | 22631 | −2.18 | Control of T cell apoptosis |
In the list of genes with the greatest differences in expression levels when comparing oldest to youngest groups, there were many genes with published associations with the immune system. The vast majority are downregulated (italicized). Changes 1.5-fold or greater are in bold and changes −1.5-fold or less are in italics.
Fig. 2.
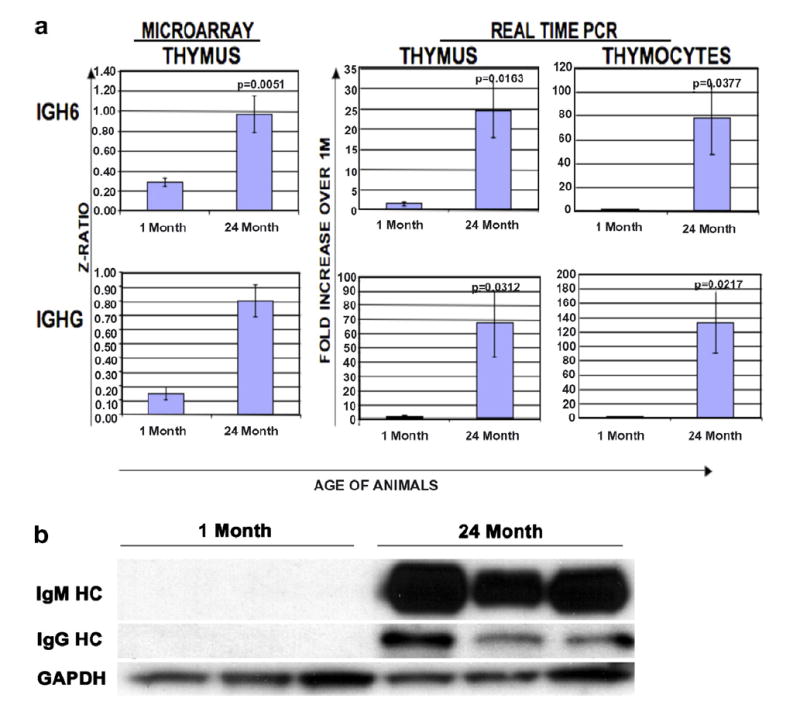
Confirmation of age-associated changes in gene expression data in thymic tissue. (a) Side-by side comparison of the array results for the genes for the heavy chains, IgM and IgG and the results when comparing young and old thymocyte and thymus samples using real-time PCR. (b) Age-associated differences in immunoglobulin protein expression are evident when comparing protein lysates derived from the total thymocytes of young (1 month) and old (24 months) mice.
3.2. Caloric restriction prevents gene expression changes observed within aging murine thymi
Laboratory animals placed on a caloric-restricted diet have exhibited an extended lifespan directly correlated with the degree of restriction [51,52]. Caloric restriction also delays the onset and reduces the incidence of age-associated diseases and complications [52-54]. In light of these observations, it was of interest to examine how calorie restriction affected the gene expression profiles of aging mouse thymi (Table 4). The data in Fig. 3a show the gene expression profile for thymi derived from mice on a CR diet compared to mice on an AL diet. Surprisingly, in all groups, changes associated with the aging profile are prevented by a CR diet by 24 months of age, reflecting the profile observed in the youngest AL mice. There were 472 genes expressed in the thymus that demonstrated significantly different levels of expression when the 24-month-old mice on a CR diet were compared to their age-matched controls on an AL diet. Dramatic changes in gene expression were observed between mice 1-month-old AL mice versus 6-month-old AL (8 genes changed; 5 up and 3 down) and CR mice (476 genes changed; 341 up and 135 down) and those of the AL (0 genes changed) and CR (165 genes changed; 97 up and 68 down) of 12 months of age or 24-month-old AL (278 genes changed; 153 up and 125 down) and CR (472 genes changed; 222 up and 250 down) (Fig. 3b). Many of the proteins encoded by these genes are fairly equally divided between the nucleus and the cytoplasm, with fewer gene products associated with the plasma membrane (data not shown). The most common genes affected by CR included transcription regulators such as BCL2-associated transcription factor 1 (+2.1, data not shown) and histone deacetylase 1 (+1.66, data not shown) and enzymes such as fatty acid desaturase 1 (+2.32) and thioredoxin (+1.68, data not shown). Moreover, genes encoding transporter proteins (including transportin 1 [−1.5], not shown) and kinases (including Pak2 [+3.61] and PTK2 [+2.19]) also demonstrated significant changes with age and diet. These proteins perform a wide variety of functions within the cells but are most commonly associated with cell death, gene expression, cancer growth and development and cell cycle.
Table 4.
Age-associated differences in expression when comparing the oldest CR mice to the oldest AL group
| Gene | ACC | Name | 6 months AL | 16 months AL | 24 months AL | 6 months CR | 16 months CR | 24 months CR |
|---|---|---|---|---|---|---|---|---|
| (A) | ||||||||
| EST | AW537151 | Mus musculus cDNA clone G0112H12 | −1.22 | −3.62 | −2.82 | −0.85 | 1.77 | 4.46 |
| Hspa8 | BG087426 | Heat-shock 70 kDA protein 8 | −0.68 | −2.95 | −2.79 | −0.49 | 2.43 | 4.22 |
| Hsp86-1 | BG064773 | Heat-shock protein, 86 kDa 1 | −0.89 | −1.56 | −2.76 | −0.68 | −1.14 | 3.95 |
| Ctsl | BG078496 | Cathepsin L | −2.53 | −4.44 | −5.70 | −2.85 | −1.70 | 3.85 |
| Pkn2 | BG078640 | Protein Kinase N2 | −0.93 | −2.93 | −3.13 | −2.14 | −1.71 | 3.79 |
| Hsp84-1 | BG079631 | Heat-shock protein, 84 kDa 1 | −1.78 | −3.53 | −3.28 | −1.81 | −0.80 | 3.74 |
| EST | AU023376 | Mus musculus cDNA clone J0431A10 | −1.39 | −2.88 | −2.77 | −1.31 | −0.05 | 3.71 |
| Slc25a5 | BG078462 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | −1.46 | −2.93 | −3.57 | −2.12 | −1.03 | 3.64 |
| Pak2 | BG065345 | p21 (CDKN1A)-activated kinase 2 | −0.50 | −3.33 | −2.90 | −2.18 | −0.68 | 3.61 |
| Hmgn2 | BG078806 | High mobility group nucleosomal binding domain 2 | −2.29 | −4.45 | −4.95 | −4.43 | −1.91 | 3.50 |
| EST | BG071906 | Mus musculus cDNA clone H3104E04 | 0.46 | 1.22 | 2.88 | 2.85 | 3.76 | −3.21 |
| AA209988 | AW555450 | Mus musculus cDNA clone L0253F11 | 0.22 | 1.20 | 3.36 | 2.49 | 3.03 | −3.23 |
| EST | BG068086 | ESTs, Weakly similar to GNMSLL retrovirus-related reverse transcriptase homolog | 0.00 | 1.40 | 2.86 | 2.27 | 1.68 | −3.25 |
| D1Ertd218e | BG066358 | DNA segment, Chr 1, ERATO Doi 218, expressed | −0.29 | 1.18 | 2.63 | 2.61 | 1.02 | −3.25 |
| Igfbp7 | BG068536 | Insulin-like growth factor binding protein 7 | 0.48 | 1.65 | 2.62 | 2.48 | 1.32 | −3.28 |
| EST | BQ551169 | Mus musculus cDNA clone H4007G03 | 1.00 | 1.68 | 4.17 | 2.97 | 2.71 | −3.33 |
| EST | BG070998 | Mus musculus cDNA clone H3093E11 | −0.59 | 1.04 | 2.47 | 2.92 | 1.70 | −3.35 |
| EST | BG067761 | Mus musculus cDNA clone H3057H05 | −0.26 | 3.47 | 3.98 | 3.45 | 1.44 | −3.41 |
| EST | BQ551211 | Mus musculus cDNA clone H4008A02 | 1.00 | 1.67 | 4.06 | 2.55 | 3.24 | −3.90 |
| EST | BG070193 | Mus musculus cDNA clone H3084F12 | 0.15 | 1.38 | 4.27 | 2.57 | 5.44 | −4.02 |
| (B) | ||||||||
| Ptpns1 | BQ551724 | Mus musculus protein tyrosine phosphatase, non-receptor type substrate 1 (Ptpns1) | −1.39 | −0.24 | −2.86 | −1.67 | −0.50 | 5.20 |
| EST | BQ551457 | Mus musculus cDNA clone H4009D09 | −4.38 | 1.73 | −2.56 | 2.47 | −2.23 | 2.94 |
| EST | BG082760 | Mus musculus cDNA clone H3079H08 | −0.47 | −1.30 | −1.37 | −1.25 | −0.34 | 2.52 |
| Metap2 | BG086961 | Methionine aminopeptidase 2 | −0.22 | 0.61 | −2.18 | −0.54 | −0.78 | 2.42 |
| Fads1 | BG078755 | RIKEN cDNA 0710001O03 gene | −0.25 | 1.19 | −1.26 | −1.32 | −3.42 | 2.32 |
| EST | BG086658 | Mus musculus cDNA clone H3129A03 | 0.64 | −0.55 | −1.95 | −1.21 | −0.22 | 2.32 |
| Zfp87 | NA | Zinc finger protein 87 | −0.21 | −0.25 | −2.25 | −0.53 | −0.59 | 2.30 |
| AU021304 | BG069921 | Expressed sequence AU021304 | −0.96 | 2.22 | −1.36 | −0.66 | −4.43 | 2.28 |
| Hspa5 | BG078795 | Heat-shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | −0.51 | −2.16 | −1.08 | −0.52 | 1.64 | 2.26 |
| EST | NA | ESTs, Weakly similar to SCP3 mouse synaptonemal complex protein 3 | −0.56 | −0.14 | −1.10 | −0.39 | −1.44 | 2.19 |
| C78452 | BG066208 | Expressed sequence C78452 | −1.74 | 1.50 | 2.41 | 3.89 | 2.71 | −2.96 |
| EST | BG069906 | Mus musculus cDNA clone H3081D07 | −4.00 | −5.77 | −1.52 | −4.17 | −4.16 | −3.10 |
| EST | BQ551932 | Mus musculus cDNA clone H4012D07 | −1.20 | −0.54 | 0.71 | 0.60 | 0.03 | −3.20 |
| EST | AU015698 | Mus musculus cDNA clone J0715C06 | −1.10 | 1.39 | 2.51 | 2.98 | 1.69 | −3.22 |
| EST | BG066049 | ESTs, Weakly similar to I49130 reverse transcriptase—mouse [Mus musculus] | −1.02 | 1.98 | 2.63 | 3.21 | 1.27 | −3.26 |
| EST | BQ552641 | Mus musculus cDNA clone H4017B10 | −1.19 | 1.57 | 2.60 | 4.11 | 2.24 | −3.29 |
| EST | BG068246 | Mus musculus cDNA clone H3063C12 | 0.79 | 1.75 | 2.96 | 2.56 | 2.81 | −3.53 |
| Vps41 | NA | Vacuolar protein sorting 41 (yeast) | −0.48 | 1.22 | 2.27 | 0.89 | 1.55 | −3.65 |
| EST | BG068614 | Mus musculus cDNA clone H3067D11 | 0.62 | 2.77 | 3.10 | 2.87 | 1.91 | −3.65 |
| Cenpa | BG082881 | Centromere autoantigen A | 0.04 | 2.76 | 1.47 | −0.10 | −3.55 | −4.33 |
(A) The data presented are statistically significant at the 24-month age group comparing animals under different dietary regiments. (B) The data presented are statistically significant differences in gene expression affected by a CR diet independent of age. Values for other age groups are shown for complete information but may not necessarily be substantial or statistically significant values. Changes 1.5-fold or greater are in bold and changes −1.5-fold or less are in italics.
