Abstract
Expressed protein ligation (EPL) is an intein-based approach that has been used for protein engineering and biophysical studies of protein structures. One major problem of the EPL is the low yield of final ligation product, primarily due to the complex procedure of the EPL, preventing EPL from gaining popularity in the research community. Here we report an efficient on-column EPL strategy, which focuses on enhancing the expression level of the intein-fusion protein that generates thioester for the EPL. We applied this EPL strategy to human apolipoprotein E (apoE) and routinely obtained 25–30 mg segmental, triple-labeled apoE from 1-L cell culture. The approaches reported here are general approaches that are not specific for apoE, thus providing a general strategy for a highly efficient EPL. In addition, we also report an isotopic labeling scheme that double-labels one domain and keeps the other domain of apoE deuterated. Such an isotopic labeling scheme can only be achieved using the EPL strategy. Our data indicated that the segmental triple-labeled apoEs using this labeling scheme produced high-quality, simplified NMR spectra, facilitating NMR spectral assignment. For large proteins, such as apoE, perdeuterated protein samples have to be used to reduce the linewidth of NMR signals, causing a major problem for the NOE-based NMR method, since perdeuterated proteins lack protons for NOE measurement. The new labeling strategy solves this problem and provides 13C/15N double-labeled, protonated protein domains, allowing for determination of high-resolution NMR structure of these large proteins.
Keywords: expressed protein ligation, high-level protein expression, segmental isotope labeling, apolipoprotein E, NMR spectroscopy, nuclear Overhauser enhancement, NMR structure determination
Expressed protein ligation (EPL), pioneered by Muir and his coworkers (Muir et al. 1998; Muir 2003), is an intein-based approach for semisynthesis of proteins. This approach utilizes a protein splicing mechanism to generate α-thioester derivatives of a recombinant fragment of a target protein in a controlled manner by a thiol reagent. The α-thioester derivatives of a recombinant fragment subsequently react with the free sulfhydryl group of the N-terminal Cys1 of the second fragment, followed by a N-S acyl shifting, to ligate both fragments of the target protein together with a native peptide bond (Muir 2003). This technique permits chemoselective and regioselective assembly of a protein from smaller synthetic and/or recombinant pieces, which offers researchers an ability to incorporate noncoded amino acids (Ayers et al. 1999; Wang and Cole 2001), biophysical probes (Cotton and Muir 2000; Scheibner et al. 2003; Maag et al. 2005), stable isotopes (Cowburn et al. 2004; Vitali et al. 2006) and post-translational modifications (Zhang et al. 2003; Thompson et al. 2004) into specific locations within a protein, providing a powerful tool to study the structure and function of proteins (Schwarzer and Cole 2005; Muralidharan and Muir 2006). For NMR structural determination of large proteins, this technique is of particular interest, since it allows for segmental labeled large proteins, which will significantly reduce the complexity of NMR spectra and simplify the spectral assignment, allowing for NMR structural determination of these large proteins (Camarero et al. 2002; Yagi et al. 2004). In addition, a segmental-labeled domain of a large protein will only display NMR spectra of this labeled domain, providing a powerful tool to study protein domain–domain interactions (Xu et al. 1999; Vitali et al. 2006; Zhang et al. 2007).
Two commercial bacterial expression vectors, pTYB and pTWIN from New England Biolabs, are available for generation of α-thioester derivatives of recombinant proteins by thiolysis of mutated intein fusions (Chong et al. 1997; Xu and Evans Jr. 2001). These two expression vectors are widely used for one-step recombinant protein purification and made it possible for EPL as a general laboratory technique. However, one major problem with EPL is the low yield of ligation product, which prevents many laboratories from using this strategy routinely to prepare properly labeled protein samples at certain defined positions for protein biophysical research. Indeed, despite its potential power in studying protein structure and function, the EPL has not gained popularity in the research community and only a small number of laboratories have successfully performed research using the EPL strategy. Several reasons contribute to this problem. First, the protein yield of intein-containing vectors is generally lower than other high-level expression vectors, such as pET expression vectors. We recently found that the lower protein yield of the pTWIN vector was due to an in vivo self-cleavage between the target protein and intein fusion during bacterial expression (Cui et al. 2006). This self-cleavage removes the affinity tag from the target protein, which flows through the chitin-bead column without binding to the beads and is unable to be recovered during purification; thus, the protein yield is dramatically reduced. We prepared a mutation of the intein (T3C) in the pTWIN vector that completely eliminates this self-cleavage, significantly enhancing protein yield by up to 10-fold (Cui et al. 2006). Another main reason is the complex procedure of the EPL, which requires many steps to achieve the final ligation product. An accumulation factor of this complex procedure results in a general overall low yield of the final ligation product. This is especially true for making segmental triple-labeled protein for NMR studies, since protein expression in D2O usually results in a low yield production of labeled proteins (Cowburn et al. 2004; Vitali et al. 2006). To make EPL practical for general laboratory application by nonexperts, this major problem of a low yield of final ligation product has to be solved.
Here we report an efficient on-column protein ligation strategy that significantly increases the final yield of the target protein (25–30 mg/L). This strategy focused on enhancement of protein yield of α-thioester derivatives of recombinant N-terminal fragment of the target protein and carried out native chemical ligation on a chitin column. A high yield of α-thioester derivatives is critical for a high yield production of final ligation product, while an on-column protein ligation simplifies the ligation procedure and enhances the efficiency of the ligation reaction. We applied this strategy to human apolipoprotein E (apoE). We prepared two apoE fragments: apoE(1–214)/pTYB1 and apoE(C215–299)/pET. Our initial result showed that we only obtained <1 mg protein production from 1 L cell culture of apoE(1–214)/pTYB1. With this low yield, we were not able to obtain any ligation product. Using our strategy, we significantly increased the yield of apoE(1–214) expression and finally obtained a yield of 25–30 mg segmental, triple-labeled apoE from 1 L of bacterial expression of recombinant apoE(1–214). We describe the details of our optimization procedure, explaining how to achieve a high yield production of ligation product. Human apoE (299 residues) is a lipid transport protein that is critical to several major human diseases, including heart disease and Alzheimer's disease (Weisgraber 1994; Huang et al. 2004; Hatters et al. 2006). ApoE is a two-domain protein that contains a 22-kDa N-terminal domain (residues 1–191) and a 10-kDa C-terminal domain (residues 216–299), linked by a protease-sensitive hinge region. The X-ray crystal structure of the apoE N-terminal domain in the lipid free state reveals a globular up-and-down four-helix bundle (Wilson et al. 1991). No structure is available to date for the apoE C-terminal domain and full-length apoE. A domain–domain interaction is hypothesized for apoE that seems to regulate its biological functions (Hatters et al. 2006). Since apoE is a 299-residue α-helical protein, its NMR spectra are significantly overlapped. To reduce NMR spectral complexity for a complete spectral assignment, segmental labeled apoEs, in which one domain is 13C/15N labeled whereas the other domain is deuterated, will be critical for NMR structural determination and verification of apoE's domain–domain interactions.
