Abstract
The cysteine protease dipeptidyl peptidase I (DPPI) activates granule-associated immune-cell serine proteases. The in vivo activator of DPPI itself is unknown; however, cathepsins L and S are candidates because they activate pro-DPPI in vitro. In this study, we tested whether cathepsins L and S activate pro-DPPI in vivo by characterizing DPPI activity and processing in cells lacking cathepsins L and S. DPPI activity, and the relative size and amounts of DPPI heavy and light chains, were identical in mast cells from wild-type and cathepsin L/S double-null mice. Furthermore, the activity of DPPI-dependent chymase was preserved in tissues of cathepsin L/S double-null mice. These results show that neither cathepsin L nor S is required for activation of DPPI and suggest that one or more additional proteases is responsible.
Keywords: chymase, cysteine protease, mast cell, papain
The lysosomal cysteine protease dipeptidyl peptidase I (DPPI), also known as cathepsin C, has numerous biological functions. In addition to a proposed degradative role as a lysosomal exopeptidase, DPPI is essential for activating granule-associated serine proteases in cytoxic T-lymphocytes (granzymes), neutrophils (neutrophil elastase, cathepsin G, proteinase 3), and mast cells (chymase) (Pham and Ley, 1999; Wolters et al., 2001; Adkison et al., 2002). The importance of serine protease activation by DPPI is illustrated in studies using DPPI-/- mice, which revealed that DPPI regulates inflammation in a collagen-induced model of arthritis (Adkison et al., 2002) and survival from sepsis (Mallen-St. Clair et al., 2004). In humans, loss-of-function defects in the DPPI gene cause Papillon-Lefèvre syndrome, which is clinically manifest as severe periodontitis and dermatitis (Hart et al., 1999; Toomes et al., 1999).
DPPI is a structurally unique member of the C1 family of cysteine proteases because it exists as a tetramer and retains a portion of the pro-peptide in its active form (Dolenc et al., 1995; Wolters et al., 1998; Turk et al., 2001). DPPI activation requires proteolytic cleavage at three sites: two in the pro-peptide and a third within the mature enzyme. In humans, these processing events generate a 163-aa heavy chain and a 68-aa light chain. Cleavage of the 206-aa pro-peptide releases an 87-aa fragment, leaving behind 119 amino acids of the N-terminal portion of pro-peptide termed the exclusion domain (Turk et al., 2001) which partially blocks the active site, rendering the enzyme a dipeptidase (Turk et al., 2001).
The mechanisms of activation of many lysosomal cysteine proteases have yet to be defined in vivo. In vitro, several cathepsins (K, L, and S) are capable of autoactivation upon exposure to low pH (McQueney et al., 1997; Menard et al., 1998; Vasiljeva et al., 2005). In contrast, studies using recombinant human pro-DPPI suggest that pro-DPPI cannot activate itself (Dahl et al., 2001). However, pro-DPPI can be activated in vitro by cathepsin L or S (Dahl et al., 2001). To establish if these in vitro findings correspond to in vivo functions, we studied DPPI activation in C57BL/6 mice deficient in cathepsins L and S generated by crossing cathepsin L-/- (Potts et al., 2004) with cathepsin S-/- (Shi et al., 1999) mice.
We used mast cells to study DPPI activation because they are an abundant source of DPPI (Wolters et al., 1998) and are convenient to derive in cell culture. First, we sought to determine if cathepsin L and cathepsin S are expressed in mast cells. RT-PCR of bone marrow mast cell (BMMC) RNA using specific primer pairs for cathepsin L and cathepsin S generated single-band amplimers of the predicted sizes (Figure 1A). Primer specificity was demonstrated by the failure to generate amplimers on BMMC RNA obtained from cathepsin L/S-/- mice. However, RNA from these cells was intact, as primers specific for G3PDH produced an amplimer of the predicted size (data not shown).
Figure 1. Mast cells express cathepsin L and cathepsin S.