Fig. 3.
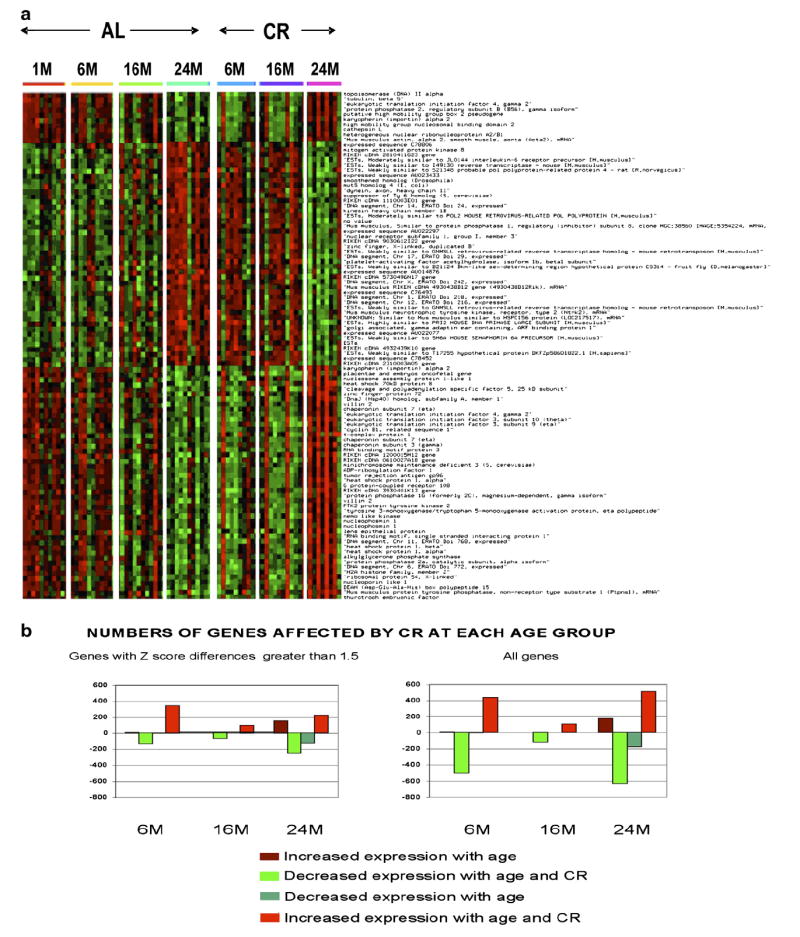
Expression profile of genes exhibiting the greatest differences when comparing the oldest CR mice to the oldest AL mice. (a) This profile was created using the Ingenuity program for the genes that exhibited the greatest degree of change when comparing the oldest (24 months) CR group to the oldest AL group. The brighter the red, the higher the expression level above the mean for all genes, and the brighter the green the lower the expression level. Expression levels are statistically significant at the 24-month age group. The other age groups are included to exhibit expression trends and may or may not be statistically significant. (b) Comparison of the total numbers of genes affected at each age group.
The most significantly affected genes that changed with age and were also affected by CR in each of the top gene categories, appear to be transcription regulators, enzymes, kinases and transporters, while additional diet-associated gene expression differences are observed in several smaller categories. Aging seemed to affect a greater proportion of peptidases, transmembrane receptors, ion-channel genes and cytokines, while a CR diet influences a greater proportion of ligand-dependent nuclear receptors and growth factors. These data are summarized in Fig. 4a. Fig. 4b shows that many of the genes affected by aging tended to also be involved in cellular growth and proliferation and protein synthesis, while a greater percentage of the genes affected by a CR diet are involved in development and survival. Focusing on the genes in these latter categories may provide some insight into the mechanism by which a CR diet exerts its beneficial effects.
Fig. 4.
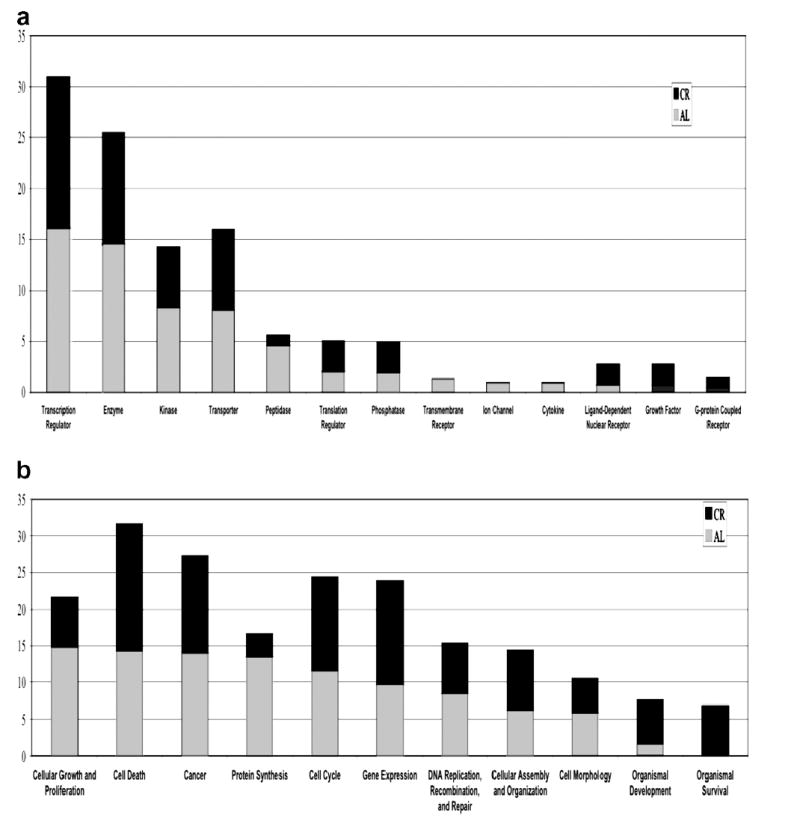
Relative percentages of genes that changed with age and were affected by CR in each of the top categories of gene type (a) and gene function (b). In each graph, the percentage of age and CR-affected genes are shown together for a direct comparison.
The results in Table 4 demonstrate that several of the genes which increase or decrease by the greatest margins were age-associated when comparing the oldest CR mice to the oldest AL mice (Table 4). Immediately evident in this list are several heat-shock proteins that are more highly expressed in aged CR mice compared to age-matched AL-fed mice. These genes were found to decrease in expression within the thymus with age; however, mice placed on CR diets demonstrated heat shock protein levels similar to those seen in the youngest AL mice. Similar reversals were observed in quite a number of genes including PTPNS1 (Table 4), cathepsin L and the glucocorticoid receptor.
