Abstract
The human tissue kallikrein (KLK) family contains 15 secreted serine proteases that are expressed in a wide range of tissues and have been implicated in different physiological functions and disease states. Of these, KLK1 has been shown to be involved in the regulation of multiple physiological processes such as blood pressure, smooth muscle contraction, and vascular cell growth. KLK6 is overexpressed in breast and ovarian cancer tissues and has been shown to cleave peptide derived from human myelin protein and Aβ amyloid peptide in vitro. Here we analyzed the substrate specificity of KLK1 and KLK6, by substrate phage display using a random octapeptide library. Consistent with earlier biochemical data, KLK1 was shown to exhibit both trypsin- and chymotrypsin-like selectivities with Tyr/Arg preferred at site P1, Ser/Arg strongly preferred at P1′, and Phe/Leu at P2. KLK6 displayed trypsin-like activity, with the P1 position occupied only by Arg and a strong preference for Ser in P1′. Docking simulations of consensus peptide provide information on the identity of the enzyme residues that are responsible for substrate binding. Bioinformatic analysis suggested several putative KLK6 protein substrates, such as ionotropic glutamate receptor (GluR) and synphilin.
Keywords: human kallikrein, phage display, substrate specificity, molecular modeling
The human tissue kallikrein locus comprises 15 structurally similar genes which represent the largest contiguous cluster of serine protease genes within the human genome. Human tissue kallikreins (KLKs) are expressed in a wide range of tissues and have been implicated in many physiological and pathological functions (Diamandis et al. 2000; Yousef and Diamandis 2001; Paliouras and Diamandis 2006). KLK1 was the first kallikrein to be discovered and was detected at high levels in the human pancreas. Together with KLK2 and KLK3, they constitute the classical kallikreins. The primary physiological function of KLK1 is the cleavage of low molecular weight kininogen at Arg–Ser and Met–Lys bonds to release lysyl-bradykinin (kallidin) (Sueiras-Diaz et al. 1994). Kallidin binding to its receptors B1 and B2 leads to a signaling cascade that regulates multiple physiological processes, such as blood pressure, smooth muscle contraction, and vascular cell growth (Bhoola et al. 1992; Borgono et al. 2004). In addition to its primary function in the release of kallidin, KLK1 has also been shown to cleave kallistatin, somatostatin, pro-insulin, low-density lipoprotein, the precursor of atrial natriuretic factor, prorenin, vasoactive intestinal peptide, procollagenase, and angiotensinogen (Bhoola et al. 1992; Yousef and Diamandis 2001). It has been suggested that KLK1 exhibits different physiological functions in different tissues, but it is not known how its activity is regulated. The enzyme was shown to exhibit both trypsin- and chymotrypsin-like activities. Its trypsin-like activity is manifest in the cleavage of low molecular weight kininogen at Arg–Ser (Fiedler and Leysath 1979), while the chymotrypsin-like activity is evident in the cleavage of kallistatin (human kallikrein-binding protein) and somatostatin, after a Phe–Phe dipeptide (Zhou et al. 1992; Pimenta et al. 1997).
KLK6 was called zyme, protease M, neurosin, or PRSS9 prior to its current nomenclature (Little et al. 1997; Yamashiro et al. 1997; Yousef et al. 1999). Though the physiological function of KLK6 is not clear, KLK6 was reported as myelencephalon-specific protease, which may play a key role in the regulation of myelin turnover and in demyelinating disease (Scarisbrick et al. 1997). Many proteins have been shown to be cleaved by KLK6 in vitro, including human myelin basic protein (MBP), Aβ amyloid peptide, plasminogen, myelin, and α-synuclein. Notably, KLK6 has been implicated in neurodegenerative diseases such as Alzheimer's, multiple sclerosis, and Parkinson's (Bernett et al. 2002; Iwata et al. 2003; Magklara et al. 2003). Furthermore, the KLK6 gene has been shown to be up-regulated in ovarian cancer, and therefore KLK6 constitutes a potential serum biomarker for diagnosis and prognosis of ovarian cancer (Diamandis et al. 2003). Also, its expression is up-regulated in primary breast tumors and down-regulated at metastatic breast cancer sites. Since KLK6 has been shown to cleave extracellular matrix peptides derived from laminin and fibronectin, it may be involved in tissue remodeling (Anisowicz et al. 1996; Yousef et al. 2003; Ghosh et al. 2004). Finally, in a recent study, Oikonomopoulou et al. (2006) reported that KLK6 cleaves protease-activated receptors (PARs), a family of G-coupled protein receptors that are activated via an N-terminal cleavage proteolytic event.
