Abstract
Nitric oxide synthase (NOS)2, an inducible enzyme that produces NO during inflammation, is transcriptionally regulated. Our goal was to determine whether high mobility group (HMG)A1 contributes to human (h)NOS2 gene regulation. Using a small molecule inhibitor of HMGA1 binding to DNA, or a dominant-negative form of HMGA1, we blunted the induction of hNOS2 by pro-inflammatory stimuli. Binding of HMGA1 in the region −3506 to −3375 of the hNOS2 promoter, a region not previously known to be involved in hNOS2 regulation, contributed to the induction of hNOS2 promoter in conjunction with upstream enhancer regions. We demonstrate a previously unknown role for HMGA1 in the regulation of hNOS2.
Keywords: nitric oxide synthase 2, high mobility group A1, pro-inflammatory cytokines
1. Introduction
Nitric oxide (NO) is a free radical gas that is produced by a family of NO synthases (NOS) [1]. During inflammatory disease processes, including sepsis, NO is produced via the inducible NOS2 pathway [2]. The molecular mechanisms of NOS2 gene regulation are complex, and occur predominantly at the level of gene transcription. NOS2 gene regulation was first characterized in murine cells [3,4], and NO production is striking in rodents, particularly when assessing the effects of lipopolysaccharide (LPS) and pro-inflammatory cytokines [5]. NO production is increased in human sepsis [6], and a combination of LPS and pro-inflammatory cytokines or exposure to infectious agents [7] is effective in generating NO in human cultured cells.
While the exact sequences of the upstream cytokine-responsive elements of the murine and human NOS2 promoters are not conserved, an analogous pattern of cytokine induction of transcription factor binding has been demonstrated in the two promoters. It is well described that LPS or interleukin (IL)-1β induces nuclear factor (NF)-κB binding, and interferon (IFN)-γ induces IFN regulatory factor (IRF)-1 and signal transducer and activator of transcription (Stat)-1α binding to the murine NOS2 promoter, resulting in a synergistic induction of NOS2 in macrophages and vascular smooth muscle cells treated with LPS or IL-1β plus IFN-γ [8–12]. Geller and colleagues [13,14] have reported that in a similar fashion, IL-1β or TNF-α induces NF-κB binding (−5.5, −5.8, and −6.1 kb) and IFN-γ induces Stat-1α binding (−5.2 and −5.8 kb) to the human NOS2 promoter, which may facilitate synergistic induction of the human NOS2 promoter by a cytokine mixture in human lung epithelial cells. Recently, it has also been demonstrated that −5 and −6 kb promoter regions act as a classical transcriptional enhancer, and evidence was provided for in vivo binding of NF-κB and Stat-1 in these regions under cytokine stimulation [15]. Others have shown that 5′-flanking regions contributing to cytokine responsiveness include AP-1 sites [16,17], with the interaction of c-Fos and Stat-1 in the −5 kb enhancer [18], and a NF-κB site extending out to −8.2 kb [16,17].
Beyond the interaction of trans-acting factors with cis-acting elements, architectural transcription factors mediate gene transcription by controlling DNA conformation [19,20]. The high mobility group (HMG) proteins are a family of architectural proteins that have been recognized to play a role in the regulation of gene transcription, and an important member of this family is the HMGA1 protein. HMGA1 binds to AT-rich regions in the minor groove of DNA, and HMGA1 facilitates the assembly of functional nucleoprotein complexes by inducing changes in DNA structure and by recruiting nuclear proteins into enhancer complexes [20–22]. We have previously demonstrated a role for HMGA1 in regulation of the murine NOS2 promoter [23,24], yet the role of HMGA1 in human NOS2 expression under inflammatory conditions is not known. The present report will focus on the ability of HMGA1 to regulate human (h)NOS2 gene expression and promoter activity.
2. Materials and methods
2.1. Cell culture and reagents
The human type II alveolar epithelial (A549) cell line (American Type Culture Collection) was cultured, as described [25]. Lipopolysaccharide from Escherichia coli (LPS, serotype O26:B6) and Distamycin A (Dist A) were purchased from Sigma. Human IFN-γ and IL-1β were purchased from PeproTech.
2.2. RNA isolation and Northern blot analysis
Cells were initially grown in medium containing 10% fetal bovine serum (FBS), and then changed to 0.4% FBS for 48h prior to stimulation. Dist A (0–40 µM) was administered to A549 cells 30 minutes prior to a cytokine mixture (CM) of LPS (2 µg/ml), IFN-γ (100 U/ml), and IL-1β(10 ng/ml. Vehicle for CM was phosphate buffered saline (PBS), while Dist A was dissolved in 25% dimethyl sulfoxide (DMSO) solution. In the experiments using CM or Dist A, an appropriate control using the same amount of vehicle was included. RNA was extracted 12 hours later, using the RNeasy mini RNA isolation kit (Qiagen). Northern blot analysis, using a radiolabeled hNOS2 probe (provided by Dr. Stella Kourembanas, Boston, MA), was performed as previously described [23].