3.3. Sex-specific transcriptional changes were also noted within the thymus and over progressive aging
Statistically significant changes in thymic gene expression were observed between male and female mice at specific ages with 158 genes (38 up and 120 down) demonstrating a change at 6 months, 159 genes (113 up and 46 down) demonstrating a change at 16 months and 215 genes (151 up and 64 down) demonstrating a change at 24 months. Fig. 5a shows the expression profile of statistically significant gene expression differences between males and females at 24 months of age. Values for other age groups in this figure are not statistically significant, but aging trends are quite evident. The gene list can be separated into three groups, namely genes that increase with age, decrease with age or genes that decrease with age in males but are highly expressed throughout the females’ lifetime. In addition, in the first two groups, the thymic expression pattern between males and females is quite similar; however, the degree of such expression changes is more subtle in the females. Interestingly, many of these gender-associated changes in gene expression were found to be related to cancer development and growth.
Fig. 5.
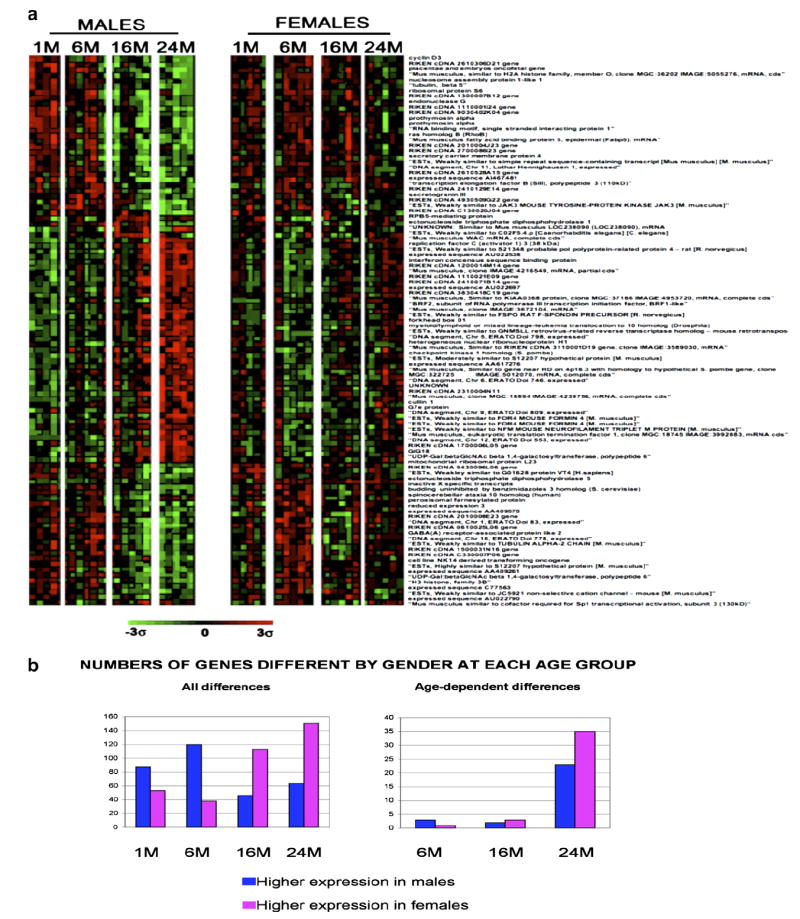
Expression profiles of genes that demonstrate expression differences with gender. (a) This profile was created using the Ingenuity program for the genes that exhibited the greatest degree of change when comparing the oldest female ad libitum group to the oldest male ad libitum group. The brighter the red, the higher the expression level above the means for all genes, and the brighter the green the lower the expression level. Expression levels are statistically significant for the 24-month age group. The other age groups are included to display expression trends and may or may not be statistically significant. (b) Comparison of the total numbers of genes affected in each gender in the context of age.
In addition, the differences observed between male and female mice in the context of thymic aging revealed approximately 532 genes significantly different between the oldest males and females. Although trends are evident in Fig. 5a, only four statistically significant changes in gene expression were observed between 1- and 6-month-old male vs. female mice (1 up, 3 down), many with only subtle changes in expression (+1.07- to −1.34-fold). The same is true for 1- and 16-month-old male vs. female mice (5 genes changed; 3 up and 2 down), although the differences are much larger (+3.87 to −3.33). However, very pronounced differences begin to develop when comparing the 1- and 24-month-old male vs. female mice (58 genes changed; 35 up and 23 down) (Fig. 5b). The majority of these genes code for proteins that function within the nucleus, while several others code for cytoplasmic proteins. The most common differences in gene expression encode transcription regulators and enzymes as well as genes involved in a variety of functions within the cells, including cell cycle, DNA replication, recombination and repair and cancer development. The genes that demonstrate the largest differences between males and females are listed in Table 5.
Table 5.
Gender- and age-associated differences in gene expression
| Gene symbol | Gene name | Entrez ID | Overall
|
Female vs male
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 6 months | 16 months | 24 months | 1 month | 6 months | 16 months | 24 months | |||
| (A) | |||||||||
| Xist | Inactive X specific transcripts | 22438 | 2.48 | 1.71 | 1.49 | 3.55 | 6.52 | 3.22 | 3.66 |
| Ablim1 | Actin-binding LIM protein 1 | 107346 | 0.41 | −2.38 | −2.17 | −1.07 | −0.13 | 2.16 | 3.40 |
| Igfbp5 | Insulin-like growth factor binding protein 5 | 98292 | 1.46 | −0.16 | −0.43 | −0.66 | −0.02 | −1.48 | 3.04 |
| H3f3b | H3 histone, family 3B | 15081 | −0.26 | −2.10 | −1.92 | −0.55 | 1.36 | 2.17 | 2.19 |
| Rnpc2 | RNA-binding region (RNP1, RRM) containing 2 | 170791 | 0.17 | −1.07 | −1.02 | −1.65 | 0.20 | 0.89 | 2.06 |
| ln | Leaden | 16920 | 0.63 | 0.31 | 0.76 | −0.09 | 0.27 | −0.53 | 1.93 |
| Wasl | Wiskott–Aldrich syndrome-like (human) | 73178 | 0.21 | 0.57 | −0.35 | −1.40 | −0.37 | −0.71 | 1.91 |
| Adrm1 | Adhesion regulating molecule 1 | 56436 | −0.33 | −0.96 | −1.09 | 0.34 | 1.21 | 0.85 | 1.86 |
| Shyc | Selective hybridizing clone | 20430 | 0.06 | −0.59 | −0.47 | −0.74 | 1.08 | 0.46 | 1.85 |
| Bcas3 | Breast carcinoma amplified sequence 3 | 104502 | 0.56 | −0.21 | −0.10 | −0.31 | −0.18 | −0.22 | 1.84 |
| D5Ertd798e | DNA segment, Chr 5, ERATO Doi 798, expressed | 52409 | −0.45 | 0.57 | 2.56 | 2.48 | 0.67 | −0.03 | −3.67 |
| Pla2g7 | Phospholipase A2 group VII | 27226 | −0.71 | 1.67 | 1.34 | 0.27 | 1.19 | −1.40 | −3.70 |
| Brf2 | BRF2, subunit of RNA polymerase III transcription initiation factor | 66653 | −0.85 | 2.72 | 3.07 | 2.90 | −1.65 | −1.76 | −3.80 |
| Hat1 | Histone aminotransferase 1 | 67376 | −0.58 | 3.17 | 3.26 | 1.69 | −0.93 | −2.42 | −4.15 |
| Oog3 | Oogenesin 3 | 100012 | −0.50 | 2.59 | 2.63 | 1.89 | −1.01 | −0.91 | −4.34 |
| Chek1 | Checkpoint kinase 1 homolog (Schizosaccharomyces pombe) | 12649 | −0.33 | 3.40 | 3.63 | 3.01 | 0.39 | −1.94 | −4.56 |
| 1200014M14Rik | RIKEN cDNA 1200014M14 gene | 67463 | −0.21 | 1.13 | 3.51 | 2.94 | −1.26 | −0.64 | −4.57 |
| No value | Mus musculus, clone IMAGE:4216549, mRNA, partial cds | No value | −0.80 | 3.74 | 3.98 | 4.21 | −1.20 | −1.73 | −5.07 |
| Cul1 | Cullin 1 | 26965 | −0.18 | 1.12 | 4.00 | 3.42 | 0.75 | −0.37 | −5.27 |
| No value | Mus musculus, clone IMAGE:3672104, mRNA | No value | −0.02 | 2.00 | 4.50 | 3.24 | −0.98 | −0.96 | −5.33 |
| (B) | |||||||||
| Adrm1 | Adhesion regulating molecule 1 | 56436 | −0.33 | −0.96 | −1.09 | 0.34 | 1.21 | 0.85 | 1.86 |
| Arhb | Ras homolog B (RhoB) | 11852 | 0.17 | −0.97 | −0.94 | −0.94 | 0.09 | 0.80 | 1.81 |
| No value | ESTs, Weakly similar to G01628 protein VT4 (Homo sapiens) | No value | −0.80 | −1.28 | −1.19 | 0.81 | −0.52 | 0.15 | 1.79 |
| No value | Mus musculus, clone IMAGE:4504748, mRNA | No value | −0.43 | −1.86 | −1.21 | −0.60 | 1.26 | 2.29 | 1.77 |
| Fabp5 | Mus musculus fatty acid binding protein 5, epidermal (Fabp5) | 16592 | −0.05 | −1.09 | −1.11 | −0.64 | 0.66 | 1.64 | 1.70 |
| Bzw2 | Basic leucine zipper and W2 domains 2 | 66912 | −0.55 | −1.58 | −1.42 | −1.21 | 0.04 | 0.05 | 1.65 |
| Tubb5 | Tubulin, β 5 | 22154 | −0.23 | −1.18 | −1.40 | −0.83 | 0.69 | 0.37 | 1.63 |
| Mki67ip | Mki67 (FHA domain) interacting nucleolar phosphoprotein | 67949 | −0.27 | −0.10 | −1.10 | 0.20 | −0.26 | −0.61 | 1.62 |
| Nap1l1 | Nucleosome assembly protein 1-like 1 | 53605 | −0.89 | −0.79 | −1.27 | −1.33 | 0.33 | 0.22 | 1.57 |
| Rfc3 | Replication factor C (activator 1) 3 (38 kDa) | 69263 | −0.53 | 2.58 | 2.56 | 2.17 | −0.39 | −1.48 | −3.47 |
| D5Ertd798e | DNA segment, Chr 5, ERATO Doi 798, expressed | 52409 | −0.45 | 0.57 | 2.56 | 2.48 | 0.67 | −0.03 | −3.67 |
| Brf2 | BRF2, subunit of RNA polymerase III transcription initiation factor | 66653 | −0.85 | 2.72 | 3.07 | 2.90 | −1.65 | −1.76 | −3.80 |