Results and Discussion
Human apoE
ApoE is well known to aggregate, forming a mixture of different oligomers at a low concentration (Weisgraber 1994). This property hinders structural studies of full-length apoE, either by X-ray crystallography or by the NMR technique. Previous data indicated that apoE aggregation was caused by its C-terminal domain (Weisgraber 1994). Using protein-engineering techniques, we prepared a monomeric, biologically active apoE C-terminal domain (apoEC-J) (Fan et al. 2004) and full-length apoE (apoE-J) (Zhang et al. 2007). In both monomeric apoEC-J and apoE-J, five bulky hydrophobic residues in the C-terminal domain were replaced by either Ala or polar residues. We demonstrated that these monomeric apoE proteins maintained the structure and stability of wild-type apoE and were biologically active. In particular, we showed that full-length apoE-J is monomeric even at a 20 mg/mL concentration. ApoE-J not only adopts identical secondary and tertiary structures, but also displays a characteristic two-transition denaturation curve identical to that of the wild-type apoE (Wetterau et al. 1988). More importantly, this monomeric apoE displays lipoprotein-binding and LDL–receptor-binding activities identical to those of wild-type apoE (Zhang et al. 2007). Using this biologically active monomeric apoE, we collected high-quality NMR spectra. Since apoE is a 299-residue α-helical protein, its NMR spectra are severely overlapped. Thus, a segmental labeled apoE, in which one domain is labeled and the other domain is not labeled, will significantly simplify the spectral assignment and allow us to study domain–domain interaction of apoE. In this report, we used apoE-J as the template for segmental labeled apoE.
An efficient on-column EPL protocol
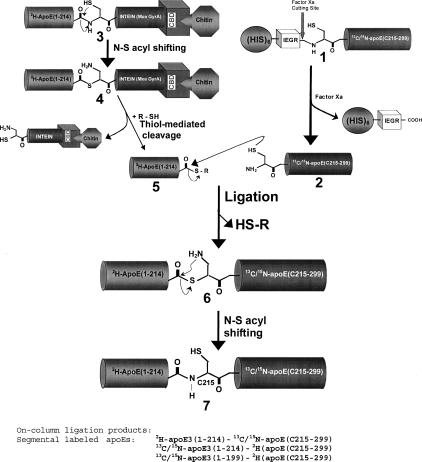
The protocol for preparing segmental, triple-labeled apoE is summarized in Figure 1, which involves the expression and isotope-labeling of apoE(1–214)/pTYB1 and apoE(215–299)/pET30a, respectively. The apoE(215–299)/pET30a expression generated the apoE C-terminal domain (residues 215–299) with an N-terminal long his-tag. To achieve native chemical ligation, we replaced residue R215 by a Cys. We further engineered the pET30a vector by including a Factor Xa cleavage site between the long his-tag and the N-terminal C215 residue (1, Fig. 1). Factor Xa cleaves the long his-tag to generate mature apoE C-terminal domain, apoE(C215–299) with an N-terminal Cys residue (2, Fig. 1). This step was carried out first to prepare the ligation-ready apoE(C215–299). We then carried out expression of the apoE(1–214)/pTYB1 vector that produced the apoE N-terminal domain (residues 1–191) and a hinge domain that linked between the N- and C-terminal domains (residues 192–214). In the apoE(1–214)/pTYB1 expression, the expressed apoE(1–214) was directly linked with an intein protein plus a CBD domain in the C terminus (3, Fig. 1). This fusion protein 3 was loaded on a chitin bead column and purified with binding buffer. After purification, the fusion protein 3 remained on the column and was mixed with an excess of ligation-ready apoE(C215–299). A thiol reagent was also added and mixed well with the chitin bead mixtures, containing both apoE(1–214) fusion protein 3 and ligation-ready apoE(C215–299) protein 2. Under these conditions, the purified fusion protein 3 would first undergo thiolysis catalyzed by the thiol reagent, generating apoE(1–214)-COSR (5, Fig. 1), which was released from the chitin column, whereas the intein-CBD fusion remained on-column. Since we have excess reactive ligation-ready apoE(C215–299) in solution, the newly generated apoE(1–214)-COSR reacts directly with the ligation-ready apoE(C215–299) to initiate the native chemical ligation. During native chemical ligation, the sulfide group of C215 attacks the carbonyl group of the C-terminal thiol ester in apoE(1–214)-COSR, resulting in the original “-SR” group leaving apoE(1–214) to form a new thiol ester apoE(1–214)-COS-apoE(C215–299) (6, Fig. 1). This new thiol ester is not stable and a simultaneous N-S acyl shift quickly happens to form the final ligation product (7, Fig. 1).
Figure 1.
Schematic diagram of the protocol of on-column expressed protein ligation for preparation of the triple, segmental-labeled apoE.
The advantage of this on-column ligation is that the newly generated apoE(1–214)-COSR immediately reacts with the excess apoE(C215–299) to form a final ligation product, segmental labeled apoE, thus eliminating the possibility of hydrolysis. In addition, the EPL procedure is also simplified, overall significantly enhancing the ligation efficiency and the final yield of ligation product. This “one pot” approach has been used by several other groups for the EPL recently (Welker and Scheraga 1999; Sydor et al. 2002; Anderson et al. 2005), to either prepare segmental labeled proteins for NMR studies or introduce unnatural amino acids into proteins or introduce post-translational modification at specific sites of proteins.