2.3. Plasmid constructs and mutagenesis of the human NOS2 promoter
The human dominant-negative HMGA1 cDNA construct (mutant HMGI(mII,mIII)) lacks the ability to bind AT-rich DNA sequences in vitro but retains its ability for specific protein-protein interactions with other transcription factors [26]. This mutant construct (provided by Dr. Raymond Reeves, Pullman, WA) was subcloned into the HindIII/KpnI sites of the pCMVFlag expression vector (Sigma) with optimization of the Kozak consensus sequence, resulting in generation of the final dnHMGA1 plasmid construct. The hNOS2 luciferase reporter plasmid (−8296/+168) in the pGL3-Basic vector (Promega) was kindly provided by Dr. Joel Moss, Bethesda, MD. Deletion constructs −5496/+168, −3658/+168, and −2950/+168 were generated by a PCR method using Pfu Turbo polymerase (Stratagene) followed by KpnI digestion, using plasmid −8296/+168 as a template. Also, site-directed mutagenesis of plasmid −8296/+168 was performed using Pfu polymerase to create an internal deletion of bp −3658 to −3094 (Δ−3658/−3094), bp −3506 to −3375 (Δ−3506/−3375), and bp −3348 to −3119 (Δ−3348/−3119). All constructs were confirmed by sequencing.
2.4. Transient transfection assays
The hNOS2 reporter-promoter constructs (WT and mutant constructs, 500 ng/well) and dnHMGA1 expression plasmid or empty vector (500 ng/well) were transiently transfected into A549 cells using FuGENE 6 transfection reagent (Roche Applied Science), as described previously [25]. Twelve hours following transfection of hNOS2 and dnHMGA1 constructs, cells were changed to medium with 0.4% FBS and treated with CM. In experiments using Dist A, −8296 construct was transiently transfected into A549 cells, and Dist A (10–40 µM) or vehicle equivalent was administered 30 minutes prior to a cytokine mixture (CM). β-galactosidase expression vector was co-transfected with hNOS2 reporter-promoter constructs (500 ng/well), to normalize for luciferase activity. In transient transfection experiments, cells were harvested 6 hours following CM treatment and assayed for luciferase (Promega luciferase assay system) [25].
2.5. Protein-DNA binding assays
Electrophoretic mobility shift assays were performed as described [24], using a series of 32P-labeled double-stranded oligonucleotide probes encoding AT-rich regions in the hNOS2 promoter (between bp −3658 and −2944) and a 43-amino acid synthetic peptide (Peptide Synthesis Core Facility, Tufts University, Boston, MA) encompassing the second and third DNA-binding regions of the HMGA1 protein (MEVPTPKRPRGRPKGSKNKGAAKTRKTTTTPGRKPRGRPKKLE, termed HMGA1(2/3)), as described previously [27]. Binding reactions were run on a 6% polyacrylamide gel, and 500 ng of peptide was used per binding reaction.