| Hat1 | Histone aminotransferase 1 | 67376 | −0.58 | 3.17 | 3.26 | 1.69 | −0.93 | −2.42 | −4.15 |
| Oog3 | Oogenesin 3 | 100012 | −0.50 | 2.59 | 2.63 | 1.89 | −1.01 | −0.91 | −4.34 |
| Chek1 | Checkpoint kinase 1 homolog (Schizosaccharomyces pombe) | 12649 | −0.33 | 3.40 | 3.63 | 3.01 | 0.39 | −1.94 | −4.56 |
| 1200014M14Rik | RIKEN cDNA 1200014M14 gene | 67463 | −0.21 | 1.13 | 3.51 | 2.94 | −1.26 | −0.64 | −4.57 |
| No value | Mus musculus, clone IMAGE:4216549, mRNA, partial cds | No value | −0.80 | 3.74 | 3.98 | 4.21 | −1.20 | −1.73 | −5.07 |
| Cul1 | Cullin 1 | 26965 | −0.18 | 1.12 | 4.00 | 3.42 | 0.75 | −0.37 | −5.27 |
| No value | Mus musculus, clone IMAGE:3672104, mRNA | No value | −0.02 | 2.00 | 4.50 | 3.24 | −0.98 | −0.96 | −5.33 |
(A) This table shows the genes with the greatest age-associated differences in expression when comparing the oldest female ad libitum age group to the oldest male ad libitum age group. All data presented is statistically significant at the 24-month age group. (B) Age-associated differences between males and females on an AL diet. These genes not only showed differences in expression levels between males and females but also had statistically significant differences when comparing the oldest to the youngest age group as a whole. Values for other age groups are shown for complete information but may not necessarily be substantial or statistically significant values. Changes 1.5-fold or greater are in bold and changes −1.5-fold or less are in italics.
Some genes were expected to change, lending validation to the array data, such as the Xist gene, which is involved in the silencing of the second X chromosome [55], and was higher in females than males. Other genes such as Tmsb10, which decreased in expression overall with age as mentioned before (Table 2), decreased more acutely in males than females. However, on a caloric restricted diet, the oldest mice exhibited Tmsb10 levels equivalent to those seen in young mice on an unrestricted diet, regardless of gender. Interestingly, thymosin beta 10 upregulation has been shown to be associated with carcinogenesis [56]. Another carcinogenesis-related gene on this list was checkpoint kinase 1, which is regulated by tumor suppressor protein p53 [57] and may also be involved in carcinogenesis. In fact, of the top 100 identifiable genes showing differential expression levels between the oldest males and females, 53 are tumor-associated (Table 6).
Table 6.
Tumor-associated genes which showed the greatest differences in expression levels at age 24 months when female values were compared to male values
| Gene symbol | Gene name | 1 month | 6 months | 16 months | 24 months |
|---|---|---|---|---|---|
| Xist | Inactive X specific transcripts | 3.55 | 6.52 | 3.22 | 3.66 |
| Ablim1 | Actin-binding LIM protein 1 | −1.07 | -0.13 | 2.16 | 3.40 |
| Igfbp5 | Insulin-like growth factor binding protein 5 | −0.66 | −0.02 | −1.48 | 3.04 |
| Tmsb10 | Thymosin, β 10 | −0.46 | 3.31 | 3.26 | 2.44 |
| Ptma | prothymosin α | −1.23 | 0.77 | 4.00 | 2.11 |
| Wasl | Wiskott–Aldrich syndrome-like (human) | −1.40 | −0.37 | −0.71 | 1.91 |
| Bcas3 | Breast carcinoma amplified sequence 3 | −0.31 | −0.18 | −0.22 | 1.84 |
| Rhob | Ras homolog gene family, member B | −0.94 | 0.09 | 0.80 | 1.81 |
| Col2a1 | Procollagen, type II, α 1 | −0.37 | −0.24 | 0.87 | 1.79 |
| Fabp5 | Mus musculus fatty acid binding protein 5, epidermal (Fabp5), mRNA | −0.64 | 0.66 | 1.64 | 1.70 |
| Pem | Placentae and embryos oncofetal gene | −0.28 | −0.81 | 0.96 | 1.65 |
| Bzw2 | Basic leucine zipper and W2 domains 2 | −1.21 | 0.04 | 0.05 | 1.65 |
| Ncor1 | Nuclear receptor co-repressor 1 | −0.56 | 1.12 | 1.26 | 1.64 |
| Sp3 | Trans-acting transcription factor 3 | 0.52 | −0.17 | 0.36 | 1.57 |
| Nap1l1 | Nucleosome assembly protein 1-like 1 | −1.33 | 0.33 | 0.22 | 1.57 |
| Got2 | Glutamate oxaloacetate transaminase 2, mitochondrial | −1.98 | 0.17 | −0.41 | 1.56 |
| Fancl | Fanconi anemia, complementation group L | −0.59 | −1.22 | 0.42 | 1.56 |
| Hif1a | Hypoxia inducible factor 1, alpha subunit | 0.07 | 0.65 | 1.00 | 1.51 |
| Clic4 | Chloride intracellular channel 4 (mitochondrial) | −0.70 | 1.01 | 0.41 | 1.50 |
| Edf1 | Endothelial differentiation-related factor 1 | −0.41 | 2.65 | 2.97 | 1.46 |
| Sdcbp | Syndecan binding protein | −0.58 | −0.11 | −0.07 | 1.46 |
| Ldb1 | LIM domain binding 1 | 0.57 | −0.21 | −0.77 | 1.45 |
| Rab8a | RAB8A, member RAS oncogene family | −0.50 | 0.95 | −0.13 | 1.44 |
| Atp2a2 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 | −0.59 | −3.52 | −0.29 | 1.40 |
| Entpd1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 0.00 | −0.13 | −0.93 | 1.38 |
| Rps16 | Ribosomal protein S16 | −0.75 | 0.58 | 2.55 | 1.37 |
| Cdc25a | Cell division cycle 25 homolog A (Saccharomyces cerevisiae) | −0.57 | −0.38 | −0.78 | 1.37 |
| Bcl9 | B-cell CLL/lymphoma 9 | 1.51 | −2.02 | −0.50 | −1.03 |
| Mad1l1 | Mitotic arrest-deficient 1-like 1 | 0.92 | −1.15 | 1.24 | −1.06 |
| Ptprg | Protein tyrosine phosphatase, receptor type, G | −1.40 | 0.29 | 1.05 | −1.09 |
| Bcas1 | Breast carcinoma amplified sequence 1 | 0.31 | −0.40 | 0.51 | −1.11 |
| Jak2 | Janus kinase 2 | 0.92 | 0.79 | −1.06 | −1.26 |
| Foxo1 | Forkhead box O1 | 1.06 | −0.40 | −1.62 | −1.38 |
| Rgs2 | Regulator of G-protein signaling 2 | 2.09 | 0.48 | −1.69 | −1.43 |
| Lancl2 | LanC (bacterial lantibiotic synthetase component C)-like 2 | 1.88 | −0.69 | 0.13 | −1.46 |
| Tde2 | Tumor differentially expressed 2 | −0.79 | 0.54 | 2.21 | −1.74 |
| Eps8 | Epidermal growth factor receptor pathway substrate 8 | −0.01 | −1.59 | −0.82 | −1.75 |
| Icsbp | Interferon concensus sequence binding protein | 2.31 | −1.07 | −0.95 | −1.77 |
| Lgals8 | Lectin, galactose binding, soluble 8 | 0.11 | 1.11 | −0.59 | −1.80 |
| Llglh | Lethal giant larvae homolog | −0.38 | 0.80 | 0.50 | −1.82 |
| Mdm4 | Transformed mouse 3T3 cell double minute 4 | 0.71 | 3.74 | −0.96 | −1.90 |
| G3bp2 | Ras-GTPase-activating protein (GAP<120>) SH3-domain binding protein 2 | 1.02 | −0.98 | −0.23 | −2.10 |
| Ccnd1 | Cyclin D1 | −0.35 | −1.79 | −1.69 | −2.23 |
| Greb1 | Gene regulated by estrogen in breast cancer protein | 2.87 | −0.56 | −1.04 | −2.29 |
| Mllt10 | Myeloid/lymphoid or mixed lineage-leukemia translocation to 10 homolog (Drosophila) | 1.64 | −1.13 | −0.81 | −2.32 |
| Ube2n | Ubiquitin-conjugating enzyme E2N | 1.31 | −0.88 | −0.55 | −2.56 |
| Rassf5 | Ras association (RalGDS/AF-6) domain family 5 | 1.87 | −0.37 | 0.27 | −2.73 |
| Cbl | Casitas B-lineage lymphoma | 1.88 | −1.23 | −0.91 | −3.44 |
| Pla2g7 | Phospholipase A2 group VII (platelet-activating factor acetylhydrolase, plasma) | 0.27 | 1.19 | −1.40 | −3.70 |
| Brf2 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like | 2.90 | −1.65 | −1.76 | −3.80 |
| Hat1 | Histone aminotransferase 1 | 1.69 | −0.93 | −2.42 | −4.15 |
| Chek1 | Checkpoint kinase 1 homolog (Schizosaccharomyces pombe) | 3.01 | 0.39 | −1.94 | −4.56 |
| Cul1 | Cullin 1 | 3.42 | 0.75 | −0.37 | −5.27 |
All data presented are statistically significant in at least one age group. Changes 1.5-fold or greater are in bold and changes −1.5-fold or less are in italics.