A R217S mutation of apoE(C215–299) for specific Factor Xa digestion
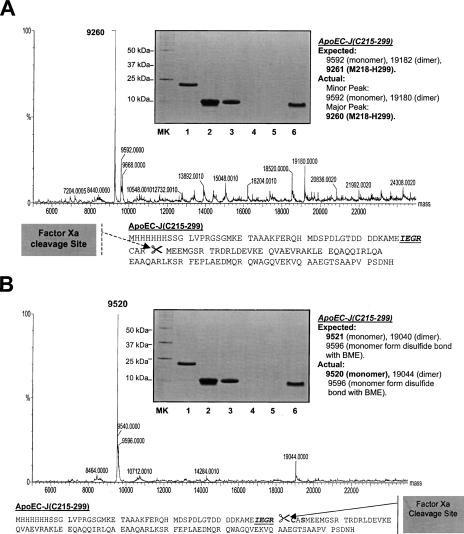
Using a newly developed bacterial expression method (A. Sivashanmugam, V. Murray, C. Cui, Y. Yang, J. Wang, and Q. Li, in prep.), we routinely produced a very high yield of his-tag-apoE(C215–299)-J (300 mg/L in D2O and 400 mg/L in H2O). However, we encountered some difficulty when we used Factor Xa to generate ligation-ready apoE(C215–299). A broad band was observed in the SDS-PAGE after purification of the Factor Xa digestion product with both Xarrest agarose for Factor Xa removal and the Ni-column for his-tag and uncut protein removal (inset, Fig. 2A). Under a reduced condition using DTT, this broad band was at a molecular weight of ∼10 kDa. Table 1 lists the calculated and mass spectroscopic observed molecular weights for apoE(C215–299). The mass spectral analysis indicated two peaks around 10 kDa. One minor peak was at a molecular weight of 9592, which was the correct molecular weight of apoE(C215–299), suggesting that the Factor Xa cleaves at the correct position before C215 (Fig. 2A). Another major peak was at a molecular weight of 9260. Molecular weight calculation scanned apoE(C215–299) sequence and found that apoE(M218–299) had a molecular weight of 9261. This suggests that the major Factor Xa cleavage site was nonspecific at position R217 (Fig. 2A). In addition to the monomeric peaks, several dimeric peaks were also observed which matched the above calculation (Table 1). Figure 2A also shows many other minor peaks, suggesting other possible minor Factor Xa cleavage sites. To eliminate these nonspecific cleavages, we prepared a mutation of R217S for apoE(C215–299). SDS-PAGE showed a sharp band of purified apoE(C215–299)-R217S after Factor Xa cleavage (inset, Fig. 2B), suggesting a possibility of elimination of the nonspecific cleavage sites by Factor Xa. This suggestion was confirmed by mass spectroscopic analysis (Table 1). Mass spectroscopic data indicated that only one major peak was observed for the apoE(C215–299)-R217S mutant at a molecular weight of 9520 Da, which was the calculated value for this mutant (9521 Da, Fig. 2B). The dimeric peaks also confirmed this result. In addition, Figure 2B shows a much cleaner mass spectrum of apoE(C215–299)-R217S as compared with that of apoE(C215–299), suggesting elimination of the other nonspecific Factor Xa cleavage sites. Thus, the generated apoE C-terminal domain using apoE(C215–299)-R217S is a ligation-ready apoE C-terminal domain.
Figure 2.
(A) Mass spectroscopic data of apoE(C215–299) after Factor Xa cleavage. Based on the observed mass spectroscopic data, the potential Factor Xa cleavage site is suggested in the bottom of A. In the sequence, the Factor Xa cleavage site IEGR is italic and underlined. (B) Mass spectroscopic data of apoE(C215–299)-R217S after Factor Xa cleavage. Based on the observed mass spectroscopic data, the Factor Xa cleavage site is at the correct position as shown in the bottom of B. In the sequence, the Factor Xa cleavage site IEGR is italic and underlined. In both panels, the inset is a SDS-PAGE of the Ni-column purification of the mixture of Factor Xa digestion and Xarrest capture. MK: Molecular marker. (Lane 1) Long his-tag-apoE(C215–299), before Factor Xa cut. (Lane 2) The mixture of Factor Xa reaction before loading on Ni-column. (Lane 3) Flow-through. (Lane 4) Binding buffer washing (5 mM imidazole). (Lane 5) Washing buffer washing (50 mM imidazole). (Lane 6) Elution buffer (1 M imidazole) to elute his-tag.
Table 1.
Calculated and mass spectroscopic observed molecular weights of different apoE(C215–299) mutants
Optimization of bacterial expressions of apoE(1–214)/pTYB1
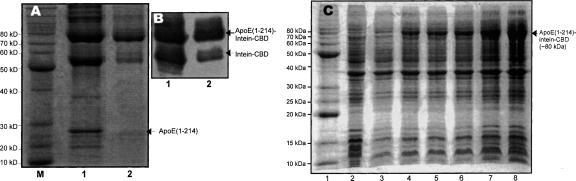
For the EPL, one bottleneck for achieving high-level production of the ligation product is the expression levels of two segments. This is especially important for apoE(1–214)/pTYB1 expression in D2O, since a low yield of this expression will directly result in a low yield of final ligation product. In contrast, in the presence of excess ligation-ready apoE(C215–299), a high concentration of reactive apoE(1–214)-COSR will significantly enhance the ligation efficiency, thus increasing the final yield of ligation product. For apoE(1–214)/pTYB1, our initial expression only produced a very low yield (<1 mg/L). Indeed, SDS-PAGE of cell lysate of this expression with or without IPTG showed no difference, and only a Western blot could detect the expression. In addition, we frequently observed inconsistencies in expression level, especially in D2O, since sometimes we could detect the protein expression using a Western blot and sometimes we could not. With such a low yield, it is impossible to pursue native chemical ligation. Therefore, we put forth a major effort in optimizing bacterial expression of apoE(1–214)/pTYB1. We developed a double colony selection protocol, which allowed us to select high-level expression colonies of apoE(1–214)/pTYB1 especially in D2O. This double colony selection procedure was also used to optimize expression using a pTWIN vector (A. Sivashanmugam, V. Murray, C. Cui, Y. Yang, J. Wang, and Q. Li, in prep.). We found that this double colony selection procedure is critical for reliable high-level apoE(1–214)/pTYB1 expression, since expression yield was dramatically enhanced (50–100-fold) after double colony selection, resulting in 10–15 mg apoE(1–214)-intein-CBD production from a 1-L cell culture in D2O. More importantly, we routinely obtained a consistent high yield of apoE(1–214)/pTYB1 expression. To further enhance this expression, we did several optimizations. First, we mutated the third residue Ala in the intein of pTYB1 vector to a Cys residue to eliminate the potential self-cleavage between the intein and apoE(1–214). We previously found that this self-cleavage was one of the main reasons for a significant reduction in protein yield for the pTWIN vector, although the overall protein expression level was pretty high. Since this self-cleavage removes the affinity tag, the target protein flows through the column without binding to chitin beads and is unable to be recovered during the purification procedure. By introduction of a T3C mutation on the second intein of the pTWIN vector, we were able to eliminate this self-cleavage (Cui et al. 2006). The pTYB vector utilizes a different intein from the Saccharomyces cerevisiae VMA1 gene, which contains an Ala at position 3 of the intein (Chong et al. 1998). We prepared an A3C mutation in the VMA1 intein and suggested that this Cys residue at position 3 may form a disulfide bond with the Cys residue at position 1 of the intein. This potential disulfide bond may block the free sulfhydryl group of Cys1 in the intein, thus eliminating self-cleavage by protecting the intein from the N-S shift. Figure 3A (lane 1) shows that ∼50% of the expressed apoE(1–214)/pTYB1 was self-cleaved after purified apoE(1–214)-intein-CBD on the chitin beads was stored in a −20°C freezer for 48 h. However, this self-cleavage was significantly inhibited by the A3C mutation of the intein in pTYB1 vector and only <5% expressed apoE(1–214)/pTYB1-A3C was self-cleaved under the same experimental condition (Fig. 3A, lane 2). This result was confirmed by the Western blot using an antibody specifically against the CBD domain (Fig. 3B), thus significantly enhancing the yield of apoE(1–214) protein. Second, we optimized glucose usage for the bacterial expression of apoE(1–214)/pTYB1. For isotopic labeling, we usually used 0.2%–0.4% labeled glucose to minimize the cost. This is especially true for triple-labeling, since 13C/2H-glucose is quite expensive. However, we found that 0.2%–0.4% glucose only produced an intermediate yield of apoE(1–214). An optimization of glucose amount indicated that 1.0% glucose significantly increased apoE(1–214) protein yield by twofold. Figure 3C shows this result of apoE(1–214)/pTYB1 expression in D2O, demonstrating that 1.0% glucose gave the highest yield of apoE(1–214) production, which was about twofold higher than the protein yield with 0.4% glucose. With these optimizations, we routinely obtain a high yield production of apoE(1–214) protein at ∼40 mg from 1 L of cell culture, ensuring a high level production of segmental labeled apoE.