3. Results
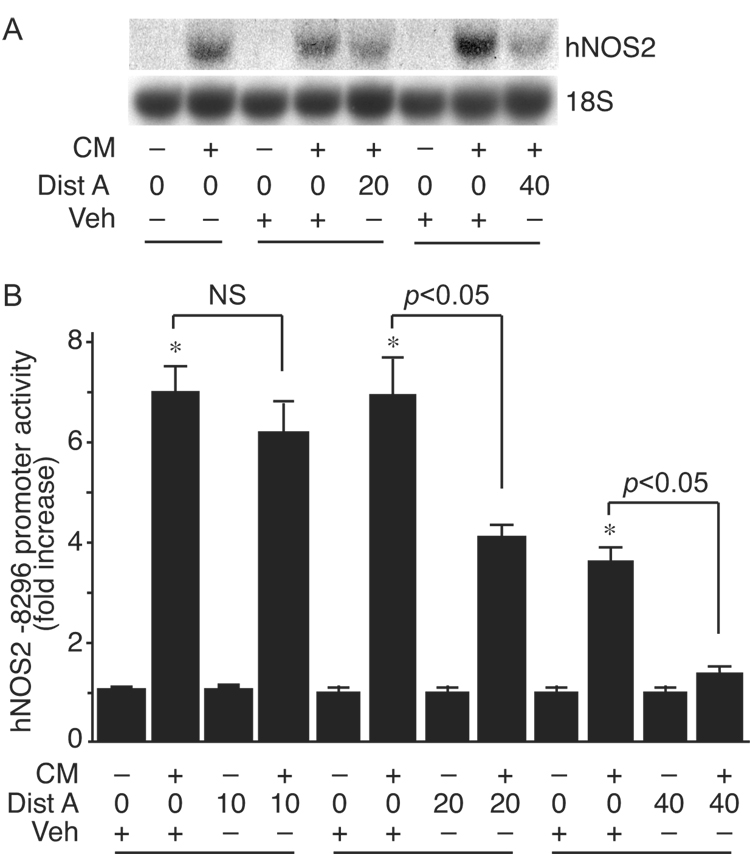
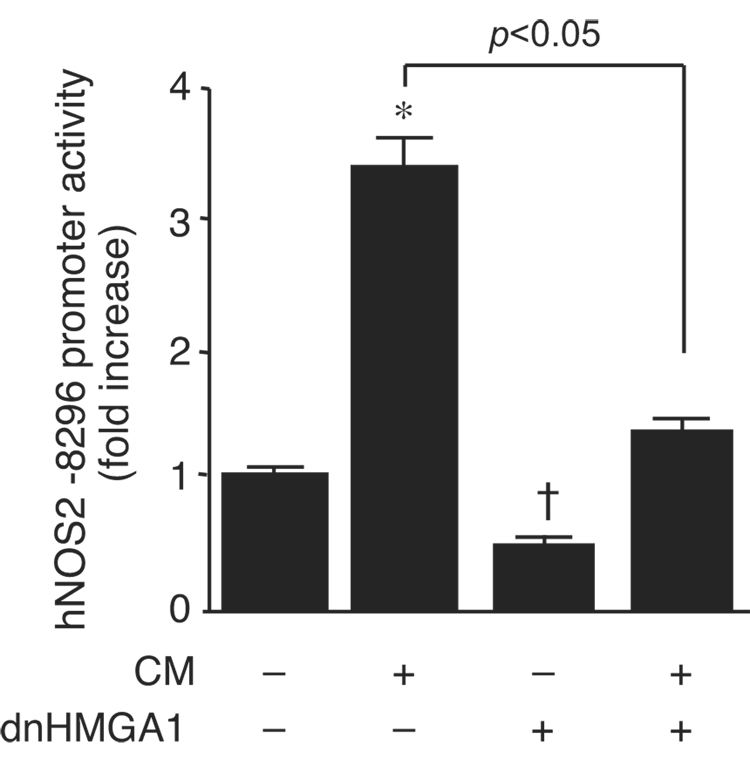
We administered the minor groove binding drug Dist A to human alveolar epithelial cells (A549) in the presence or absence of CM. Dist A is a small molecule that interferes with binding of HMGA1 to AT-rich regions of DNA [28]. Dist A suppressed induction of hNOS2 message by CM in A549 cells (Fig. 1A). To confirm this occurred at the level of gene transcription, Dist A was administered to A549 cells after transfection with the −8296 hNOS2 promoter construct, in the presence or absence of CM. Dist A suppressed cytokine inducible hNOS2 promoter activity (Fig. 1B), mimicking its effects on endogenous levels of hNOS2 mRNA (Fig. 1A). Similar to the Dist A effect, overexpression of dnHMGA1 suppressed cytokine induction of the hNOS2 promoter (Fig. 2). In addition, dnHMGA1 suppressed basal hNOS2 promoter activity, suggesting HMGA1 may play a role under basal and inflammatory conditions.
Fig. 1.
Dist A blunts induction of hNOS2 mRNA and promoter activity by CM. (A) A549 cells were treated with CM or PBS, in the presence of increasing doses of Dist A (µM) or vehicle solution (25% DMSO). Total RNA was extracted, and Northern blot analyses were performed using 32P-labeled hNOS2 and 18S rRNA probes. This experiment was performed 2 independent times. (B) Luciferase reporter plasmid hNOS2 −8296 (500 ng/well) was transiently co-transfected along with β-galactosidase expression plasmid (500 ng/well) into A549 cells. After transfection, cells were treated with CM or PBS, in the presence or absence of increasing doses of Dist A (µM) or vehicle solution (25% DMSO). Cells were harvested for luciferase activity normalized for β-galactosidase, and plotted as fold induction of hNOS2 promoter activity from cells receiving no CM and no Dist A. n=6 in each group. *, p < 0.05 versus Vehicle alone group; NS, not significant.