3.4. Age-associated changes in gene expression levels reflect changes in thymic cellularity and thymocytes
Given that the thymus involutes with age and a significant loss in thymocyte numbers occurs, it is important to determine whether the observed changes in the array samples were due to the relative decrease in thymocyte number or due to intrinsic changes within the thymocytes and/or thymic tissue. Table 7 shows the array results that were confirmed by quantitative real-time PCR using the total thymus samples used for the array. These same genes were then examined using purified age-matched thymocytes to determine whether these confirmed changes could be attributed simply to a diminished thymic cellularity or changes within the thymocytes themselves. Several of these genes demonstrated changes with age in the purified thymocytes, reflecting the changes observed in the thymic arrays. In contrast to the thymic array data, other genes including several heat-shock proteins failed to demonstrate any changes in expression in thymocytes at various ages. Clearly, both thymocyte-specific and architectural changes appear to be important contributors to thymic involution. The involution process could be influenced by increases in gene expression attributed to the infiltration of fat cells in the thymus and/or decreases in gene expression due to changes in the thymocyte number and profile.
Table 7.
Summary of real-time PCR confirmation of array results and comparison to thymocyte gene expression
| Symbol | Gene name | Entrez ID | Changes with age in:
|
||
|---|---|---|---|---|---|
| Array | Thymus | Thymocytes | |||
| CCT3 | Chaperonin subunit 3 (γ) | 12462 | D | D* | D |
| CTSL | Cathepsin L | 13039 | D | D | D* |
| Dock7 | Dedicator of cytokinesis 7 | 67299 | U | U | U |
| EIF3S10 | Eukaryotic translation initiation factor 3, subunit 10 (θ) | 13669 | D | D | NC |
| EIF4G2 | Eukaryotic translation initiation factor 4, γ 2 | 13690 | D | D* | D* |
| ETFB | Electron transferring flavoprotein, β polypeptide | 110826 | U | U* | NC |
| HSPA8 | Heat-shock 70 kDa protein 8 | 15481 | D | D | NC |
| HSPCA | Heat-shock protein 1, α | 15519 | D | D | D* |
| HSPCB | Heat-shock protein 1, β | 15516 | D | D | NC |
| IGH6 | Immunoglobulin heavy chain 6 (heavy chain of IgM) | 16019 | U | U* | U* |
| IGHG | Immunoglobulin heavy chain (γ polypeptide) | 380794 | U | U* | U* |
| NR1I3 | Nuclear receptor subfamily 1, group I, member 3 | 12355 | U | U | NC |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 | 14815 | D | D* | D |
| SMARCD2 | SWI/SNF-related, matrix associated, actin-dependent regulation of chromatin, subfamily d, 2 | 83796 | D | D | D* |
| VIL2 | Villin 2 | 22350 | D | D* | D |
Data represent comparison of results using six samples from each of the 1- and 24-month age groups. D, downregulated; U, upregualted; NC, no change. Asterisk indicates data with a p value ≤ 0.05.
3.5. Control of protein translation in aging thymocytes by elongation initiation factors
Another group of genes which changed expression with age and whose age-associated expression levels were reversed by a restricted diet were the elongation initiation factors (eIFs). The eIFs are a group of proteins that control translation within the cell, and are important in protein synthesis and cell proliferation [58]. Both the eIF3 and the eIF4 families act as scaffolding proteins during ribosomal complex assembly and translation initiation [59,60], and have been associated with sarcopenia and muscle wasting during aging [61]. Importantly, a recent proteomics study showed that eIF is decreased in aging brain tissue, and that its expression can be reversed by caloric restriction [62]. Our data validated these observations in thymic tissue, where several members of the IF family, specifically, eIF3S2, eIF3S9, eIF3S10, eIF4A1 and eIF4G2 (Fig. 3 and Table 7), all decreased within the thymus with age were restored in caloric-restricted old mice. Age-associated changes in eIF3S10 and eIF4G2 were confirmed by real-time RT-PCR on independent total thymus samples, and changes in levels of eIF4G2 were further confirmed by real-time RT-PCR on purified thymocytes (Table 7), thus indicating that these changes are intrinsic to the thymocytes and may play a role in involution. Further, Western analysis of protein lysates from young and old thymocytes using antibody to eIF4G demonstrates that there is a clear decrease in the level of eIF4G protein in the old samples when compared to the young (Fig. 6).
Fig. 6.
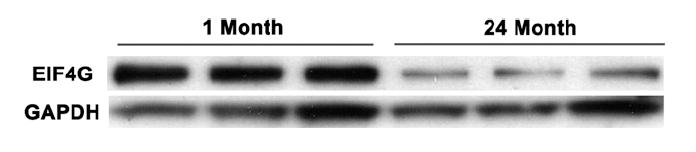
Western analysis of protein lysates from young and old thymocytes using antibody to eIF4G. In 24-month-old mice, levels of eIF4G protein are significantly decreased as compared to young mice.
3.6. Involvement of the glucocorticoid receptor in the aging of thymocytes
After validating the observed array changes using real-time PCR, pathway analysis was utilized to examine if multiple genes associated with specific signaling pathways may be altered within the aging thymus. Genes that changed were submitted to the Ingenuity Pathways Analysis program available from Ingenuity Systems (http://www.ingenuity.com), and a representative pathway is shown in Fig. 7a. This particular pathway was of great interest because it represented regulation of the glucocorticoid pathway, and glucocorticoids are produced by thymic epithelial cells and it has been postulated that GR is necessary for thymocyte development [63]. Furthermore, as mentioned in the earlier section referring to CR prevention of age-associated changes, we had also observed that the glucocorticoid receptor and HSPs display similar relationships but exhibit expression levels opposite from those seen in aging mice on an AL diet. Thus it was of great interest to us that this particular pathway was identified by gene ontology analysis of our data. At the center of the network is the nuclear receptor NR3C1, also called the glucocorticoid receptor (GR), which decreased in expression with age in both total thymus and purified thymocytes. Fig. 7b shows a network derived using the list of genes that were affected by CR. This was in concordance with the array results, where a CR diet restored the expression levels of this gene to the levels seen in the thymi of young AL mice. To confirm these results and also confirm that this was not due to decreases in thymocyte numbers, we performed Western analysis using purified thymocytes. Fig. 7c shows that the glucocorticoid receptor levels also decrease with age specifically within the thymocytes themselves. To ascertain the region in which this loss of GR was occurring, we then performed immunohistochemistry of various thymic sections and revealed that the decrease of glucocorticoid receptor protein in the thymus with age appears to be within both the cortex and medulla of the thymus (Fig. 7). Loss of GR expression in the cortex appears to be in the cytoplasm of all the cells, as well as a complete loss within the nucleus of many cells (Fig. 7d). In the thymic medulla, age-associated loss of GR appears to be cytoplasmic (Fig. 7e). Taken together, these data support the loss of the GR with thymic aging and this loss may have potential implications for the thymic involution process.
Fig. 7.