Figure 3.
Optimization of apoE(1–214)/pTYB1 expression. (A) SDS-PAGE of apoE(1–214)-intein-CBD (lane 1) and apoE(1–214)-A3C-intein-CBD (lane 2) samples. M: Molecular marker. After bacterial expression, the cells were harvested by centrifugation. The cell pellet was sonicated and apoE(1–214)-intein-CBD protein was purified using chitin beads. The purified chitin beads were stored in a −20°C freezer for 48 h and then lysis for SDS-PAGE. (B) Western blot of apoE(1–214)-intein-CBD (lane 1) and apoE(1–214)-A3C-intein-CBD (lane 2) samples. (C) SDS-PAGE of the optimization of glucose usages for expression level of apoE(1–214)-intein-CBD in D2O (lanes 2–7) and H2O (lane 8). (Lane 1) Molecular marker. (Lane 2) Without IPTG, 0.4% glucose. (Lane 3) With 0.5 mM IPTG, 0. 2% glucose. (Lane 4) With 0.5 mM IPTG, 0. 4% glucose. (Lane 5) With 0.5 mM IPTG, 0. 6% glucose. (Lane 6) With 0.5 mM IPTG, 0. 8% glucose. (Lane 7) With 0.5 mM IPTG, 1.0% glucose. (Lane 8) With 0.5 mM IPTG, 1.0% glucose in H2O.
A high-level (25–30 mg/L) production of segmental, triple-labeled apoE
Using the on-column expressed protein ligation protocol (Fig. 1), we prepared several segmental, triple-labeled apoEs, including 13C/15N-apoE3(1–199)-2H-(C215–299) [apoE(1–299)-Δ(200–214)], 2H-apoE3(1–214)-13C/15N-(C215–299), and 13C/15N-apoE3(1–214)-2H-(C215–299). The segmental labeled apoE(1–299)-Δ(200–214) was prepared to study the potential role played by the hinge domain between the N- and C-terminal domains of apoE (residues 192–215) on apoE's domain–domain interaction, since our recent NMR data suggested that this hinge domain might play a key role in modulating this domain–domain interaction (Zhang et al. 2007). The latter two segmental labeled apoEs were prepared for NMR structural determination of apoE.
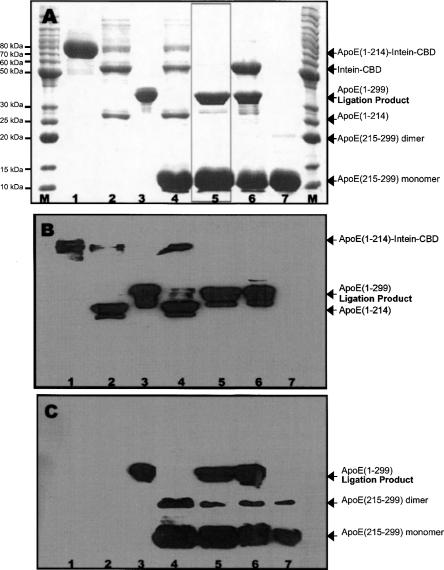
Figure 4 shows the results of native chemical ligation for preparation of segmental labeled apoE(1–299). Panel A shows SDS-PAGE of all the reactants used for ligation and the ligation reaction mixture. The reactants included apoE(1–214)-intein-CBD (∼80 kDa) on the chitin beads (lane 1) and ligation-ready apoE(C215–299)-J (10 kDa, lane 7). Lane 2 shows the apoE(1–214)-intein-CBD with 20 mM tris[2-carboxyethyl]-phosphine (TCEP), indicating the apoE(1–214)-intein-CBD (80 kDa) is readily cleaved into intein-CBD (56 kDa) and apoE(1–214) (24 kDa) in the presence of TCEP. Lane 3 is the full-length apoE-J protein (35 kDa), which serves as a control. Lanes 4–6 show ligation mixtures, including apoE(1–214)-intein-CBD on the beads, which was cleaved into intein-CBD and apoE(1–214), ligation-ready apoE(C215–299)-J, TCEP (lane 4) and thiol phenol (lanes 5,6). Lane 4 includes beads/supernatant mixture with TCEP, showing no ligation product. This suggests that no ligation reaction occurred in the ligation mixture without a thiol reagent. In contrast, lanes 5 and 6 show the supernatant only (lane 5) and supernatant/beads mixture (lane 6) at the end of ligation in the presence of thiol phenol, showing the ligation product, apoE-J (35 kDa). Lane 5 only shows two bands in the ligation supernatant: One is ligation product, apoE-J, and the other is the excess ligation-ready apoE(C215–299). Interestingly, no apoE(1–214) is observed in lane 5, suggesting that the newly produced apoE(1–214)-COR completely reacts with ligation-ready apoE(C215–299) to form ligation product. This is confirmed by lane 6, showing no band of apoE(1–214). Lanes 5 and 6 demonstrate a high ligation efficiency, since all expressed apoE(1–214) protein was converted into segmental-labeled apoE by the native ligation. Finally, lane 6 shows no band at 80 kDa, indicating that the ligation reaction is completed and all apoE(1–214)-intein-CBD reacts with ligation-ready apoE(C215–299) to produce ligation product, apoE-J. These results indicate that the reactants, both apoE(1–214)-intein-CBD and apoE(C215–299)-J, are highly reactive, and thiol reagent catalyzes native chemical ligation. The newly produced apoE(1–214)-COSR by thiol reagent immediately reacts with apoE(C215–299)-J to form ligation product, apoE(1–299)-J.
Figure 4.
The EPL results. (A) SDS-PAGE of the samples from the EPL experiment for apoE. (B) Western blot of the same samples with 1D7 (for apoE N-terminal domain). (C) Western blot of the same samples with 3H1 (for apoE C-terminal domain). M: Molecular marker. (Lane 1) Aliquot purified chitin bead (apoE[1–214]-A3C-intein-CBD, 80 kDa). (Lane 2) Aliquot purified chitin beads with TCEP. (Lane 3) ApoE3-J (35 kDa). (Lane 4) Ligation mixture (purified chitin beads, ligation-ready apoE[C215–299], and TCEP) without thiol phenol. No ligation product is observed. (Lane 5) Supernatant of the ligation mixture with thiol phenol at the end of ligation (24 h). (Lane 6) Ligation mixture with thiol phenol at the end of ligation (24 h). (Lane 7) Ligation-ready apoE(C215–299) with DTT.