Fig. 2.
dnHMGA1 suppresses cytokine induction of hNOS2 promoter activity. Luciferase reporter plasmid hNOS2 −8296 (500 ng/well) was transiently co-transfected with an expression plasmid for dnHMGA1 (500 ng/well) or empty vector into A549 cells. Cells were exposed to CM or vehicle, and then harvested for luciferase activity. Promoter activity was plotted as fold induction of hNOS2 promoter activity from cells receiving no CM and no dnHMGA1. n=30 in each group. *, increase (p < 0.05) versus no CM and no dnHMGA1; † decrease (p < 0.05) versus no CM and no dnHMGA1.
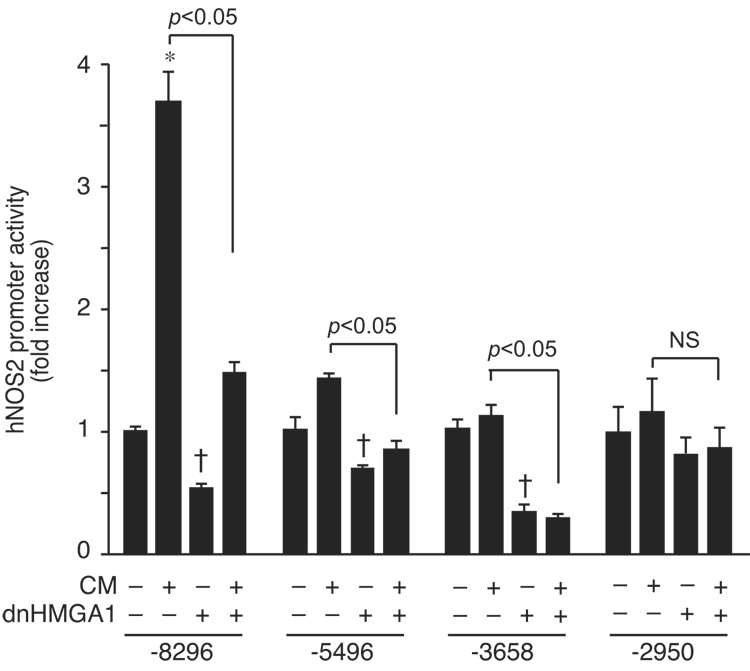
In an effort to uncover the hNOS2 promoter region(s) responsible for this HMGA1 effect, we co-transfected the dnHMGA1 expression plasmid with various deletion constructs of the hNOS2 promoter, in the presence or absence of CM. Figure 3 reveals that deletion beyond enhancer regions (−8296 to −5496) responsible for hNOS2 induction led to a loss of promoter activation by CM, but persistent suppression of hNOS2 promoter activity by dnHMGA1. The ability of dnHMGA1 to suppress hNOS2 promoter activity (both in the presence or absence of CM) remained in construct −3658, but was lost when the promoter was deleted down to construct −2950. These data suggest that the effect of HMGA1 resides in region −3658 to −2950.
Fig. 3.
Localization of HMGA1-responsive region in the hNOS2 promoter. Luciferase reporter plasmid constructs (hNOS2 −8296, −5496, −3658, and −2950) were transiently co-transfected (500 ng/well) with an expression plasmid for dnHMGA1 (500 ng/well) or empty vector into A549 cells. Cells were exposed to CM or vehicle, and then harvested for luciferase activity. Promoter activity was plotted as fold induction of hNOS2 promoter activity from cells receiving no CM and no dnHMGA1. n=6 in each experiment. *, increase (p < 0.05) versus no CM and no dnHMGA1; † decrease (p < 0.05) versus no CM and no dnHMGA1; NS, not significant.