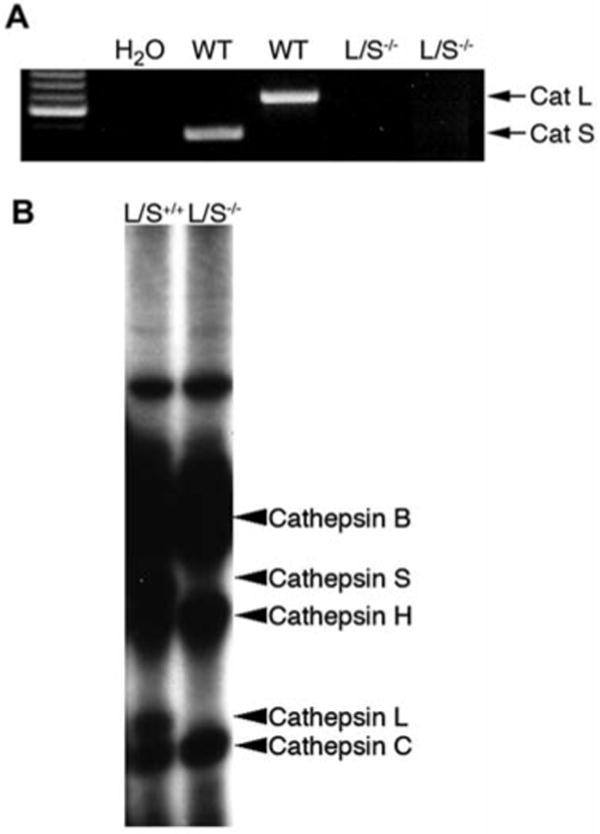
Mast cells were cultured from mouse bone marrow in 50% WEHI-3B-conditioned medium as described by Razin et al. (1984). (A) RT-PCR for cathepsin L and cathepsin S produced amplimers of predicted size on total RNA harvested from wild-type (WT) but not cathepsin L/S-/- BMMCs. Total RNA was isolated from cathepsin L/S+/+ or cathepsin L/S-/- BMMCs using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and then reverse-transcribed into single-stranded cDNA using Superscript II reverse transcriptase (BD Biosciences, Palo Alto, CA, USA) and oligo dT primers. Cathepsin L and S transcripts in this cDNA were detected by PCR using the following primers: cathepsin L: forward, 5′-GTT GTG TGA CTC CTG TGA AG-3′; reverse, 5′-CTT GCT GCT ACA GTT GGG T-3′; cathepsin S: forward, 5′-CCA AGT GGG CAT GAA CG-3′; reverse, 5′-GCC AGT AAT CTT TGC CAT CAA G-3′. (B) Active site [125I]-JPM labeling of cysteine proteases. Cathepsin L/S+/+ or cathepsin L/S-/- BMMCs (2×106 /sample) were labeled by culturing cells in the presence of [125I]-JPM ethyl ester for 24 h at 37°C as described by Liu et al. (2006). Cells were washed twice with PBS, lysed in SDS-PAGE sample buffer, and labeled proteins were analyzed by 12% SDS-PAGE and autoradiography.
To confirm cathepsin L and cathepsin S were translated into active proteins, we labeled the active site of lysosomal cysteine proteases in mast cells with the [125I]-JPM-ethyl ester (Shi et al., 1992; Liu et al., 2006) and found that mouse BMMCs contain several active cathepsins, including cathepsins B, C, H, L and S (Figure 1B). The presence of each cathepsin was established by their characteristic size (Liu et al., 2006) on the autoradiogram and the absence of cathepsin L and cathepsin S bands in cathepsin L/S-/- BMMC lysates.
Next, we compared DPPI and chymase activities in lysates of BMMCs from cathepsin L/S+/+ and cathepsin L/S-/- mice. Previous results from our laboratory demonstrated that DPPI is essential for activating mast-cell chymase (Wolters et al., 2001) and that BMMCs lacking DPPI have no active DPPI or chymases (Table 1). Thus, if cathepsin L or S is required for DPPI activation, we predicted that BMMCs from cathepsin L/S-/- mice would have no active DPPI or chymase. Instead, we found that cathepsin L/S-/- BMMCs contain levels of DPPI and chymase activity indistinguishable from those of cathepsin L/S+/+ controls (Table 1), indicating that cathepsin L and cathepsin S are not required for activation of DPPI in cultured BMMCs.
Table 1.
Protease activity in mast cell (BMMC) lysates.