To confirm this result, we carried out Western blot experiments on the same reactants and ligation mixture shown in Figure 4A using two apoE monoclonal antibodies: 1D7 (Fig. 4B) and 3H1 (Fig. 4C). The 1D7 is a monoclonal antibody that is specifically against the LDL receptor binding region of apoE in the N-terminal domain (epitope: residues 139–169) (Weisgraber et al. 1983), and 3H1 is a monoclonal antibody that is specifically against the apoE C-terminal domain (epitope: residues 243–272) (Weisgraber 1994). We anticipate that the ligation product should be recognized by both antibodies since it contains both the N- and C-terminal domains of apoE-J (35-kDa band, lane 5). In addition, 1D7 should only recognize apoE(1–214)-intein-CBD and apoE(1–214) without recognizing apoE(C215–299). Accordingly, 3H1 should only recognize apoE(C215–299)-J without recognizing apoE(1–214). As expected, Fig. 4B shows that 1D7 only recognized apoE(1–214)-intein-CBD (80-kDa band, lanes 1,2,4) and apoE(1–214) (24-kDa band, lanes 2,4), without recognition of apoE(C215–299)-J (10-kDa band, lanes 4,5,6,7). In contrast, Figure 4C shows that 3H1 did not recognize apoE(1–214)-intein-CBD (lanes 1, 2, and 4) and apoE(1–214) (lanes 2,4), but recognized apoE(C215–299) (lanes 4–7). Both antibodies were able to recognize the apoE control (lane 3) and the ligation product, segmental-labeled apoE-J (lanes 5,6). However, no antibody recognition was observed in the reactant mixture without thiol reagent for full-length apoE-J (lane 4), confirming that a thiol reagent catalyzed the reaction for native chemical ligation.
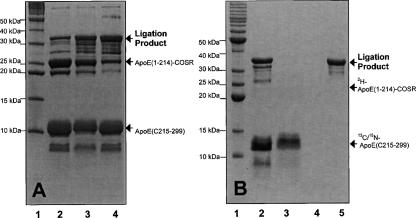
For the best ligation reaction efficiency, we carried out a time course for the ligation reaction. Our data indicated that the reaction time of 16–22 h at room temperature gave the best ligation efficiency and purity of ligation product. During the ligation reaction, we usually added fivefold excess of ligation-ready apoE(C215–299)-J, compared to apoE(1–214)-intein-CBD, to ensure completion of the ligation reaction. We observed that this would also provide excess apoE(C215–299)-J that immediately reacted with newly generated apoE(1–214)-COR; thus no remaining thiolester was left at the end of ligation. This significantly enhanced ligation efficiency and increased the final yield of ligation product. We further tested several different thiol reagents and found that thiol phenol was the best thiol reagent for native chemical ligation (Fig. 5A). In this case, we prepared apoE(1–214)-COSR using 2-mercaptoethanesulfonic acid (MESNA) first and then mixed with apoE(C215–299)-J and different thiol reagents to start ligation reaction. This way, we would have the same starting concentration of apoE(1–214)-COSR and ligation-ready apoE(C215–299) for the ligation reaction. We could also monitor an increase in the concentration of the ligation product, apoE(1–299)-J, which is accompanied with the decrease in the concentration of apoE(1–214)-COSR. As an example, Figure 5A indicated that thiol phenol gave the best ligation yield (lane 4), with the weakest band of apoE(1–214)-COSR. MESNA gave the second best ligation efficiency (lane 3), whereas DTT gave the worst ligation efficiency (lane 2). We also tested different thiol reagents on the efficiency of catalysis of intein splicing and found that these thiol reagents gave a similar efficiency of production of apoE(1–214)-COSR under the same experimental conditions. This result agrees with the previously published data (Muir et al. 1997; Muir 2003), suggesting that these thiol reagents have the same catalysis capability of intein splicing to form apoE(1–214)-COR. However, the leaving group “-R” is important to the ligation efficiency, dictating the final yield of segmental-labeled apoE-J (Muir 2003).
Figure 5.
Optimization of the EPL experiments. (A) SDS-PAGE of the optimization of different thiol reagents for the EPL of apoE. The ligation mixture contains apoE(1–214)-COSR and apoE(C215–299) and MESNA. (Lane 1) Molecular marker. (Lane 2) Ligation mixture with DTT. (Lane 3) Ligation mixture with MESNA. (Lane 4) Ligation mixture with thiol phenol. (B) Purification of ligation mixture of 2H-apoE(1–214)-13C/15N(C215–299) using a heparin Sepharose CL-6B column. (Lane 1) Molecular marker. (Lane 2) Ligation mixture before loading on the CL-6B column. (Lane 3) Flow-through. (Lane 4) Washing. (Lane 5) Elution. Only 2H-apoE(1–214)-13C/15N-(C215–299) was observed in the elution.
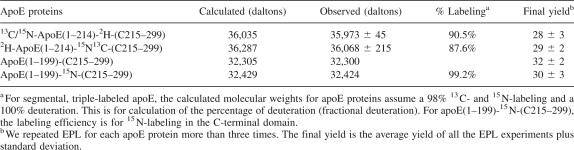
To obtain purified ligation product, we performed chromatographic purification using a heparin Sepharose CL-6B column. This purification procedure utilized the property of the positive charges in the apoE N-terminal domain, binding to heparin Sepharose CL-6B column (Fisher et al. 1997), whereas the apoE C-terminal domain did not. During the ligation reaction, the newly created apoE(1–214)-COSPh was quickly reacting with the ligation-ready apoE(C215–299)-J. The cleaved intein-CBD remained on the chitin beads and would not be released into the ligation solution. In addition, our data indicated that the expressed apoE(1–214)-intein-CBD was completely cleaved and reacted with ligation-ready apoE(C215–299)-J to form ligation product (Fig. 4). Thus, only two major proteins were in the ligation solution (Fig. 4A, lane 5): the ligation product and excess apoE(C215–299)-J. Figure 5B shows that a heparin Sepharose CL-6B column can effectively purify the ligation product, 2H-apoE(1–214)-13C/15N-(C215–299), from 13C/15N-apoE(C215–299)-J (lane 5). The other ligation products, 13C/15N-apoE(1–214)-2H-(C215–299) and 13C/15N-apoE(1–199)-2H-(C215–299), could also be purified using this method. Mass spectroscopic analysis indicated that the final purification resulted in a single mass spectroscopic peak at the expected molecular weights. For the two segmental, triple-labeled apoEs, the deuteration level of one domain is ∼90%, if we assume that the 13C/15N labeling of the other domain is 98%. Table 2 lists these mass spectroscopic results and the yields of final ligation products, indicating that our on-column expressed protein ligation protocol indeed produces a very high yield of segmental, triple-labeled apoEs at 25–30 mg from 1-L expression of apoE(1–214)/pTYB1.
Table 2.