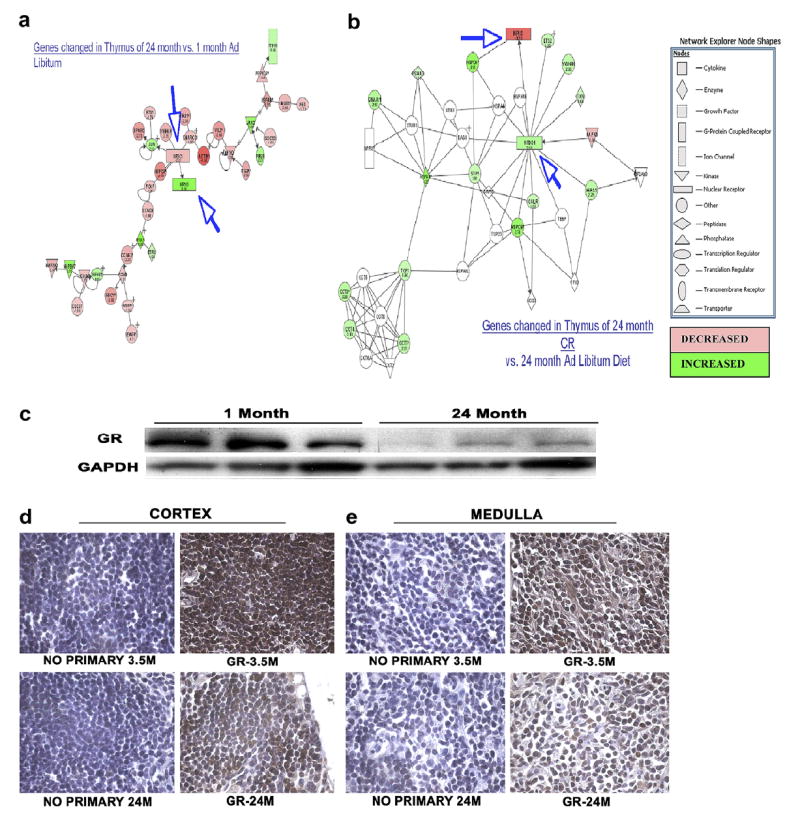
Possible pathways for cellular senescence (a) and CR rescue (b) involving the glucocorticoid receptor, androstane receptor and heat-shock proteins. These pathways were designed by the Ingenuity program by compiling published associations between genes in the lists whose expression levels changed with age and were then reversed by a CR diet. (c) Western blot analysis of protein lysates derived from thymocytes of C57BL/6 mice of designated ages confirm that GR is changed in aging thymocytes. (d,e) Whole thymus was also analyzed for GR expression by immunohistochemistry. Sections were stained with a nuclear DAPI stain (blue) and a mouse anti-GR following a standard protocol for DAB staining of paraffin sections. The images (100× magnification) clearly show the GR staining in the young thymus is much darker than that in the old thymus in both the cortex (d) and medulla (e). Data are representative of three experiments, each with one sample from each age.
4. Discussion
The specific aim of the current study was to create an atlas of changes in thymus gene expression with age and assess those affected by a CR diet. There have been numerous array papers published that reveal profiles of age-associated gene expression in various tissues. However, there are no data on global changes in gene expression associated specifically with thymic aging and involution. Many genes decreased in age in the thymus, and were restored to their original levels by a CR diet, indicating that these may play a role in thymocyte death and involution. These included housekeeping genes such as actin and tubulin, heat- and stress-related genes such as HSP90 and HSC70, and the glucocorticoid receptor, as well as elongation initiation factors. Decreased production of these types of proteins in the aging thymus could adversely affect cell function and contribute to involution within the thymus. Indeed, eIF4G2, for example, has been specifically associated with apoptosis and is referred to as a death associated protein (DAP) [64,65]. Decreased production of eIF4G2 specifically has been associated with inhibition of cell proliferation, and has been proposed to be involved in the switching of the cell from generalized translation to translation related to stress response [66], implicating other proteins previously mentioned such as the HSPs. Further scrutiny of these pathways is required to examine their roles in thymic involution. An important note is that, although our initial analyses were restricted to C57/Bl mice, we have expanded and validated these observations in Balb/C mice as well.
Regardless of whether the comparison was by age, diet or sex, the most common types of genes that changed expression levels were transcription regulators, enzymes, transporters and kinases. These genes code for proteins that perform a variety of functions within cells. Strikingly, genes involved in DNA repair and recombination were significantly downregulated, suggesting that aging thymocytes have a decreased ability to repair DNA damage, a common feature of both aging and cancer cells. The largest functional group of genes that changed with age is involved in cellular growth and proliferation, while the largest functional group of genes affected by a caloric-restricted diet is involved in cellular death. With regard to immune function, CR has been shown to reduce inflammatory cytokine expression in young and aged subjects and mediates an enhancement of immune cell proliferation and activation in response to mitogens and antigen [21-23,67,68]. However, the literature on the effects of CR on immune and thymic function is both sparse and contradictory. Several researchers have reported that CR induces a significant decrease in splenic cellularity and an elevation in the percentage of splenic CD4+ T cells compared with CD8+ T cells [69], while others have not observed any subset changes in CR mice vs. AL controls [70]. The studies by Chen and co-workers suggest that CR may preserve cortical thymocytes and may directly influence thymopoiesis and thymocyte survival. It would be interesting to speculate that these changes may relate to the possible effects of a caloric-restricted diet on thymocyte survival and delay of the aging process within the cells. Also, given the dramatic changes in gene expression in aged CR thymi compared to their AL counterparts and the similarities to young AL thymic gene expression, studies are underway attempting to further elucidate the effects of CR on thymocytes and the involution process. Of specific interest were genes known to be associated with T cell function and aging. Cathepsin L, which decreased with age in our arrays, is critical for MHC II processing and thymocyte survival [49]. Age-specific decreases in this gene may thus be responsible for the lowered maturation and survival of thymocytes with age. Other processes known to be altered in aging T cells also included actin polymerization and altered MHC processing, and genes known to be involved in these processes are indeed altered in our current array data (Table 2).
Using published data, the ingenuity pathways program designed possible networks of interactions between genes in our lists. The networks shown in Fig. 6 highlight the interaction between the glucocorticoid receptor, androstane receptor and heat shock proteins, which change with age and are reversed by a CR diet. Western blot and immunohistochemical analysis demonstrate that significant alterations in GR protein expression can be observed with age and supports our gene expression analysis. Glucocorticoids, acting through the glucocorticoid receptor (GR), potently modulate immune function and are a mainstay of therapy for treatment of inflammatory conditions, autoimmune diseases, leukemias and lymphomas [71-74]. Removal of systemic glucocorticoids, by adrenalectomy in animal models or adrenal insufficiency in humans, has shown that endogenous glucocorticoid production is required for regulation of physiologic immune responses [75,76]. These effects have been attributed to suppression of cytokines, although the crucial cellular and molecular targets remain unknown. In addition, considerable controversy remains as to whether glucocorticoids are required for thymocyte development [76,77] GR knockout mice are lethal perinatally but mice with a T-cell-specific disruption of the GR gene show no impairment in thymocyte development or T-cell populations [78]. In a recent paper by Brewer and co-workers [78], they demonstrate in T-cell-specific GR KO mice that T-cells are a critical cellular target of glucocorticoid receptor signaling, as immune activation in these mice resulted in significant mortality. This lethal activation is rescued by the presence of cyclooxygenase-2 (COX-2) inhibition but not glucocorticoid administration or cytokine neutralization. Thus, it would appear that glucocorticoid receptor suppression of COX-2 is crucial for curtailing lethal immune activation, and suggests new therapeutic approaches for regulation of T-cell-mediated inflammatory diseases. Enhanced expression of the GR increases the cells’ resistance to stress [79]. Also, mice with an increased sensitivity to glucocorticoids exhibit delayed involution of the thymus [80]. Perhaps GR may not be necessary for thymocyte development, but its presence may make it easier for the cells to survive under stress conditions associated with aging. Thus, decreased levels and activity of the GR could contribute to thymocyte death and thymic involution. Our data demonstrate that the GR practically disappears in thymus and thymocytes derived from old mice and that mice maintained under a CR regimen demonstrate restored GR levels to those of young AL-fed mice. This age-associated decrease in GR expression occurs within the thymocytes themselves. This implies that there could be a critical balance between GR and the HSPs required for thymocyte survival. Further study of the role of GR in thymocyte survival and thymic involution is planned.
Overall, this study was performed in combination with a larger study being conducted at the Gerontology Research Center of the National Institute on Aging called the “AGEMAP Project”. This project aims to generate a database of gene expression patterns associated with aging and caloric restriction in many of the mammalian organ systems, which may provide a valuable source of information about genetic influences involved in aging including those unique to specific organ systems and others that may be universally associated with aging. Additional studies are currently underway examining the spleens and bone marrow cells of these mice. In our current report, the data have demonstrated that microarray analysis can robustly identify significant differences in the profiles of aging thymi, and also isolate sex-specific and dietary effects upon this process. It will be interesting to attempt to define roles and manipulate pathways indicated by the array data that are involved in thymic aging. Many more genes from this array will need to be identified and tested for their contributions to the cellular aging process as well as their ability to be rescued by a CR diet. Further studies are under way in order to create a clearer picture of the dynamics involved in thymic involution and aging within the immune system.
Acknowledgments
We thank Ms. Angela Feehley for assistance with preparation of the manuscript. We gratefully acknowledge the excellent suite of in-house microarray tools from the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD. This work was supported by the Intramural Research Program of the National Institute on Aging, Baltimore, MD, USA.