Due to its physiological significance, the biochemical properties and substrate specificity of KLK6 have attracted significant attention. Crystal structures for both proteins are available (Bernett et al. 2002; Laxmikanthan et al. 2005). Even though the physiological role of KLK1 has been studied in detail, relatively little is known about its extended substrate specificity. Fluorogenic peptides derived from a preferred KLK1 substrate indicated that Arg, Lys, Phe, Tyr, and Met are accepted in the S1 subsite, Ser is strongly preferred in S1′ which can also accommodate Arg to a lesser extent, and finally, Phe and Leu are preferred in S2 (Pimenta et al. 1997). The substrate specificity of KLK6 has been investigated by positional scanning using two different approaches: Debela et al. (2006) employed peptide substrates of the general structure acetyl-P4-P3-P2-P1–7-aminomethylcoumarin and reported that KLK6 exhibits strong preference for Arg or Lys in S2 whereas the S1 subsite accepts not only Arg but also Lys, Ala, or Met. These results were in conflict with the data of Angelo et al. (2006) who examined the substrate preference of KLK6 using the same set of fluorescence resonance energy transfer (FRET) peptides that had been used for the analysis of the specificity of KLK1 mentioned above. Specifically, Angelo et al. reported that only Arg is accepted in the S1 subsite of KLK6 whereas S2 strongly prefers Phe and, to a lesser extent Leu, over all other amino acids (Angelo et al. 2006).
The determination of protease specificity using positional libraries of FRET or fluorogenic peptide substrates suffers from two drawbacks. First, interactions between the fluorescent dyes and the enzymes can bias substrate preference. This can be particularly true when the dye occupies the P1′ position, as is the case with the 7-aminomethylcoumarin substrates used by Debela et al. (2006). Second, positional scanning methods cannot take into account neighboring effects whereby the occupancy of one subsite by a particular amino acid affects the preference of the adjacent subsites.
Substrate phage display provides a means for discerning the extended amino acid specificity of proteases without the need for chemical labeling (Deperthes 2002; Diamond 2007). It relies on the selective cleavage of specific peptide sequences sandwiched between the gene pIII minor coat protein of fd bacteriophage and an affinity tag. The phage is immobilized on a solid support via the affinity tag, and, following treatment with a purified protease of interest, clones containing susceptible peptide sequences are cleaved, which in turn releases them from the support, allowing amplification. Consensus substrate sequences are isolated after several rounds of panning. The substrate specificity of over 20 bacterial and mammalian proteases including KLK2, KLK3, and KLK14 have been analyzed by substrate phage (Cloutier et al. 2002; Felber et al. 2005; Ferrieu-Weisbuch et al. 2006). In an effort to address discrepancies in earlier studies of KLK1 and KLK6 fine specificity, here we used substrate phage with a random octapeptide library. Cleavage and enrichment were performed in buffers with physiological pH and salt concentrations. Selected peptides were synthesized, their respective cleavage sites were determined by MS, and the kinetics of hydrolysis were determined. KLK1 showed its dual activity, both trypsin- and chymotrypsin-like, while KLK6 only showed trypsin-like activity with strong preference of Arg at P1 site. Both KLK1 and KLK6 efficiently cleaved the synthesized peptides which were derived from phage panning sequences. Docking studies with selected peptides were employed to provide information on the KLK1 and KLK6 residues that form the S1 and S1′ subsites and interact with side chains on the substrate.
Results
Analysis of the specificity of KLK6
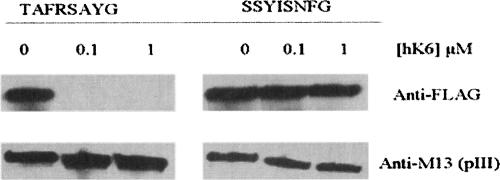
We first examined whether phage encoding a putative KLK6 sequence inserted between the pIII protein and the Flag peptide epitope can be recognized and cleaved following incubation with protease. Phages containing the octapeptide sequence TAFRSAYG from the panning and SSYISNYG which contains no putative cleavage sites, were constructed, purified, and incubated with 0, 0.1, or 1 μM KLK6 at 37°C for 45 min. Cleavage of the phage was then analyzed by Western blotting using anti-Flag and anti-pIII antisera. The TAFRSAYG sequence was efficiently cleaved under these conditions, resulting in loss of the Flag tag. In contrast, phage encoding SSYISNYG remained intact (Fig. 1). Thus, KLK6 does not cleave pIII nor does it affect the integrity of the phage, but it can recognize substrate sequences inserted between pIII and the Flag epitope. Similarly, KLK1 does not cleave either pIII or the intact phage (data not shown).