Calculated and mass spectroscopic observed molecular weights of different apoE ligation products
NMR spectral analysis of segmental, triple-labeled apoEs
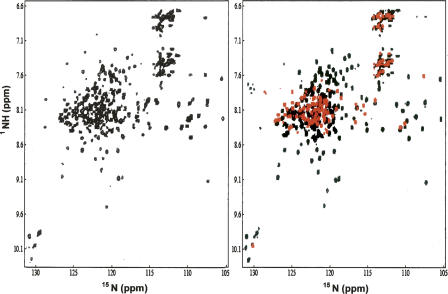
With two segmental, triple-labeled apoEs, we collected 1H-15N HSQC-TROSY spectra and compared them with that of the uniformly triple-labeled apoE. Figure 6A shows a 1H-15N HSQC-TROSY spectrum of uniformly, triple-labeled apoE, indicating a well-dispersed spectrum, even though significant spectral overlaps are observed in the center of the spectrum. This is not surprising since apoE is a 299-residue α-helical protein, which usually gives smaller backbone NH chemical shift dispersion. As a comparison, Figure 6B shows two 1H-15N HSQC-TROSY spectra plotted on top of each other. One spectrum is from an NMR sample of 13C/15N-apoE(1–214)-2H-(C215–299) (black) and the other is from a NMR sample of 2H-apoE(1–214)-13C/15N-(C215–299) (red). Since a 1H-15N HSQC-TROSY spectrum correlates 1HN with 15N atoms, whereas 1HN with 14N atoms in a protein do not produce any signals in this spectrum, the black spectrum in Fig. 6B only records 13C/15N-apoE(1–214), and 2H-apoE(C215–299) is transparent. Similarly, the red spectrum in Fig. 6B only records 13C/15N-apoE(C215–299), whereas 2H-apoE(1–214) is transparent. This way, each segmental, triple-labeled apoE gives the NMR spectrum of one individual domain in the context of full-length apoE.
Figure 6.
(Left panel) 1H-15N HSQC-TROSY of uniformly triple-labeled apoE with 15N, 13C, and 2H. (Right panel) Superposition of two 1H-15N HSQC-TROSY spectra of segmental-labeled apoE samples, including 2H-apoE(1–214)-13C/15N-(C215–299) (red) and 13C/15N-apoE(1–214)-2H-(C215–299) (black). The apoE samples are in 25 mM phosphate buffer, containing 25 mM NaCl, 5 mM EDTA, and 0.1 mM NaN3 (pH 6.9) in 5%D2O/95%H2O. The NMR data were collected on a Varian INOVA 600 MHz NMR instrument with a cyrogenic probe at 30°C.
The purpose of deuteration of one apoE domain while the other domain is doubly 13C/15N labeled is to reduce the relaxation rate and spin diffusion by the deuterated domain, while the double-labeled domain contains 100% proton. Such a labeling scheme ensures that we collect high-quality NMR data. Indeed, apoE is a 299-residue protein that requires deuteration for NMR structural studies. In addition, each apoE domain is less than 215 residues which is well within the size range of the traditional NMR structural determination of proteins using doubly 13C/15N labeled protein samples. Instead of triple labeling, we only double labeled one apoE domain while we kept the other domain deuterated on purpose, so that we have 100% proton concentration in one domain for high-quality NMR data collection. This will be especially important for the NOESY spectral collection. For NMR structural studies of large proteins, triple-labeled NMR samples are required for the recently developed TROSY technique (Pervushin et al. 1997; Salzmann et al. 1998). A triple-labeled NMR sample replaces protons that attach carbon atoms with deuteron. This causes a major problem for the NOE-based NMR structural determination method that depends on NOEs between protons. To solve this problem, a specific labeling strategy was developed by Kay's laboratory in which the protein was perdeuterated with the methyl group protonated, allowing for the collection of NOE correlations of the methyl groups in proteins (Rosen et al. 1996; Tugarinov et al. 2006). This method has been widely used to study large proteins for determination of the global folds of these proteins, due to a small number of NOE cross-peaks from methyl protons (Tugarinov and Kay 2005; Tugarinov et al. 2005). Other NMR techniques, such as site-directed spin-labeling (Battiste and Wagner 2000) and residual dipolar coupling (Bax 2003) are proposed for use in refining protein structures of large proteins, along with methyl labeling technique. The segmental labeling strategy we reported here allows us to produce segmental-labeled proteins in which one domain is protonated and the other domain is perdeuterated. This arrangement of segmental-labeled protein may solve the problem of the lack of protons in the perdeuterated protein NMR sample for large protein structural determination using modern NMR techniques.
Materials and Methods
Materials and chemicals
The thiol reagents, including DTT, β-mercaptoethanol, thiophenol, and TCEP, were purchased from Sigma. Ampicillin and kanamycin were purchased from Fisher Scientific. The pET30a vector was purchased from Novagen and pTYB vector was purchased from New England BioLabs. Factor Xa and Xarrest were purchased from New England BioLabs. Heparin Sepharose CL-6B was purchased from GE Healthcare. Isotopes, including 13C-glucose, 15NH4Cl, and D2O, were purchased from Sigma. Vitamin mixture was purchased from Sigma.
Molecular cloning
For apoE(1–214)/pTYB1 and apoE(1–199)/pTYB1 DNA constructs, we inserted apoE(1–199) and apoE(1–214) DNAs into the pTYB1 vector using NdeI and SapI enzymes in the multiple cloning sites of pTYB1 vector; thus, the intein fusion was in the C terminus of apoE(1–199) and apoE(1–214). For apoE(C215–299)/pET30a DNA construct, we inserted apoE(C215–299) DNA into the pET30a vector using NcoI and HindIII enzymes in the multiple cloning sites of pET30a vector. During the subcloning procedure, we introduced a Factor Xa site between the long his-tag and apoE(C215–299) for apoE(C215–299)/pET30a. The expression vectors were transformed into BL-21(DE3) bacterial strains.
Bacterial expression and protein purification
For apoE(1–214)/pTYB1 and apoE(1–199)/pTYB1, the expression started with LB medium. Glycerol stock was made after double colony selection and was added into 300 mL LB with 1 mM ampicillin. The expression grew at 37°C until OD600 reached 2.5–3.0. The bacteria were gently spun down and transferred into 300 mL M9 medium that contained 1% glucose, 1/100 × Vitamin, pH 8.0. The expression was carried out at 20°C for 1.5 h, and 0.5 mM IPTG was then added to induce protein expression. The protein expression was further carried out at 20°C for another 22 h before harvesting the cells. With this expression method, the final OD600 before harvest usually reached 7–9, ensuring a high-level production of apoE(1–214) and apoE(1–199). The expression of apoE(C215–299)/pET30a essentially followed the same method, except kanamycin replaced ampicillin. In addition, in the M9 medium, we added 0.4% 13C-glucose for double labeling and 1% unlabeled glucose for unlabeled apoE(C215–299). Protein purifications essentially followed both pET30a and pTYB manuals. Specifically, for a 500-mL expression of apoE(C215–299)/pET30a, the cell pellet was dissolved into 50 mL binding buffer and sonicated three times. The combined supernatants were loaded on a His • Bind resin column at room temperature and washed with 250 mL binding buffer first and 400 mL washing buffer containing 30 mM imidazole. The protein was eluted down from the column with 200 mL elution buffer that contained 1 M imidazole. The elution was dialyzed against water containing 20 mM ammonium bicarbonate and lyophilized. With this method, we routinely produced 150–200 mg apoE(C215–299) from a 500-mL cell culture. For a 300-mL expression of apoE(1–199)/pTYB1 and apoE(1–214)/pTYB1, the cell pellet was dissolved into 50 mL binding buffer and sonicated three times. The combined supernatants were loaded on a Chitin Bead column in a cold room and washed with 500 mL binding buffer. The purified chitin beads that bind either apoE(1–199)-intein-CBD or apoE(1–214)-intein-CBD were ready for ligation.