| Protease | Cathepsin L/S+/+ BMMCs
(ΔA410/h 80×106 cells) |
Cathepsin L/S-/- BMMCs
(ΔA410/h 80×106 cells) |
DPPI-/- BMMCs
(ΔA410/h 80×106 cells) |
|---|---|---|---|
| DPPI | 0.48±0.06 | 0.48±0.05 | 0.00±0.00 |
| Chymase | 0.01±0.00 | 0.01±0.00 | 0.00±0.00 |
BMMCs (20×106) were solubilized in 250 μl of lysis buffer (100 mm Na acetate, pH 5.0, 300 mm NaCl, 1 mm EDTA). DPPI activity was measured spectrophotometrically by monitoring hydrolysis of l-Ala-Ala-p-nitroanilide (Bachem, King of Prussia, PA, USA). Aliquots of 50 μl of cell lysates were incubated for 5 min in 500 μl of activation buffer (100 mm Na2HPO4 buffer, 20 mm NaCl, 1 mm EDTA, 4 mm cysteine, pH 6), followed by the addition of 400 μl of substrate buffer (100 mm Na2HPO4, 20 mm NaCl, 1 mm EDTA, 125 μm l-Ala-Ala-p-nitroanilide, 1% dimethylsulfoxide, pH 6). Chymase activity was measured by the addition of 50 μl of cell lysate to 1 ml of 0.45 m Tris-HCl (pH 8.0) containing 1 mm succinyl-l-Ala-Ala-Pro-Phe-p-nitroanilide (Sigma-Aldrich, St. Louis, MO, USA), 1.8 mm NaCl, and 1% dimethylsulfoxide. The release of free nitroaniline was measured spectrophotometrically at 410 nm for 5–10 min in all assays.
BMMCs are differentiated in vitro and do not fully reflect the phenotype of various mast-cell subtypes in vivo. Thus, it is possible that cathepsin L or S is required for DPPI activation in mast cell subtypes in vivo. DPPI is required for chymase activation (McCoy et al., 1988; Wolters et al., 2001). Therefore, if cathepsin L and cathepsin S are essential for activating DPPI, cathepsin L/S-/- mice should recapitulate the DPPI-/- phenotype and lack chymase activity using the enzyme histochemical chloroacetate esterase stain (Wolters et al., 2001). To test this possibility, we examined sections of tongue, trachea, lung, spleen, stomach, and skin from cathepsin L/S-/- mice for chloroacetate esterase activity and found positive cells in all of these tissues, but not tissues obtained from DPPI-/- mice (Figure 2, and data not shown). Esterase-positive cells had a distribution, number and morphology typical for mast cells identified in wild-type mice (Wolters et al., 2001). The presence of esterase activity in these tissues demonstrates that active chymase and, by extension, active DPPI are present in tissue mast cells of cathepsin L/S-/- mice.
Figure 2. Enzyme histochemical detection of active mast-cell chymase in mouse tissues.
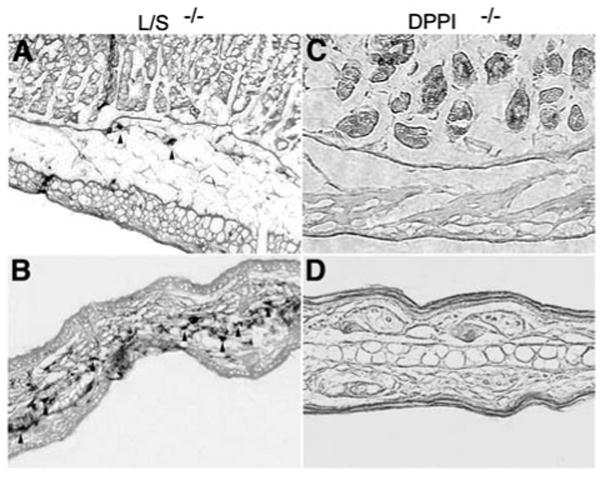
Sections of mouse stomach (A,C) and ear (B,D) obtained from cathepsin L/S-/- (A,B) or DPPI-/- (C,D) mice. Harvested tissues were fixed for 6–18 h in PBS containing 4% paraformaldehyde. Tissues were embedded directly in Tissue-Tek OCT compound (Miles, Elkhart, IN, USA), frozen at -80°C, and then sectioned (5 μm). Prior to use, these sections were equilibrated in PBS. To detect active chymase in tissue sections, chloroacetate esterase histochemistry was performed as described by Leder (1979). Slides were then washed with water, counterstained in 0.5% eosin Y, mounted, and photographed. Note the esterase-positive mast cells (arrowheads) in tissue sections obtained from cathepsin L/S-/- (A,B) but not DPPI-/- mice (C,D). Images were obtained using a 20× objective lens and are representative of observations made in six animals.
Finally, we investigated whether, despite maintaining activity, DPPI was processed at different sites in cathepsin L/S-/- mice. Normally, three proteolytic processing events, yielding a heavy chain, a light chain and an exclusion domain, are required for DPPI activation (Dahl et al., 2001; Turk et al., 2001). If cathepsin L and cathepsin S were involved in processing, we predicted we would detect the absence or a change in the size of some or all of these domains in cathepsin L/S-/- BMMCs. To investigate these possibilities, we probed immunoblots of wild-type and cathepsin L/S-/- BMMC lysates with rabbit anti-bovine DPPI. Immunoblots showed that the antibody recognized both heavy and light chains of mouse DPPI (Figure 3). Furthermore, the size of the heavy and light chains were the same in wild-type and cathepsin L/S-/- BMMC lysates, indicating they were processed to their expected size in the absence of cathepsins L and S.