Figure 1.
Cleavage of putative substrate phage and non-substrate phage by KLK6 by Western blotting with anti-Flag and anti-pIII. Purified phage was incubated with 0, 0.1, or 1 μM of KLK6 in 50 mM Tris-HCl, 150 mM NaCl containing 1 mM EDTA (pH 7.5) at 37°C for 45 min, and analyzed by Western blotting. Phage containing the TAFRSAYG octapeptide is a substrate for KLK6 whereas, phage containing the SSYISNFG sequence is not.
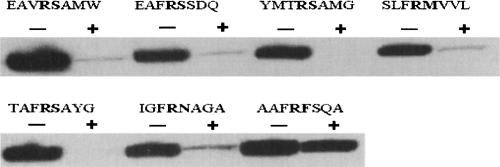
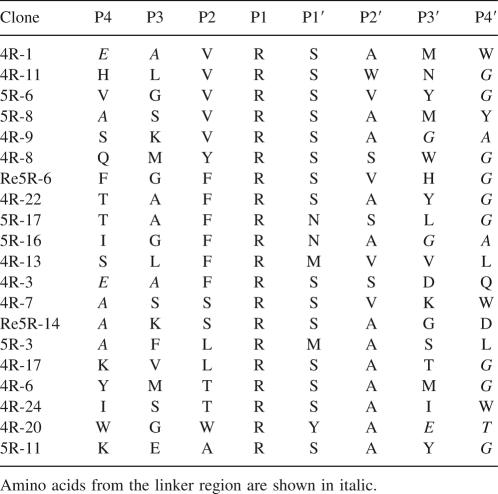
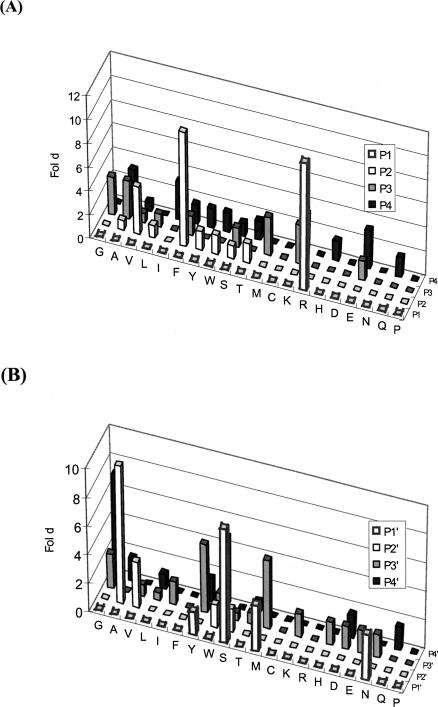
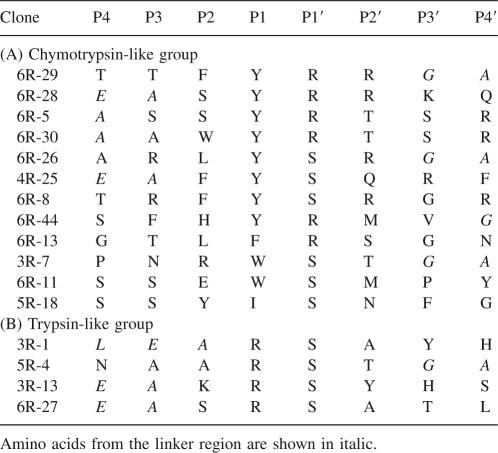
A random octapeptide library was constructed as described earlier (Hwang et al. 2007). The diversity of the library was 1.1 × 108 cfu (colony forming unit), and therefore it encoded only a small fraction of the possible 8-mer amino acid sequences (2.56 × 1010); nonetheless, it should be sufficient to capture the key properties of amino acids preferred by the protease (Kerr et al. 2005). The phage library was immobilized on anti-Flag conjugated beads and subjected to panning with purified recombinant KLK1 or KLK6. Cleaved phage clones were amplified in E. coli K91BK and used as input in subsequent panning experiments. After each round of panning, phage clones were picked at random and sequenced. The sequencing results of KLK6 panning revealed that six out of 12 clones in the third round contained sequences encoding an Arg–Ser dipeptide sequence while 10 out of 12 clones in the fourth round contained Arg–Ser dipeptide consistent with the expected cleavage of this sequence. Individual isolated clones from the third to the fifth rounds were amplified; phage was purified and subjected to digestion by KLK6, and loss of the Flag epitope was examined by Western blotting. For comparison, we also constructed phage in which the random octapeptide in fUSE55 was replaced with the sequence encoding AAFRFSQA. This peptide had been reported to exhibit the maximum rate of cleavage by KLK6 in vitro (Angelo et al. 2006). Six phage clones isolated after five rounds showed a greater degree of cleavage relative to phage containing the AAFRFSQA peptide (Fig. 2). All 20 isolated phage clones examined showed KLK6-specific cleavage. The selected sequences that were cleaved by KLK6 are shown in Table 1. Consistent with the expected cleavage preference of the enzyme, the sequences were aligned so that Arg is positioned in the P1 site. Figure 3 shows a histogram of the observed amino acid frequencies between sites P4 and P4′ normalized relative to the codon usage in the respective NNK library position. Only four different amino acids were found in the P1′ position with a strong preference for Ser (eightfold greater than the frequency of Ser that would be expected based on the NNK randomization scheme). We also