Factor Xa cleavage to remove his-tag from apoE(C215–299)
A sample of his-tag-ApoE(C215–299) at 5 mg/mL was prepared using Factor Xa cleavage buffer containing 5 mM β-mercaptoethanol. Factor Xa was then added at a ratio of 1000 μg: 1 unit = his-tag-apoE(C215–299): Factor Xa. The reaction was carried out at room temperature for 1 h and stopped. For a 100-mg apoE(C215–299)/Factor Xa reaction, Factor Xa was removed using Xarrest agarose and uncut protein and his-tag were removed by passing through a 10 mL His • Bind column. The column was washed using 50 mL binding buffer twice. All flow-through and washing were combined and dialyzed against 20 mM ammonium bicarbonate and lyophilized. The lyophilized powder is ligation-ready apoE(C215–299).
Site-directed mutagenesis
Site-directed mutagenesis was carried out using the QuikChange mutagenesis kit from STRATAGENE. Primers containing the desired mutations were annealed to the denatured DNA expression vector harboring human apoE gene, which was then extended using PfuTurbo DNA polymerase to generate nicked, circular strands. The methylated, nonmutated parental DNA was digested using DpnI. The circular, nicked, double-stranded DNA containing the mutation was then transformed into ER2566 cells. The mutations were confirmed by DNA sequencing.
On-column native chemical ligation
Typically, with 300 mL expression of apoE(1–214)/pTYB1, we used a 6-mL chitin bead column to bind and purify apoE(1–214)-intein-CBD protein. The sonication supernatants were combined and passed through the chitin bead column twice to ensure that all apoE protein bound to the column. The column was washed with 500 mL binding buffer (20 mM Tris-HCl, 250 mM NaCl, 1 mM EDTA, pH 8.0) in a cold room. The ligation-ready chitin beads were then transferred from the column into a small bottle and the supernatant was carefully removed. In addition, 40 mg ligation-ready apoE(C215–299) powder was dissolved into 2 mL (20 mg/mL) ligation buffer, containing 20 mM Tris-HCl, 250 mM NaCl, 1 mM EDTA, 0.15 M TCEP, pH 8.0, and mixed well with 6 mL chitin beads. The ligation reaction was initiated by addition of 240 μL of thiol phenol. The ligation reaction was maintained at room temperature for 20–24 h with a gentle stir. At 20–24 h, an aliquot of the chitin bead mixture was taken out, mixed with 4 × SDS loading buffer, and loaded on SDS-PAGE to ensure that the ligation reaction was complete. Once the ligation reaction was complete, the ligation mixture was gently spun down and the chitin beads were washed with 5 × 5 ml of loading buffer. All supernatants were combined and mixed with 500 μL of new chitin beads to remove any intein-CBD in the supernatant. The supernatant was diluted to 100 mL with loading buffer (25 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and then loaded on a Heparin Sepharose CL-6B column. The CL-6B column was washed with 100 mL loading buffer and then eluted with elution buffer containing different NaCl concentrations (20 mL 175 mM NaCl, 20 mL 200 mM NaCl, 10 mL 225 mM NaCl, 10 mL 250 mM NaCl, 5 mL 300 mM NaCl). A 12% SDS-PAGE was carried out to check all the fractions of purification. The fractions that contained purified ligation product were all pooled together and dialyzed against water containing 20 mM ammonium bicarbonate and then lyophilized.
NMR methods
The NMR samples contain 0.3–1.0 mM uniformly triple-labeled apoE and segmentally labeled apoE proteins, in a buffer of 25 mM sodium phosphate, 25 mM NaCl, 5 mM EDTA, and 0.1 mM NaN3, pH 6.9 in 95%H2O/5%D2O. The chemical shift was referenced using DSS. All NMR experiments were performed at 30°C on a Vavian INOVA 600-MHz spectrometer equipped with a cyrogenic probe. The 2D 1H-15N HSQC-TROSY spectra were collected using a sensitivity-enhanced mode with 1024 points at the proton dimension and 256 complex points at the 15N dimension. The acquisition times for both dimensions were 64 ms. The NMR data processing was achieved using nmrPipe and nmrDraw software (Delaglio et al. 1995) and analyzed with PIPP.
Acknowledgments
This work was supported by a RO1 grant from the NIH (HL74365 to J.W.), a grant from the American Health Assistant Foundation (J.W.), and a start-up grant provided by Wayne State University. We thank Victoria Murray for critical reading of the manuscript.
Footnotes
Reprint requests to: Qianqian Li, Department of Biochemistry and Molecular Biology, School of Medicine, Wayne State University, Detroit, MI 48201, USA; e-mail: qil@med.wayne.edu; fax: (313) 577-8836; or Jianjun Wang, School of Medicine, Wayne State University, Room 5113, Scott Hall, 540 East Canfield Avenue, Detroit, MI 48201, USA; e-mail: jjwang@med.wayne.edu; fax: (313) 577-8836.
Abbreviations: apoE, apolipoprotein E; apoEC-J, the monomeric mutant of apoE C-terminal domain; apoE-J, the monomeric apoE mutant; CBD, chitin-binding domain; DSS, 2,2-dimethyl-2-silapentane5-sulfonic acid; DTT, dithiothreitol; EDTA, ethylene-diamine-tetra-acetic acid; EPL, expressed protein ligation; HSQC, heteronuclear single quantum coherence; IPTG, isopropyl-beta-D-thiogalactopyranoside; LDL, low-density lipoprotein; MESNA, 2-mercaptoethanesulfonic acid; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser enhancement spectroscopy; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TCEP, tris[2-carboxyethyl]-phosphine; TROSY, transverse relaxation-optimized spectroscopy.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073383708.