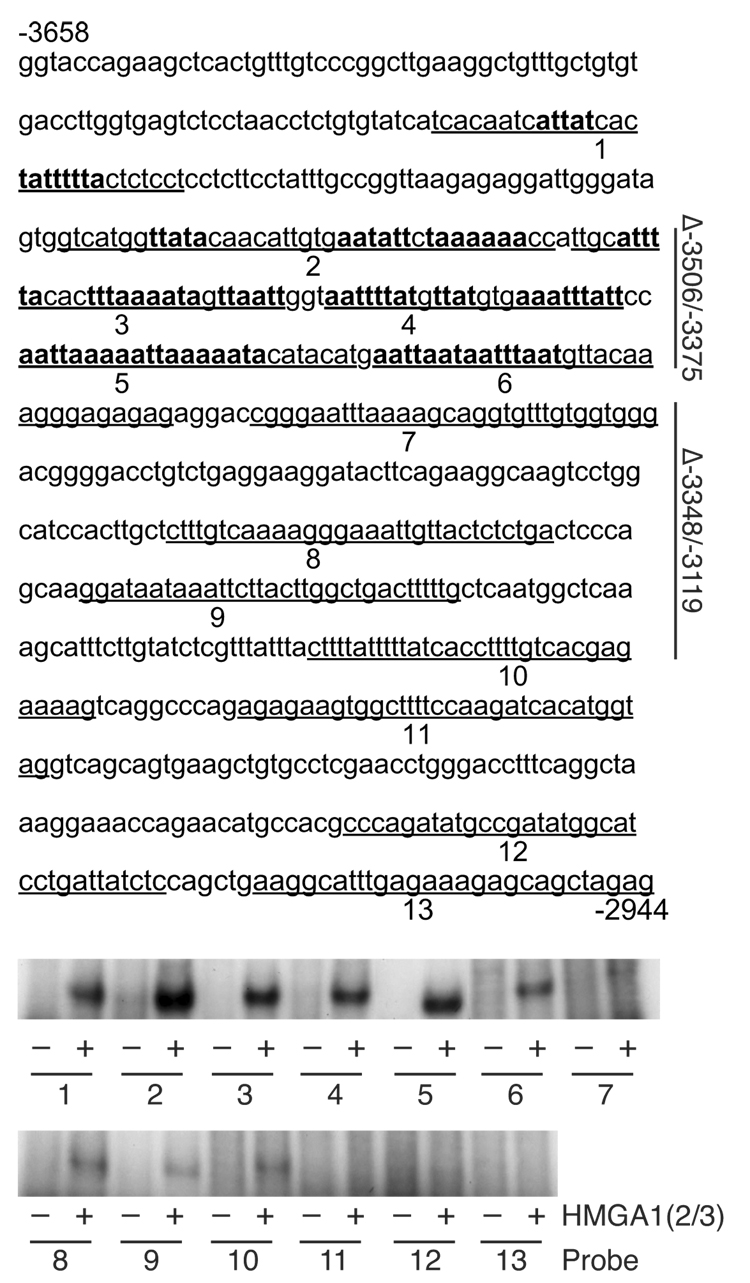
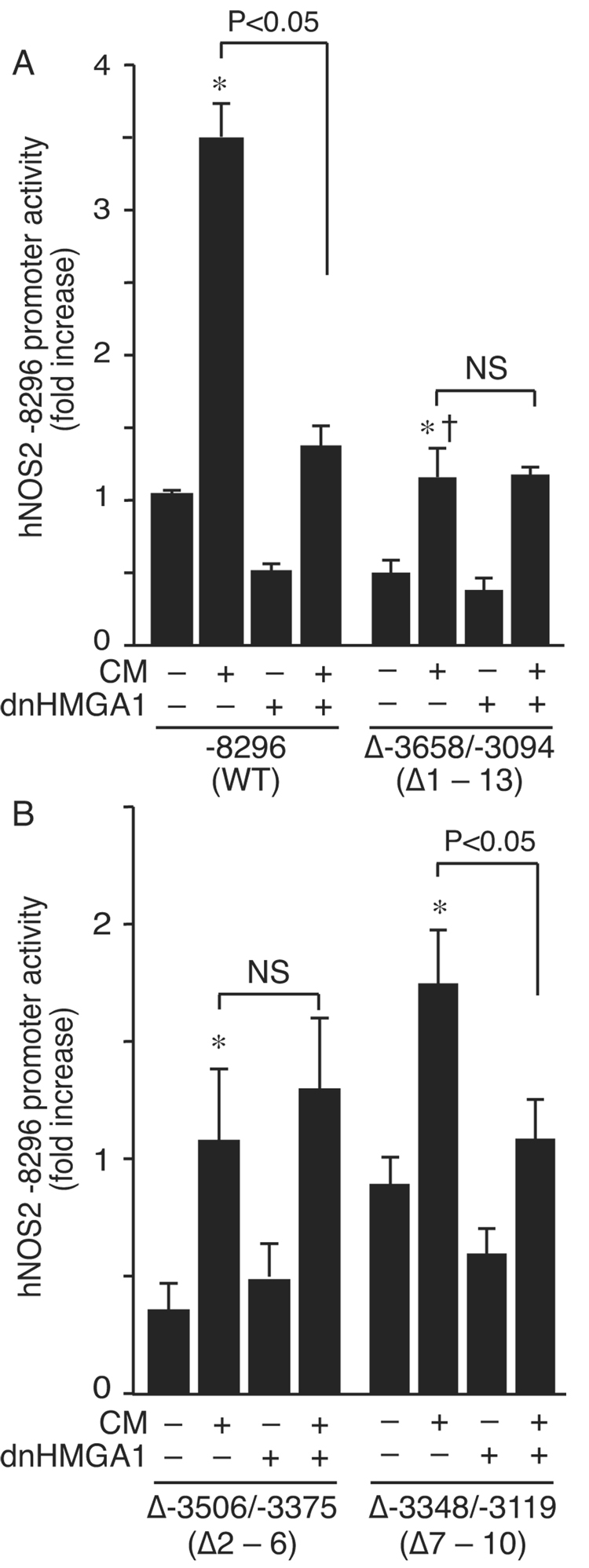
To further elucidate which AT-rich regions of the hNOS2 promoter were capable of binding HMGA1, protein-DNA binding assays were performed using 13 DNA sequences spanning major AT-rich regions (between −3658 and −2944) as probes (Fig. 4). HMGA1 bound more to probes 1 – 6, and less to probes 7 – 13. To confirm functionality of these regions, deletion of −3658 to −3094 within the −8296 construct (Δ−3658/−3094) was performed. This construct allowed hNOS2 promoter transactivation by CM, though to a lesser degree than the wild-type promoter (Fig. 5A). Notably, expression of the dnHMGA1 plasmid did not suppress promoter activity in construct Δ−3658/−3094. Deletion of region −3506 to −3375 (Δ−3506/−3375) also completely abolished suppression of the hNOS2 promoter by dnHMGA1, while deletion of region −3348 to −3119 (Δ−3348/−3119) did not alter hNOS2 promoter suppression by dnHMGA1 (Fig. 5B). Together, these data localize the AT-rich regions from −3506 to −3375 as critical for modulation of hNOS2 promoter activity by HMGA1.
Fig. 4.
HMGA1 binds to the hNOS promoter. The hNOS2 5′-flanking sequence between −3658 and −2944 is depicted, with 13 DNA sequences (underlined and numbered) containing AT-rich regions used as probes for protein-DNA binding assays. The bottom panels show protein-DNA binding reactions using these probes after radiolabeling, in the presence (+) or absence (−) of synthetic HMGA1 peptide (HMGA1(2/3), 500 ng). Reaction mixtures of the probes and HMGA1(2/3) were then subjected to electrophoresis. Potential HMGA1 binding sites are denoted by bold type. A vertical line in the right margin designates regions −3506/−3375 and −3348/−3119, which are deleted (Δ) and tested functionally in Figure 5.
Fig. 5.
Deletion of the HMGA1-responsive region suppresses cytokine induction of the hNOS2 promoter. (A) Luciferase reporter plasmid hNOS2 −8296 and an internal deletion construct from −3658 to −3094 (Δ−3658/−3094) were transiently co-transfected (500 ng/well) with an expression plasmid for dnHMGA1 (500 ng/well) or empty vector into A549 