observed an increased frequency of Val or Phe at the P2 position (four- and 9.6-fold, respectively), whereas Ala is strongly preferred at the P2′ position (9.6-fold enrichment over background). KLK6 appears to accept a variety of amino acids at P3, P4, P3′, and P4′ positions. Residues derived from the linker sequence in the octapeptide library showed up at P3, P4 or P3′, and P4′ and thus do not affect the significance of the analysis described above. All Gly on P4′ are from linker, which is not considered as significant.
Figure 2.
Cleavage of selected phage clones by KLK6. Purified phages were incubated with 0 or 0.1 μM KLK6 in 50 mM Tris-HCl, 150 mM NaCl containing 1 mM EDTA (pH 7.5) at 37°C for 45 min. The reaction mixture was separated with 4%–20% gradient SDS-PAGE and then analyzed with anti-Flag antibody by Western blotting. The label “+” or “−” indicates with or without enzyme, respectively. The octapeptide sequences are shown above the respective band. AAFRFSQA has been reported to exhibit the maximum rate of cleavage by KLK6 in vitro (Angelo et al. 2006).
Table 1.
Alignment of sequences selected by substrate phage display with KLK6
Figure 3.
Normalized frequency of occurrence of amino acids at P1–P4 (A) and P1′–P4′ (B) of KLK6 substrates. Data represent the observed amino acid frequencies between P4 to P4′ normalized relative to the codon usage in the respective NNK library position. The Y-axis shows the ratio of the observed frequency to the theoretical frequency of the respective amino acids in the NNK library.
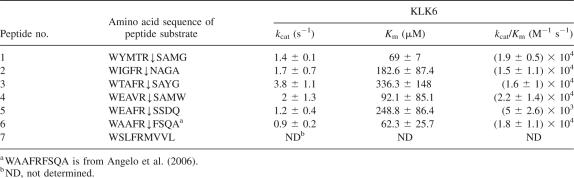
Six peptides derived from selected phages were synthesized by solid-phase synthesis. Each peptide possessed an N-terminal Trp to facilitate detection of the intact peptide and the hydrolysis product by monitoring absorbance at 280 nm. Initial rates of hydrolysis determined by measuring product formation relative to the initial substrate concentration via HPLC, and the kinetic parameters k cat and K m were evaluated and fitted by nonlinear regression to the Michaelis-Menten equation (Table 2). Six/seven peptides were cleaved by KLK6 with k cat/K m values between 5 ± 2.6 × 103 M−1s−1 and 2.2 ± 1.4 × 104 M−1s−1. ESI/MS analysis revealed that all peptides were cleaved at a single site following the Arg residue. No secondary cleavage products could be detected. Even though phage encoding SLFRMVVL was efficiently cleaved by KLK6, the corresponding synthetic peptide showed very little cleavage even after 4 h of incubation with 100 μM KLK6. The lack of cleavage of WSLFRMVVL was at least partly due to the lower solubility of this peptide relative to the other sequences tested. The Flag sequence in the phage encoding SLFRMVVL may have affected the solubility so that it was cleaved by the enzyme efficiently.