Footnotes
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
References
- 1.Louria DB, Sen P, Sherer CB, Farrer WE. Infections in older patients: a systematic clinical approach. Geriatrics. 1993;48:28–34. [PubMed] [Google Scholar]
- 2.Pinner RW, Teutsch SM, Simonsen L, Klug LA, Graber JM, Clarke MJ, Berkelman RL. Trends in infectious diseases mortality in the United States. JAMA. 1996;275:189–193. [PubMed] [Google Scholar]
- 3.Parsons HK, Dockrell DH. The burden of invasive pneumococcal disease and the potential for reduction by immunisation. Int J Antimicrob Agents. 2002;19:85–93. doi: 10.1016/s0924-8579(01)00491-5. [DOI] [PubMed] [Google Scholar]
- 4.Neuzil KM. Influenza: new insights into an old disease. Curr Infect Dis Rep. 2000;2:224–230. doi: 10.1007/s11908-000-0039-3. [DOI] [PubMed] [Google Scholar]
- 5.Schmader K. Herpes zoster in older adults. Clin Infect Dis. 2001;32:1481–1486. doi: 10.1086/320169. [DOI] [PubMed] [Google Scholar]
- 6.Castle SC. Clinical relevance of age-related immune dysfunction. Clin Infect Dis. 2000;31:578–585. doi: 10.1086/313947. [DOI] [PubMed] [Google Scholar]
- 7.Yancik R. Cancer burden in the aged: an epidemiologic and demographic overview. Cancer. 1997;80:1273–1283. [PubMed] [Google Scholar]
- 8.Ershler WB, Longo DL. Aging and cancer: issues of basic and clinical science. J Natl Cancer Inst. 1997;89:1489–1497. doi: 10.1093/jnci/89.20.1489. [DOI] [PubMed] [Google Scholar]
- 9.Burns EA, Leventhal EA. Aging, immunity, and cancer. Cancer Control. 2000;7:513–522. doi: 10.1177/107327480000700603. [DOI] [PubMed] [Google Scholar]
- 10.Miller RA. The aging immune system: primer and prospectus. Science. 1996;273:70–74. doi: 10.1126/science.273.5271.70. [DOI] [PubMed] [Google Scholar]
- 11.Ginaldi L, De Martinis M, D’Ostilio A, Marini L, Loreto MF, Quaglino D. Immunological changes in the elderly. Aging (Milano) 1999;11:281–286. doi: 10.1007/BF03339801. [DOI] [PubMed] [Google Scholar]
- 12.Franceschi C, Bonafe M, Valensin S. Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine. 2000;18:1717–1720. doi: 10.1016/s0264-410x(99)00513-7. [DOI] [PubMed] [Google Scholar]
- 13.Malaguarnera L, Ferlito L, Imbesi RM, Gulizia GS, Di Mauro S, Maugeri D, Malaguarnera M, Messina A. Immunosenescence: a review. Arch Gerontol Geriatr. 2001;32:1–14. doi: 10.1016/s0167-4943(00)00086-8. [DOI] [PubMed] [Google Scholar]
- 14.Effros RB. Ageing and the immune system. Novartis Found Symp. 2001;235:130–139. doi: 10.1002/0470868694.ch12. discussion: 139–45, 146–149. [DOI] [PubMed] [Google Scholar]
- 15.Wayne SJ, Rhyne RL, Garry PJ, Goodwin JS. Cell-mediated immunity as a predictor of morbidity and mortality in subjects over 60. J Gerontol. 1990;45:M45–M48. doi: 10.1093/geronj/45.2.m45. [DOI] [PubMed] [Google Scholar]
- 16.Franceschi C, Monti D, Sansoni P, Cossarizza A. The immunology of exceptional individuals: the lesson of centenarians. Immunol Today. 1995;16:12–16. doi: 10.1016/0167-5699(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 17.Utsuyama M, Hirokawa K. Age-related changes of splenic T cells in mice—a flow cytometric analysis. Mech Ageing Dev. 1987;40:89–102. doi: 10.1016/0047-6374(87)90037-6. [DOI] [PubMed] [Google Scholar]
- 18.Hulstaert F, Hannet I, Deneys V, Munhyeshuli V, Reichert T, De Bruyere M, Strauss K. Age-related changes in human blood lymphocyte subpopulations. II. Varying kinetics of percentage and absolute count measurements. Clin Immunol Immunopathol. 1994;70:152–158. doi: 10.1006/clin.1994.1023. [DOI] [PubMed] [Google Scholar]
- 19.Berzins SP, Uldrich AP, Sutherland JS, Gill J, Miller JF, Godfrey DI, Boyd RL. Thymic regeneration: teaching an old immune system new tricks. Trends Mol Med. 2002;8:469–476. doi: 10.1016/s1471-4914(02)02415-2. [DOI] [PubMed] [Google Scholar]
- 20.Mackall CL, Bare CV, Granger LA, Sharrow SO, Titus JA, Gress RE. Thymic-independent T cell regeneration occurs via antigen-driven expansion of peripheral T cells resulting in a repertoire that is limited in diversity and prone to skewing. J Immunol. 1996;156:4609–4616. [PubMed] [Google Scholar]
- 21.Joncourt F, Kristensen F, De Weck AL. Ageing and immunity in outbred NMRI mice: lack of correlation between age-related decline of the response to T cell mitogens, the antibody response to a T-dependent antigen and lifespan in outbred NMRI mice. Clin Exp Immunol. 1981;44:270–277. [PMC free article] [PubMed] [Google Scholar]
- 22.Joncourt F, Bettens F, Kristensen F, de Weck AL. Age-related changes of mitogen responsiveness in different lymphoid organs from outbred NMRI mice. Immunobiology. 1981;158:439–449. doi: 10.1016/S0171-2985(81)80014-9. [DOI] [PubMed] [Google Scholar]
- 23.Hobbs MV, Ernst DN, Torbett BE, Glasebrook AL, Rehse MA, McQuitty DN, Thoman ML, Bottomly K, Rothermel AL, Noonan DJ et al. Cell proliferation and cytokine production by CD4+ cells from old mice. J Cell Biochem. 1991;46:312–320. doi: 10.1002/jcb.240460406. [DOI] [PubMed] [Google Scholar]
- 24.Haynes BF, Sempowski GD, Wells AF, Hale LP. The human thymus during aging. Immunol Res. 2000;22:253–261. doi: 10.1385/IR:22:2-3:253. [DOI] [PubMed] [Google Scholar]
- 25.Mackall CL, Gress RE. Thymic aging and T-cell regeneration. Immunol Rev. 1997;160:91–102. doi: 10.1111/j.1600-065x.1997.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 26.Haynes BF, Markert ML, Sempowski GD, Patel DD, Hale LP. The role of the thymus in immune reconstitution in aging, bone marrow transplantation, and HIV-1 infection. Annu Rev Immunol. 2000;18:529–560. doi: 10.1146/annurev.immunol.18.1.529. [DOI] [PubMed] [Google Scholar]
- 27.Butler RN, Sprott R, Warner H, Bland J, Feuers R, Forster M, Fillit H, Harman SM, Hewitt M, Hyman M, Johnson K, Kligman E, McClearn G, Nelson J, Richardson A, Sonntag W, Weindruch R, Wolf N. Biomarkers of aging: from primitive organisms to humans. J Gerontol A Biol Sci Med Sci. 2004;59:B560–B567. doi: 10.1093/gerona/59.6.b560. [DOI] [PubMed] [Google Scholar]
- 28.Sell C. Caloric restriction and insulin-like growth factors in aging and cancer. Horm Metab Res. 2003;35:705–711. doi: 10.1055/s-2004-814156. [DOI] [PubMed] [Google Scholar]
- 29.Masoro EJ. Possible mechanisms underlying the antiaging actions of caloric restriction. Toxicol Pathol. 1996;24:738–741. doi: 10.1177/019262339602400617. [DOI] [PubMed] [Google Scholar]
- 30.Gredilla R, Barja G. Minireview: The role of oxidative stress in relation to caloric restriction and longevity. Endocrinology. 2005;146:3713–3717. doi: 10.1210/en.2005-0378. [DOI] [PubMed] [Google Scholar]
- 31.Cefalu WT, Bell-Farrow AD, Wang ZQ, Sonntag WE, Fu MX, Baynes JW, Thorpe SR. Caloric restriction decreases age-dependent accumulation of the glycoxidation products, n-epsilon-(carboxymethyl)lysine and pentosidine, in rat skin collagen. J Gerontol A Biol Sci Med Sci. 1995;50:B337–B341. doi: 10.1093/gerona/50a.6.b337. [DOI] [PubMed] [Google Scholar]
- 32.Sell DR, Lane MA, Obrenovich ME, Mattison JA, Handy A, Ingram DK, Cutler RG, Roth GS, Monnier VM. The effect of caloric restriction on glycation and glycoxidation in skin collagen of nonhuman primates. J Gerontol A Biol Sci Med Sci. 2003;58:508–516. doi: 10.1093/gerona/58.6.b508. [DOI] [PubMed] [Google Scholar]
- 33.Heilbronn LK, de Jonge L, Frisard MI, DeLany JP, Larson-Meyer DE, Rood J, Nguyen T, Martin CK, Volaufova J, Most MM, Greenway FL, Smith SR, Deutsch WA, Williamson DA, Ravussin E. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295:1539–1548. doi: 10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bevilacqua L, Ramsey JJ, Hagopian K, Weindruch R, Harper ME. Long-term caloric restriction increases UCP3 content but decreases proton leak and reactive oxygen species production in rat skeletal muscle mitochondria. Am J Physiol Endocrinol Metab. 2005;289:E429–E438. doi: 10.1152/ajpendo.00435.2004. [DOI] [PubMed] [Google Scholar]
- 35.Ugochukwu NH, Figgers CL. Caloric restriction inhibits up-regulation of inflammatory cytokines and TNF-alpha, and activates IL-10 and haptoglobin in the plasma of streptozotocin-induced diabetic rats. J Nutr Biochem. 2007;18:120–126. doi: 10.1016/j.jnutbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Paoletti R, Poli A, Cignarella A. The emerging link between nutrition, inflammation and atherosclerosis. Expert Rev Cardiovasc Ther. 2006;4:385–393. doi: 10.1586/14779072.4.3.385. [DOI] [PubMed] [Google Scholar]