Figure 3. DPPI is processed normally in cathepsin L/S-/- mice.
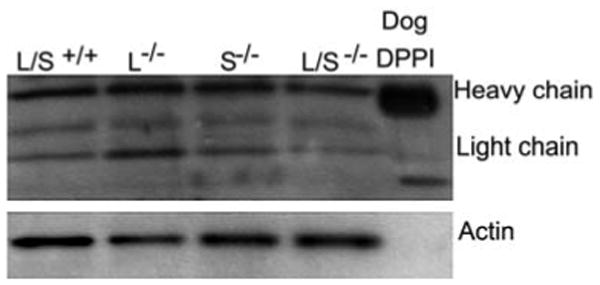
Samples of 10 μg of total protein from cell lysates obtained from cathepsin L/S+/+, cathepsin L-/-, cathepsin S-/- or cathepsin L/S-/- BMMCs and purified dog DPPI were subjected to SDS-PAGE under reducing conditions and transferred to polyvinylidine difluoride membranes (Life Sciences Products, Boston, MA, USA). The membrane was washed with 50 mm Tris-HCl containing 0.5 m NaCl, 0.01% Tween-20 (TBS; pH 7.5), and incubated for 1 h in TBS containing either a 1:1000 dilution of rabbit anti-bovine DPPI (a gift from John Hoidal, University of Utah, Salt Lake City, USA) or monoclonal anti-β-actin antibody (Sigma-Aldrich). The membrane was then washed with TBS, incubated in TBS for 30 min containing a 1:2000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG (New England Biolabs, Beverly, MA, USA) and washed again. Immunoreactivity was detected using the phototope-HRP detection kit (New England Biolabs). Note that bands of immunoreactive DPPI appear at similar sizes in both cathepsin L/S+/+ and cathepsin L/S-/- BMMC lysates.
Studies using recombinantly expressed cysteine proteases demonstrate that both cathepsins L and S activate DPPI in vitro (Dahl et al., 2001). In contrast, our data show that cathepsins L and S are not required for DPPI activation in vivo. Furthermore, given the evidence that DPPI is processed into appropriately sized heavy and light chains in the absence of cathepsins L and S, it is also unlikely that they secondarily process DPPI following activation by another protease. While these findings do not eliminate the possibility that cathepsin L or S participates in the activation of DPPI in vivo, they do suggest that, at minimum, other proteases activate DPPI and that there may be redundancy in the DPPI activation system. This redundancy would serve to ensure activation of DPPI and the proteases it activates to guarantee they maintain their activity and are able to participate in important biological processes (Pham and Ley, 1999; Wolters et al., 2001; Adkison et al., 2002).
The identity of the other protease(s) responsible for DPPI activation in vivo remains unknown. However, although we cannot rule out the possibility that BMMCs contain cathepsins at levels below the detection limit for the JPM label, in mast cells, the protease is unlikely to be a lysosomal cysteine protease because the only active cathepsins we detected in these cells are cathepsins B, C (DPPI), H, L, and S. Previous studies using recombinant DPPI demonstrated that DPPI cannot autoactivate itself in vitro (Dahl et al., 2001). As discussed, cathepsins L and S are not required to activate DPPI. Cathepsin B will not likely activate DPPI because it is preferentially a carboxypeptidase (Turk et al., 1997) and does not activate pro-DPPI in vitro (Dahl et al., 2001). Finally, cathepsin H is an aminopeptidase and therefore is not likely to be capable of generating the endoproteolytic cleavages required for DPPI activation.
In summary, these data show that cathepsins L and S are not required for activation of DPPI and that another protease(s) must be involved. Identification of the specific peptidases involved in this processing step will be important for future understanding of the unique biochemistry of DPPI. Because previous reports suggested that DPPI plays a role in the pathophysiology of arthritis (Adkison et al., 2002) and sepsis (Mallen-St. Clair et al., 2004), identification of DPPI activators may provide novel targets for treating these diseases.
Acknowledgments
This work was supported in part by grants HL60942, HL-72301, HL024136, and HL-075026 from the National Institutes of Health, and the American Lung Association of California.