Table 2.
Kinetic measurements of KLK6 catalyzed hydrolysis of substrate
Analysis of the specificity of KLK1
Phage containing octapeptide sequences that are cleaved by KLK1 were isolated after six rounds of panning. Individual phage clones were amplified and examined by Western blotting for loss of the Flag tag following incubation with 4 μM KLK1 for 45 min. Sixteen clones that showed extensive cleavage in this assay were examined further. Peptides were aligned such that either Arg or Phe and other hydrophobic amino acids occupied the P1 position, in accord with the published information on the substrate specificity of KLK1 (for review, see Paliouras and Diamandis 2006). As expected, KLK1 showed both trypsin- and chymotrypsin-like specificity (Table 3). Twelve of 16 phage clones encoded chymotrypsin-like substrate sequences, having large hydrophobic amino acids N-terminal at P1. Four out 16 phage clones encoded trypsin-like substrates containing an Arg at the P1 position. The P2′ position favored polar or charged amino acids. No specific bias for particular amino acids could be discerned for the P2–P4 and P3′, P4′ positions.
Table 3.
Alignment of sequences selected by substrate phage display with KLK1
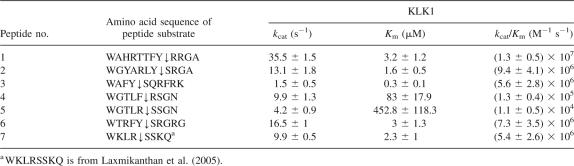
Seven peptides corresponding to selected sequences were synthesized. ESI/MS of the KLK1 hydrolysis products revealed in all cases that a single cleavage occurred N-terminal to Arg or Ser, consistent with the assignment of these amino acids to the P1′ position. WAHRTTFYRRGA was found to be the best substrate with a k cat/K m of 1.3 ± 0.5 × 107 (Table 4).
Table 4.
Kinetic measurements of KLK1 catalyzed hydrolysis of substrate peptides
Molecular modeling of peptide substrates bound to KLK6 or KLK1
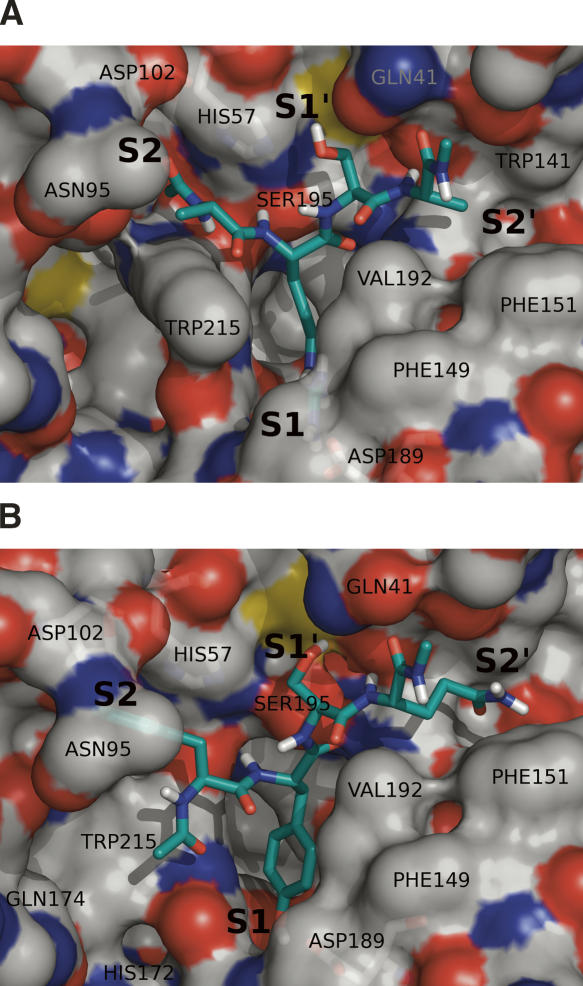
Molecular docking of the consensus peptide substrates onto the crystal structure of KLK1 or KLK6 was performed using GOLD v3.1.1 For KLK6, the cavity was defined with a radius of 20 Å centered around the OH group of Ser195. Five acetylated peptides were tested (FRSA, FRSV, VRSA, VRSV, and FRFSQ), and a total of 200 genetic algorithm (GA) simulations were performed on each peptide. Among the top five candidates from the docking calculations, Arg is shown to predominantly occupy the S1 pocket (Fig. 4), consistent with the biochemical data. The Arg guanidinium group is capable of making a number of hydrogen bonds with protein residues Asp189, Ser190, and Asn217 in the S1 pocket and with Ser214, Thr229, Asp102, and Tyr94 in the S2 pocket. The catalytic Ser195 residue makes hydrogen bond contacts to the backbone of the peptide at P1–P1′. C-terminal residues show consistent binding to the S2′ hydrophobic pocket comprising residues Gly193, Leu40, Leu41, Gly142, Trp141, and Phe151. When Phe precedes Arg, as in FRSA and FRSV, the docking also predicts among the top candidates' binding modes where Phe may reside in S1 and Arg in S2. This is likely an artifact of the scoring function of GOLD, which utilizes approximated functions describing hydrophobic forces.