- 37.Smith JV, Heilbronn LK, Ravussin E. Energy restriction and aging. Curr Opin Clin Nutr Metab Care. 2004;7:615–622. doi: 10.1097/00075197-200411000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci USA. 2001;98:10630–10635. doi: 10.1073/pnas.191313598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han E, Hilsenbeck SG, Richardson A, Nelson JF. Cdna expression arrays reveal incomplete reversal of age-related changes in gene expression by calorie restriction. Mech Ageing Dev. 2000;115:157–174. doi: 10.1016/s0047-6374(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 40.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 41.Weeraratna AT, Nagel JE, de Mello-Coelho V, Taub DD. Gene expression profiling: from microarrays to medicine. J Clin Immunol. 2004;24:213–224. doi: 10.1023/B:JOCI.0000025443.44833.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melov S, Hubbard A. Microarrays as a tool to investigate the biology of aging: a retrospective and a look to the future. Sci Aging Knowledge Environ. 2004;2004:re7. doi: 10.1126/sageke.2004.42.re7. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Mo R, Lescure PA, Misek DE, Hanash S, Rochford R, Hobbs M, Yung RL. Aging is associated with increased T-cell chemokine expression in c57bl/6 mice. J Gerontol A Biol Sci Med Sci. 2003;58:975–983. doi: 10.1093/gerona/58.11.b975. [DOI] [PubMed] [Google Scholar]
- 44.Carter MG, Hamatani T, Sharov AA, Carmack CE, Qian Y, Aiba K, Ko NT, Dudekula DB, Brzoska PM, Hwang SS, Ko MS. In situ-synthesized novel microarray optimized for mouse stem cell and early developmental expression profiling. Genome Res. 2003;13:1011–1021. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bar-Dayan Y, Afek A, Goldberg I, Kopolovic J. Proliferation, apoptosis and thymic involution. Tissue Cell. 1999;31:391–396. doi: 10.1054/tice.1999.0001. [DOI] [PubMed] [Google Scholar]
- 47.Yu FX, Lin SC, Morrison-Bogorad M, Yin HL. Effects of thymosin beta 4 and thymosin beta 10 on actin structures in living cells. Cell Motil Cytoskel. 1994;27:13–25. doi: 10.1002/cm.970270103. [DOI] [PubMed] [Google Scholar]
- 48.Wise T, Klindt J. Thymic weight changes and endocrine relationships during maturation in cattle: effects of age, sex, and castration. Growth Dev Aging. 1995;59:139–148. [PubMed] [Google Scholar]
- 49.Arudchelvan Y, Nishimura Y, Tokuda N, Sawada T, Ueyama Y, Fukumoto T. Identification and characterization of major histocompatibility complex class II compartments in cortical thymic epithelial cells. Anat Rec. 2003;274A:798–806. doi: 10.1002/ar.a.10081. [DOI] [PubMed] [Google Scholar]
- 50.Brock MA, Chrest F. Differential regulation of actin polymerization following activation of resting T lymphocytes from young and aged mice. J Cell Physiol. 1993;157:367–378. doi: 10.1002/jcp.1041570221. [DOI] [PubMed] [Google Scholar]
- 51.van Remmen H, Guo Z, Richardson A. The anti-ageing action of dietary restriction. Novartis Found Symp. 2001;235:221–230. doi: 10.1002/0470868694.ch18. Discussion, pp. 230–233. [DOI] [PubMed] [Google Scholar]
- 52.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 53.Weinert BT, Timiras PS. Invited review: theories of aging. J Appl Physiol. 2003;95:1706–1716. doi: 10.1152/japplphysiol.00288.2003. [DOI] [PubMed] [Google Scholar]
- 54.Mattson MP, Duan W, Guo Z. Meal size and frequency affect neuronal plasticity and vulnerability to disease: cellular and molecular mechanisms. J Neurochem. 2003;84:417–431. doi: 10.1046/j.1471-4159.2003.01586.x. [DOI] [PubMed] [Google Scholar]
- 55.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Santelli G, Califano D, Chiappetta G, Vento MT, Bartoli PC, Zullo F, Trapasso F, Viglietto G, Fusco A. Thymosin beta-10 gene overexpression is a general event in human carcinogenesis. Am J Pathol. 1999;155:799–804. doi: 10.1016/s0002-9440(10)65178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung KJ, Jr, Li G. Tissue-specific regulation of CHK1 expression by p53. Exp Mol Pathol. 2001;71:156–160. doi: 10.1006/exmp.2001.2398. [DOI] [PubMed] [Google Scholar]
- 58.Flynn A, Proud CG. The role of EIF4 in cell proliferation. Cancer Surv. 1996;27:293–310. [PubMed] [Google Scholar]
- 59.Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to EIF4 families. Mech Dev. 2005;122:865–876. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 60.Hinnebusch AG. Eif3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Cuthbertson D, Smith K, Babraj J, Leese G, Waddell T, Atherton P, Wackerhage H, Taylor PM, Rennie MJ. Anabolic signaling deficits underlie amino acid resistance of wasting, aging muscle. FASEB J. 2005;19:422–424. doi: 10.1096/fj.04-2640fje. [DOI] [PubMed] [Google Scholar]
- 62.Poon HF, Shepherd HM, Reed TT, Calabrese V, Stella AM, Pennisi G, Cai J, Pierce WM, Klein JB, Butterfield DA. Proteomics analysis provides insight into caloric restriction mediated oxidation and expression of brain proteins associated with age-related impaired cellular processes: mitochondrial dysfunction, glutamate dysregulation and impaired protein synthesis. Neurobiol Aging. 2006;27:1020–1034. doi: 10.1016/j.neurobiolaging.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Ashwell JD, Lu FW, Vacchio MS. Glucocorticoids in T cell development and function. Annu Rev Immunol. 2000;18:309–345. doi: 10.1146/annurev.immunol.18.1.309. [DOI] [PubMed] [Google Scholar]
- 64.Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. A novel form of dap5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved dap5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci USA. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SH, McCormick F. P97/dap5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurashima C, Utsuyama M, Kasai M, Ishijima SA, Konno A, Hirokawa K. The role of thymus in the aging of T cell subpopulations and age-associated alteration of cytokine production by these cells. Int Immunol. 1995;7:97–104. doi: 10.1093/intimm/7.1.97. [DOI] [PubMed] [Google Scholar]
- 68.Murasko DM, Weiner P, Kaye D. Decline in mitogen induced proliferation of lymphocytes with increasing age. Clin Exp Immunol. 1987;70:440–448. [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, Astle CM, Harrison DE. Delayed immune aging in diet-restricted b6cbat6 f1 mice is associated with preservation of naive T cells. J Gerontol A Biol Sci Med Sci. 1998;53:B330–B337. doi: 10.1093/gerona/53a.5.b330. discussion B338-9. [DOI] [PubMed] [Google Scholar]
- 70.Poetschke HL, Klug DB, Perkins SN, Wang TT, Richie ER, Hursting SD. Effects of calorie restriction on thymocyte growth, death and maturation. Carcinogenesis. 2000;21:1959–1964. doi: 10.1093/carcin/21.11.1959. [DOI] [PubMed] [Google Scholar]
- 71.Mathew BS, Carson KA, Grossman SA. Initial response to glucocorticoids. Cancer. 2006;106:383–387. doi: 10.1002/cncr.21583. [DOI] [PubMed] [Google Scholar]
- 72.Styczynski J, Wysocki M. Ex vivo modulation of response to prednisolone in childhood acute lymphoblastic leukaemia. Br J Haematol. 2006;133:397–399. doi: 10.1111/j.1365-2141.2006.06032.x. [DOI] [PubMed] [Google Scholar]
- 73.Schacke H, Rehwinkel H, Asadullah K, Cato AC. Insight into the molecular mechanisms of glucocorticoid receptor action promotes identification of novel ligands with an improved therapeutic index. Exp Dermatol. 2006;15:565–573. doi: 10.1111/j.1600-0625.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 74.Barnes PJ. How corticosteroids control inflammation: Quintiles prize lecture 2005. Br J Pharmacol. 2006;148:245–254. doi: 10.1038/sj.bjp.0706736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 76.Vacchio MS, Ashwell JD. Glucocorticoids and thymocyte development. Semin Immunol. 2000;12:475–485. doi: 10.1006/smim.2000.0265. [DOI] [PubMed] [Google Scholar]
- 77.Ashwell JD, Vacchio MS, Galon J. Do glucocorticoids participate in thymocyte development? Immunol Today. 2000;21:644–646. doi: 10.1016/s0167-5699(00)01758-8. [DOI] [PubMed] [Google Scholar]
- 78.Brewer JA, Khor B, Vogt SK, Muglia LM, Fujiwara H, Haegele KE, Sleckman BP, Muglia LJ. T-cell glucocorticoid receptor is required to suppress cox-2-mediated lethal immune activation. Nat Med. 2003;9:1318–1322. doi: 10.1038/nm895. [DOI] [PubMed] [Google Scholar]
- 79.Reichardt HM, Umland T, Bauer A, Kretz O, Schutz G. Mice with an increased glucocorticoid receptor gene dosage show enhanced resistance to stress and endotoxic shock. Mol Cell Biol. 2000;20:9009–9017. doi: 10.1128/mcb.20.23.9009-9017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pazirandeh A, Jondal M, Okret S. Glucocorticoids delay age-associated thymic involution through directly affecting the thymocytes. Endocrinology. 2004;145:2392–2401. doi: 10.1210/en.2003-1660. [DOI] [PubMed] [Google Scholar]