References
- Anderson, L.L., Marshall, G.R., Crocker, E., Smith, S.O., Baranski, T.J. Motion of carboxyl terminus of Gα is restricted upon G protein activation. A solution NMR study using semisynthetic Gα subunits. J. Biol. Chem. 2005;280:31019–31026. doi: 10.1074/jbc.M503690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers, B., Blaschke, U.K., Camarero, J.A., Cotton, G.J., Holford, M., Muir, T.W. Introduction of unnatural amino acids into proteins using expressed protein ligation. Biopolymers. 1999;51:343–354. doi: 10.1002/(SICI)1097-0282(1999)51:5<343::AID-BIP4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bax, A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste, J.L., Wagner, G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- Camarero, J.A., Shekhtman, A., Campbell, E.A., Chlenov, M., Gruber, T.M., Bryant, D.A., Darst, S.A., Cowburn, D., Muir, T.W. Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, S., Mersha, F.B., Comb, D.G., Scott, M.E., Landry, D., Vence, L.M., Perler, F.B., Benner, J., Kucera, R.B., Hirvonen, C.A., et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- Chong, S., Williams, K.S., Wotkowicz, C., Xu, M.Q. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J. Biol. Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- Cotton, G.J., Muir, T.W. Generation of a dual fluorescence biosensor for Crk-II phosphoryltion using solid-phase expressed protein ligation. Chem. Biol. 2000;7:253–261. doi: 10.1016/s1074-5521(00)00100-9. [DOI] [PubMed] [Google Scholar]
- Cowburn, D., Shekhtman, A., Xu, R., Ottesen, J.J., Muir, T.W. Segmental isotopic labeling for structural biological applications of NMR. Methods Mol. Biol. 2004;278:47–56. doi: 10.1385/1-59259-809-9:047. [DOI] [PubMed] [Google Scholar]
- Cui, C., Zhao, W., Chen, J., Wang, J., Li, Q. Elimination of in vivo cleavage between target protein and intein in the intein mediated protein purification systems. Protein Expr. Purif. 2006;50:74–81. doi: 10.1016/j.pep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRpipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Fan, D., Li, Q., Korando, L., Jerome, W.G., Wang, J. A monomeric human apolipoprotein E carboxyl-terminal domain. Biochemistry. 2004;43:5055–5064. doi: 10.1021/bi035958w. [DOI] [PubMed] [Google Scholar]
- Fisher, C.A., Wang, J., Francis, G.A., Sykes, B.D., Kay, C.M., Ryan, R.O. Bacterial overexpression, isotope enrichment, and NMR analysis of the N-terminal domain of human apolipoprotein E. Biochem. Cell Biol. 1997;75:45–53. [PubMed] [Google Scholar]
- Hatters, D.M., Peters-Libeu, C.A., Weisgraber, K.H. Apolipoprotein E structure: Insights into function. Trends Biochem. Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Weisgraber, K.H., Mucke, L., Mahley, R.W. Apolipoprotein E: Diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J. Mol. Neurosci. 2004;23:189–204. doi: 10.1385/JMN:23:3:189. [DOI] [PubMed] [Google Scholar]
- Maag, D., Fekete, C.A., Grycznski, Z., Lorsch, J.R. A conformational change in the eukaryotic translation preinhibition complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;20:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Muir, T.W. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Muir, T.W., Dawson, P.E., Kent, S.B. Protein synthesis by chemical ligation of unprotected peptides in aqueous solution. Methods Enzymol. 1997;289:266–298. doi: 10.1016/s0076-6879(97)89052-0. [DOI] [PubMed] [Google Scholar]
- Muir, T.W., Sondhi, D., Cole, P.A. Expressed protein ligation: A general method for protein engineering. Proc. Natl. Acad. Sci. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, V., Muir, T.W. Protein ligation: An enabling technology for the biophysical analysis of proteins. Nat. Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- Pervushin, K., Riek, R., Wider, G., Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, M.K., Gardner, K.H., Willis, R.C., Parris, W.E., Pawson, T., Kay, L.E. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- Salzmann, M., Pervushin, K., Wider, G., Senn, H., Wuthrich, K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibner, K.A., Zhang, Z., Cole, P.A. Merging fluorescence resonance energy transfer and expressed protein ligation to analyze protein–protein interactions. Anal. Biochem. 2003;317:226–232. doi: 10.1016/s0003-2697(03)00087-3. [DOI] [PubMed] [Google Scholar]
- Schwarzer, D., Cole, P.A. Protein semisynthesis and expressed protein ligation: Chasing a protein tail. Curr. Opin. Chem. Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Sydor, J.R., Mariano, M., Sideris, S., Nock, S. Establishment of intein-mediated protein ligation under denaturing conditions: C-terminal labeling of a single-chain antibody for biochip screening. Bioconjug. Chem. 2002;13:707–712. doi: 10.1021/bc025534z. [DOI] [PubMed] [Google Scholar]
- Thompson, P.R., Wang, D., Wang, L., Fulco, M., Pediconi, N., Zhang, D., An, W., Ge, Q., Roeder, R.G., Wong, J., et al. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Tugarinov, V., Kay, L.E. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- Tugarinov, V., Choy, W.Y., Orekhov, V.Y., Kay, L.E. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc. Natl. Acad. Sci. 2005;102:622–627. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov, V., Kanelis, V., Kay, L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- Vitali, F., Henning, A., Oberstrass, F.C., Hargous, Y., Auweter, S.D., Erat, M., Allain, F.H. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Cole, P.A. Protein tyrosine kinase Csk-catalyzed phosphorylation of Src containing unnatural tyrosine analogues. J. Am. Chem. Soc. 2001;123:8883–8886. doi: 10.1021/ja010540b. [DOI] [PubMed] [Google Scholar]
- Weisgraber, K.H. Apolipoprotein E: Structure–function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- Weisgraber, K.H., Innerarity, T.L., Harder, K.J., Mahley, R.W., Milne, R.W., Marcel, Y.L., Sparrow, J.T. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J. Biol. Chem. 1983;258:12348–12354. [PubMed] [Google Scholar]
- Welker, E., Scheraga, H.A. Use of benzyl mercaptan for direct preparation of long polypeptide benzylthio esters as substrates of subtiligase. Biochem. Biophys. Res. Commun. 1999;254:147–151. doi: 10.1006/bbrc.1998.9913. [DOI] [PubMed] [Google Scholar]
- Wetterau, J.R., Aggerbeck, L.P., Rall S.C., Jr, Weisgraber, K.H. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- Wilson, C., Wardell, M.R., Weisgraber, K.H., Mahley, R.W., Agard, D.A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- Xu, M.Q., Evans T.C., Jr Intein-mediated ligation and cyclization of expressed proteins. Methods. 2001;24:257–277. doi: 10.1006/meth.2001.1187. [DOI] [PubMed] [Google Scholar]
- Xu, R., Ayers, B., Cowburn, D., Muir, T.W. Chemical ligation of folded recombinant proteins: Segmental isotopic labeling of domains for NMR studies. Proc. Natl. Acad. Sci. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi, H., Tsujimoto, T., Yamazaki, T., Yoshida, M., Akutsu, H. Conformational change of H+-ATPase β monomer revealed on segmental isotope labeling NMR spectroscopy. J. Am. Chem. Soc. 2004;126:16632–16638. doi: 10.1021/ja045279o. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Zhang, Z., Ganguly, S., Weller, J.L., Klein, D.C., Cole, P.A. Cellular stabilization of the mechanism rhythm enzyme induced by nonhydrolyzable phosphonate incorporation. Nat. Struct. Biol. 2003;10:1054–1057. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Vasudevan, S., Sojitrawala, R., Zhao, W., Cui, C., Xu, C., Fan, D., Newhouse, Y., Balestra, R., Jerome, W.G., et al. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]