Figure 4.
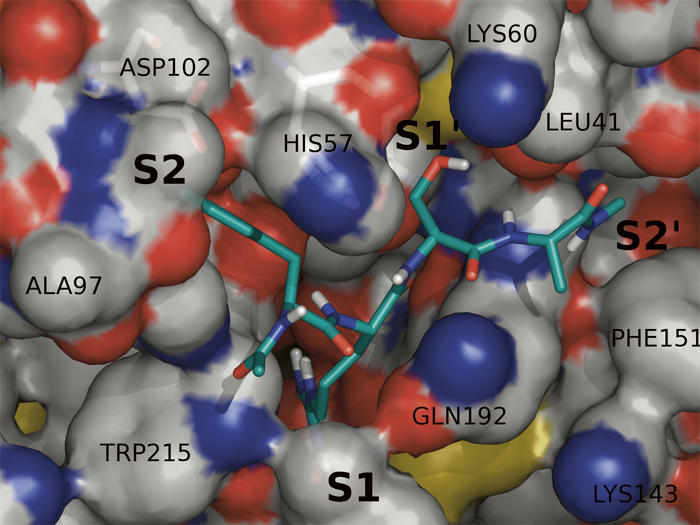
Proposed substrate binding around the active-site binding pocket of KLK6 by docking simulation using GOLD v3.1.1. The peptide FRSA exists in the active-site binding pocket of KLK6. Arg is capable of making hydrogen bonds with protein residues Asp189, Ser190, and Asn217 which compose the S1 binding pocket.
For KLK1, seven peptides were tested: FYRR, LYSR, ARSA, SRSA, KRSY, FYSQ, and FYSR. The predominant binding mode for peptides with Arg at P1 (trypsin-like substrate) into the KLK1 cavity is with Arg in S1 (Fig. 5A). When Arg is not present at P1 (chymotrypsin-like substrate), Tyr is found in S1 almost exclusively, making a hydrogen bond with ASP189 (Fig. 5B). For FYRR, the docking calculations show that either Tyr or the first Arg can occupy in the S1 pocket, indicating that the benzene ring competes with guanidinium for the S1 pocket. As KLK1 possesses both trypsin and chymotrypsin characteristics, the S1 pocket is able to recognize both Tyr and Arg.
Figure 5.
Proposed substrate binding around the active-site binding pocket of KLK1 by docking simulation using GOLD v3.1.1. (A) Peptide ARSA (trypsin-like substrate) and (B) peptide FYSQ (chymotrypsin-like substrate) bind the active-site binding pocket of KLK1.