cells. (B) Internal deletion constructs of hNOS2 −8296, from −3506 to −3375 (Δ−3506/−3375) and from −3348 to −3119 (Δ−3348/−3119), were transiently co-transfected (500 ng/well) with an expression plasmid for dnHMGA1 (500 ng/well) or empty vector into A549 cells. Probe regions (from Fig. 4) that were deleted are noted under the construct names on the x-axis. In both (A) and (B), cells were exposed to CM or vehicle, and then harvested for luciferase activity. Promoter activity was plotted as fold induction compared with hNOS2 −8296 receiving no CM and no dnHMGA1. n=9 in each group. *, increase (p < 0.05) versus no CM and no dnHMGA1; † decrease (p < 0.05) versus construct −8296 in the presence of CM; NS, not significant.
4. Discussion
NO is a molecule with a wide variety of functions depending on the specific NOS isoform that generates its production, and how this NOS isoform is being regulated during a physiologic or pathophysiologic stimulus [7,29]. The inducible isoform, NOS2, is responsible for upregulation of NO during inflammatory processes. We have previously demonstrated the ability of HMGA1 to facilitate mouse NOS2 promoter activity in vitro [24], and we also have used a small molecule inhibitor of HMGA1 binding (Dist A) in mice to blunt NOS2 induction in vivo and to improve outcome in a model of endotoxemia [30]. In this report, we advance the understanding of human NOS2 by determining whether HMGA1 plays a role in its regulation. Figure 1 demonstrates that in human lung epithelial cells, Dist A is able to blunt the induction of hNOS2 at both the level of mRNA (Fig. 1A) and promoter activity (Fig. 1B). Moreover, by overexpressing a dominant-negative form of HMGA1 (dnHMGA1), we were able to suppress hNOS2 promoter activity (Fig. 2), both at baseline and after stimulation with a mixture of LPS and pro-inflammatory cytokines (IFN-γ and IL-1β), in airway epithelial cells. These data suggest that HMGA1 plays a role in the regulation of hNOS2 not only under inflammatory conditions, but also at baseline, as human airway epithelial cells are known to express small amounts of NOS2 under basal conditions [31].