References
- Adkison AM, Raptis SZ, Kelley DG, Pham CT. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest. 2002;109:363–371. doi: 10.1172/JCI13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl SW, Halkier T, Lauritzen C, Dolenc I, Pedersen J, Turk V, Turk B. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry. 2001;40:1671–1678. doi: 10.1021/bi001693z. [DOI] [PubMed] [Google Scholar]
- Dolenc I, Turk B, Pungercic G, Ritonja A, Turk V. Oligomeric structure and substrate induced inhibition of human cathepsin C. J Biol Chem. 1995;270:21626–21631. doi: 10.1074/jbc.270.37.21626. [DOI] [PubMed] [Google Scholar]
- Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, Zhang Y, Firatli E. Mutations of the cathepsin C gene are responsible for Papillon-Lefèvre syndrome. J Med Genet. 1999;36:881–887. [PMC free article] [PubMed] [Google Scholar]
- Leder LD. The chloroacetate esterase reaction. A useful means of histological diagnosis of hematological disorders from paraffin sections of skin. Am J Dermatopathol. 1979;1:39–42. [PubMed] [Google Scholar]
- Liu J, Sukhova GK, Yang JT, Sun J, Ma L, Ren A, Xu WH, Fu H, Dolganov GM, Hu C, et al. Cathepsin L expression and regulation in human abdominal aortic aneurysm, atherosclerosis, and vascular cells. Atherosclerosis. 2006;184:302–311. doi: 10.1016/j.atherosclerosis.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Mallen-St Clair J, Pham CTN, Villalta SA, Caughey GH, Wolters PJ. Mast cell dipeptidyl peptidase I mediates survival from sepsis. J Clin Invest. 2004;113:628–634. doi: 10.1172/JCI19062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy K, Gal S, Schwartz RH, Gottesman MM. An acid protease secreted by transformed cells interferes with antigen processing. J Cell Biol. 1988;106:1879–1884. doi: 10.1083/jcb.106.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueney MS, Amegadzie BY, D'Alessio K, Hanning CR, McLaughlin MM, McNulty D, Carr SA, Ijames C, Kurdyla J, Jones CS. Autocatalytic activation of human cathepsin K. J Biol Chem. 1997;272:13955–13960. doi: 10.1074/jbc.272.21.13955. [DOI] [PubMed] [Google Scholar]
- Menard R, Carmona E, Takebe S, Dufour E, Plouffe C, Mason P, Mort JS. Autocatalytic processing of recombinant human procathepsin L. Contribution of both intermolecular and unimolecular events in the processing of procathepsin L in vitro. J Biol Chem. 1998;273:4478–4484. doi: 10.1074/jbc.273.8.4478. [DOI] [PubMed] [Google Scholar]
- Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA. 1999;96:8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts W, Bowyer J, Jones H, Tucker D, Freemont AJ, Millest A, Martin C, Vernon W, Neerunjun D, Slynn G, et al. Cathepsin L-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int J Exp Pathol. 2004;85:85–96. doi: 10.1111/j.0959-9673.2004.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin E, Ihle JN, Seldin D, Mencia-Huerta JM, Katz HR, LeBlanc PA, Hein A, Caulfield JP, Austen KF, Stevens RL. Interleukin 3: a differentiation and growth factor for the mouse mast cell that contains chondroitin sulfate E proteoglycan. J Immunol. 1984;132:1479–1486. [PubMed] [Google Scholar]
- Shi GP, Munger JS, Meara JP, Rich DH, Chapman HA. Molecular cloning and expression of human alveolar macrophage cathepsin S, an elastinolytic cysteine protease. J Biol Chem. 1992;267:7258–7262. [PubMed] [Google Scholar]
- Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, Riese R, Ploegh HL, Chapman HA. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- Toomes C, James J, Wood AJ, Wu CL, McCormick D, Lench N, Hewitt C, Moynihan L, Roberts E, Woods CG, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- Turk B, Turk V, Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol Chem. 1997;378:141–150. [PubMed] [Google Scholar]
- Turk D, Janjic V, Stern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva O, Dolinar M, Pungercar JR, Turk V, Turk B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. FEBS Lett. 2005;579:1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Raymond WW, Blount JL, Caughey GH. Regulated expression, processing, and secretion of dog mast cell dipeptidyl peptidase I. J Biol Chem. 1998;273:15514–15520. doi: 10.1074/jbc.273.25.15514. [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ, Caughey GH. Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J Biol Chem. 2001;276:18551–18556. doi: 10.1074/jbc.M100223200. [DOI] [PubMed] [Google Scholar]