Discussion
In this study we investigated the substrate specificity of kallikreins 1 and 6 using substrate phage display. The advantages of the substrate phage method over methods that rely on cleavage of fluorescently labeled synthetic substrate libraries have been reviewed recently (Sedlacek and Chen 2005). However, one potential problem with this technique is that clones that are cleaved by endogenous proteases either during phage biogenesis or upon phage purification become enriched together with sequences that are cleaved following incubation by the target protease. To avoid such artifacts, the cleavage of all isolated phage clones by either KLK1 or KLK6, as appropriate, was examined by Western blotting following incubation with or without target protease. In this manner we identified a total of 16 and 20 unique sequences cleaved by KLK1 and KLK6, respectively. Peptides corresponding to sequences isolated by substrate phage were synthesized. N-terminal tryptophan was added to provide a chromophore which allowed the kinetics of cleavage to be determined by HPLC. The products of cleavage were analyzed by ESI/MS. Synthetic peptides based on isolated sequences were found to be cleaved efficiently, further confirming that they constitute true substrates. Meanwhile, the high-efficiency cleavage of peptide substrates by KLK1 and KLK6 indicated that the N-terminal tryptophan did not interfere with enzyme activity. The one exception was the sequence WSLFRMVVL which, however, was less soluble than the other synthetic substrates and could only be tested at lower concentrations.
Analysis of the sequences from the KLK6 selection revealed the presence of an Arg at the P1 position of all 20 phage clones analyzed. P1′ showed a very strong preference for Ser, although clones with Asn, Met, and Tyr at that position were also enriched. A significant preference for Phe or Val in P2 and for Val or Ala in P2′ was also discerned (Table 1). Our results are consistent with the analysis of Angelo et al. (2006) except for the amino acid preferences at P2′ where positional scanning of peptide substrates did not detect a preference for Ala or Val. Debela et al. (2006) examined the specificity of KLK6 using positional libraries of the general structure acetyl-P4-P3-P2-P1-ACC (ACC = 7-amino-4-carbamoylmethylcoumarin) and reported that, in addition to Arg, the S1 subsite can accommodate Ala, His, Met, and Lys and that Arg and Lys preferentially occupy the S2 subsite. These results are in conflict with both the data from Angelo et al. (2006) and from the present study and are probably a consequence of the ACC moiety occupying the P1′ position. It should be noted that human pro-KLK6 is self-autoactivated to active by self-cleavage between Lys21 and Leu22 (Yoon et al. 2007). Self-autoactivation is detected after 24 h of incubation at 37°C but not after 1 h of incubation at 37°C, indicating that the rate of hydrolysis is slow. In this work we carried out incubation of the phage with protease for 30 min, to avoid the selection of slowly hydrolyzed peptides containing nonoptimal cleavage sites.
The tetrapeptide Phe/Val-Arg-Ser-Ala/Val from P2 to P2′ constitutes the consensus recognition site for KLK6. A blast search (www.ncbi.nlm.nih.gov) was performed based on sequence homology with phage display selected substrates and the consensus sequence Phe/Val-Arg-Ser-Ala/Val of KLK6. The blast search gave us hundreds of hits. However, several putative substrate candidates of KLK6 were found based on additional information from published biological studies. For example, ionotropic glutamate receptor (GluR) has been proposed to be a possible substrate of KLK6 so that KLK6 may be involved in the modulation of glutamate-mediated neuronal activity in CNS (Angelo et al. 2006). Our blast search showed that GluR N-methyl d-aspartate 2C (GRIN2C) contains the sequence VRSV that matches the consensus. Similarly, GluR N-methyl d-aspartate 2D (GRIN2D) contains the consensus sequence VRSA. This finding further supports the notion that the ionotropic glutamate receptor is a potential target of KLK6 and might interact with GRIN mediating further GluR excitation in CNS.
The major character of Parkinson syndrome is the aggregation of insoluble α synuclein and the formation of Lewy bodies in the patient's brain (Spillantini et al. 1997). KLK6 was shown to degrade α synuclein and prevent its polymerization (Iwata et al. 2003). We found that the synuclein α binding protein, synphilin, contains the sequence FRSI which exhibits strong homology with the consensus cleavage motif determined here. Synphilin has been shown to interact with α synuclein and to promote aggregation of synuclein and formation of inclusion bodies in neurons (Engelender et al. 1999). Our results suggest that KLK6 might play a role in the degradation of synphilin, thus decreasing the formation of inclusions in neurons in Parkinson's disease. However, further experiments to check the KLK6 digestion of these proteins are suggested to confirm these results.