While the major cytokine-responsive elements in the murine NOS2 promoter are located within 1 kb of the transcription initiation start site [8,9,11,12], regions of the human NOS2 promoter responsible for maximal cytokine induction are located much further upstream [13–17]. Interestingly, overexpression of dnHMGA1 was able to suppress deletion mutants of the hNOS2 promoter, even when enhancer regions (between −5 kb and −6 kb [15]) responsible for cytokine responsiveness (Fig. 3) were removed. These data point to a clear separation in promoter regions responsible for classical cytokine-inducible enhancer activity and the HMGA1 response. Protein-DNA binding studies demonstrated that HMGA1 bound at AT-rich nucleotides spanning region −3658 to −2944 (Fig. 4) of the hNOS2 promoter, with more binding in region −3506 to −3375. The functional significance of this binding was confirmed by the generation of internal deletion constructs, where deletion of region −3506 to −3375 abolished suppression of hNOS2 promoter activity by dnHMGA1, either in the presence or absence of CM (Fig. 5). Moreover, deletion of this region blunted the cytokine responsiveness of the hNOS2 promoter, confirming its potential importance. These data support the concept that binding of HMGA1 at AT-rich sequences in region −3506 to −3375 of the hNOS2 promoter is important for full promoter activity, in conjunction with upstream classical cytokine-inducible enhancer regions [15,16].
This report provides the first evidence that HMGA1 is able to modify the regulation of human NOS2. To our knowledge, the functional region responsible for HMGA1 activity (−3506 to −3375) has never been shown to regulate hNOS2 promoter activity previously. Thus, our data point to HMGA1 as working in conjunction with upstream enhancer regions to most efficiently drive hNOS2 promoter activity. We believe that the unique properties of HMGA1, including the ability to bend and alter DNA conformation [22], allow widely separated enhancer regions to interact and regulate human NOS2 expression.
Acknowledgements
This work was supported by National Institutes of Health grants GM53249 and AI061246 to M.A.P., and AI054465 to R.M.B.
Nonstandard abbreviations
- NO
nitric oxide
- NOS
nitric oxide synthase
- HMG
high mobility group
- dn
dominant-negative
- AP
activator protein
- NF
nuclear factor
- LPS
lipopolysaccharide
- IL
interleukin
- IFN
interferon
- TNF
tumor necrosis factor
- IRF
interferon regulatory factor
- Stat
signal transducer and activator of transcription
- CM
cytokine mixture
- Dist
Distamycin
- bp
base pairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nathan C, Xie Q-w. Nitric oxide synthases: Roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 2.Moncada S. Nitric oxide gas: mediator, modulator, and pathophysiologic entity. J. Lab. Clin. Med. 1992;120:187–191. [PubMed] [Google Scholar]
- 3.Lyons CR, Orloff GJ, Cunningham JM. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J Biol Chem. 1992;267:6370–6374. [PubMed] [Google Scholar]
- 4.Xie Q-w, Cho HJ, Calaycay J, Mumford RA, Swiderek KM, Lee TD, Ding A, Troso T, Nathan C. Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 5.Harbrecht BG, Di Silvio M, Demetris AJ, Simmons RL, Billiar TR. Tumor necrosis factor-alpha regulates in vivo nitric oxide synthesis and induces liver injury during endotoxemia. Hepatology. 1994;20:1055–1060. doi: 10.1002/hep.1840200439. [DOI] [PubMed] [Google Scholar]
- 6.Titheradge MA. Nitric oxide in septic shock. Biochim Biophys Acta. 1999;1411:437–455. doi: 10.1016/s0005-2728(99)00031-6. [DOI] [PubMed] [Google Scholar]
- 7.Nathan C. Inducible nitric oxide synthase: What difference does it make? J. Clin. Invest. 1997;100:2417–2423. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Q-w, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon γ and bacterial lipopolysaccharide. J. Exp. Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowenstein CJ, Alley EW, Raval P, Snowman AM, Snyder SH, Russell SW, Murphy WJ. Macrophage nitric oxide synthase gene: Two upstream regions mediate induction by interferon γ and lipopolysaccharide. Proc. Natl. Acad. Sci. USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorsbach RB, Murphy WJ, Lowenstein CJ, Snyder SH, Russell SW. Expression of the nitric oxide synthase gene in mouse macrophages activated for tumor cell killing. Molecular basis for the synergy between interferon-γ and lipopolysaccharide. J Biol Chem. 1993;268:1908–1913. [PubMed] [Google Scholar]
- 11.Teng X, Zhang H, Snead C, Catravas JD. Molecular mechanisms of iNOS induction by IL-1β and IFN-γ in rat aortic smooth muscle cells. Am. J. Physiol. (Cell Physiol.) 2002;282:C144–C152. doi: 10.1152/ajpcell.2002.282.1.C144. [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-γ-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon- and lipopolysaccharide. J. Biol. Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 13.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Morris SM, Jr, Billiar TR, Geller DA. Multiple NF-κB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J. Biol. Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 14.Ganster RW, Taylor BS, Shao L, Geller DA. Complex regulation of human inducible nitric oxide synthase gene transcription by Stat 1 and NF-κB. Proc. Natl. Acad. Sci. USA. 2001;98:8638–8643. doi: 10.1073/pnas.151239498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Z, Shao L, Du Q, Park KS, Geller DA. Identification of a classic cytokine-induced enhancer upstream in the human iNOS promoter. FASEB J. 2007;21:535–542. doi: 10.1096/fj.06-6739com. [DOI] [PubMed] [Google Scholar]
- 16.Chu SC, Marks-Konczalik J, Wu H-P, Banks TC, Moss J. Analysis of the cytokine-stimulated human inducible nitric oxide synthase (iNOS) gene: Characterization of differences between human and mouse iNOS promoters. Biochem. Biophys. Res. Commun. 1998;248:871–878. doi: 10.1006/bbrc.1998.9062. [DOI] [PubMed] [Google Scholar]
- 17.Marks-Konczalik J, Chu SC, Moss J. Cytokine-mediated transcriptional induction of the human inducible nitric oxide synthase gene requires both activator protein 1 and nuclear factor kB-binding sites. J. Biol. Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Comhair SAA, Zheng S, Chu SC, Marks-Konczalik J, Moss J, Haque J, Erzurum SC. STAT-1 and c-Fos interaction in nitric oxide synthase-2 gene activation. Am. J. Physiol. Lung Mol. Physiol. 2003;285:L137–L148. doi: 10.1152/ajplung.00441.2002. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe AP. Architectural Transcription Factors Science. 1994;264:1100–1101. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- 20.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 21.Thanos D, Maniatis T. Virus induction of human IFN-β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 22.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 23.Pellacani A, Chin MT, Wiesel P, Ibanez M, Patel A, Yet S-F, Hsieh C-M, Paulauskis JD, Reeves R, Lee M-E, Perrella MA. Induction of high mobility group-I(Y) protein by endotoxin and interleukin-1β in vascular smooth muscle cells: Role in activation of inducible nitric oxide synthase. J. Biol. Chem. 1999;274:1525–1532. doi: 10.1074/jbc.274.3.1525. [DOI] [PubMed] [Google Scholar]
- 24.Perrella MA, Pellacani A, Wiesel P, Chin MT, Foster LC, Ibanez M, Hsieh C-M, Reeves R, Yet S-F, Lee M-E. High mobility group-I(Y) protein facilitates nuclear factor-kB binding and transactivation of the inducible nitric-oxide synthase promoter/enhancer. J. Biol. Chem. 1999;274:9045–9052. doi: 10.1074/jbc.274.13.9045. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Baron RM, Zhu G, Joo M, Christman JW, Silverman ES, Perrella MA, Riese RJ, Cernadas M. PU.1 regulates cathepsin S expression in professional APCs. J. Immunol. 2006;176:275–283. doi: 10.4049/jimmunol.176.1.275. [DOI] [PubMed] [Google Scholar]
- 26.Himes SR, Reeves R, Attema J, Nissen M, Li Y, Shannon MF. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J. Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- 27.Huth JR, Bewley CA, Nissen MS, Evans JNS, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 28.Neidle S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 29.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function, and inhibition. Biochem. J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron RM, Carvajal IM, Liu X, Okabe RO, Chen Y-H, Ejima K, Layne MD, Perrella MA. Reduction of nitric oxide synthase 2 expression by distamycin A improves survival from endotoxemia. J. Immunol. 2004;173:4147–4153. doi: 10.4049/jimmunol.173.6.4147. [DOI] [PubMed] [Google Scholar]
- 31.Guo FH, De Raeve JR, Rice TW, Stuehr DJ, Thunnissen FBJM, Erzurum SC. Continuous nitric oxide synthesis by inducible nitric oxide synthase in normal human airway epithelium in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:7809–7813. doi: 10.1073/pnas.92.17.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]