In addition to kininogen, KLK1 appears to have many other physiological protein substrates including pro-insulin, prorenin, low-density lipoprotein, etc. (for review, see Yousef and Diamandis 2001; Paliouras and Diamandis 2006). Therefore, KLK1 has been proposed to exhibit different enzyme functions in specific tissues or cell types. KLK1 cleaves human low molecular weight kininogen at Met–Lys and Arg–Ser bonds (Sueiras-Diaz et al. 1994) while it can also cleave after the Phe–Phe pair of kallistatin (Zhou et al. 1992) and somatostatin (Pimenta et al. 1997). Here, phage display confirmed that KLK1 is able to accept two distinct classes of substrates and exhibits trypsin- and chymotrypsin-like activities (Table 3A,B). For the latter class of substrates, Tyr was the predominant residue in the P1 position. Further, our analysis indicates a strict preference for Ser or Arg in the P1′ position of chymotrypsin-like substrates of KLK1 (Table 3A). As expected based on the known substrates of KLK1, a variety of amino acids can be accommodated in the P2 position of the substrate, although there is a preference for aromatic residues (five out of 12 phage clones).
Substrates containing trypsin-like sites were enriched at a lower frequency relative to chymotrypsin-like substrates (Table 3B). Among the four such sequences obtained, there was a perfect consensus for Arg at P1 and Ser at P1′. Table 3B also indicates a preference for Ala in the P3 position (three/four sequences). However, given the small number of trypsin-like substrates obtained, this data should be interpreted with caution.
Five peptides corresponding to sequences isolated by substrate phage were synthesized, the kinetics of cleavage was determined by HPLC, and the products of cleavage were analyzed by ESI/MS. All five peptides were cleaved with high catalytic efficiency under the conditions used (k cat/K m > 105 M−1s−1). The best substrate was WAHRTTFYRRGA, which was hydrolyzed very rapidly, with k cat/K m of 1.3 ± 0.5 × 107 (Table 4). Interestingly, the higher rate of cleavage of this peptide was primarily due to an increased k cat. Although peptide #4 (WGTLFRSGN) contained both a putative chymotrypsin cleavage site (between Phe and Arg) and a trypsin cleavage site (between Arg and Ser), incubation with KLK1 resulted exclusively in the formation of WGTLF and RSGN. To examine whether the amino acids distal to the cleavage site determine whether KLK1 exhibits trypsin- or chymotrypsin-like activity, we synthesized a derivative of the substrate phage-derived peptide #4 (WGTLF↓RSGN) in which the P1 and P1′ were changed from Phe–Arg to Arg–Ser, respectively. This substitution resulted in a 10-fold reduction in the k cat/K m . Keeping the P2–P2′ sequence in peptide #5 fixed but changing the P3–P4 and P3′P4′residues increased the catalytic rate with peptide #7 by 500-fold.
Superposition of KLK1 and KLK6 reveals the two enzymes are very similar structurally, with RMSD of 0.88 Å for Cα. A closer examination shows that the S1 cavity in KLK6 is generally tighter and is surrounded by more polar groups. Specifically, Tyr288, Tyr172, Thr190, Gln192, Asn217, Ile218, Gly226, and Ile138 form the S1 cavity of KLK6 compared to Ala228, His172, Ser190, Val192, Try217, Val218, Ser226, and Ala138 in KLK1. Also, loop 216–226 is brought closer to the S1 cavity in KLK6. The presence of Asp189 in both KLK1 and KLK6 indicates that, unlike chymotrypsin, KLK1 enzyme employs more subtle differences around the S1 cavity to allow for chymotrypsin-like activity, namely, a larger cavity and smaller hydrophobic groups to accommodate binding of aromatic groups. The docking simulations show that Arg or Tyr fit into the S1 pocket of KLK1 equally well, which places the peptide in the binding mode consistent with the dual specificities of the enzyme. A major factor in determining the binding mode of KLK1 appears to be the residues at P1′ and P2′, as the S1′ site accepts small polar residues and S2′ prefers large hydrophobic residues almost exclusively. The S2 site, according to GOLD, appears to be largely promiscuous, capable of accepting a range of polar and aromatic residues. For KLK6, the above preferences are also true; however, since the S1 pocket of KLK6 is less tolerant to large aromatic residues, GOLD predicts that Arg binds to the S1 pocket of KLK6, facilitating a trypsin-like reaction. Comparing different binding modes, we observe that the peptide backbone stays close to catalytic Ser195, indicating that a nucleophilic attack is likely to take place between P1 and P1′.
Materials and Methods
Bacterial strains and plasmids
Escherichia coli strain MC1061 was used for cloning (Casabadan and Cohen 1980), and K91BK was used for phage infection and amplification. The polyvalent phage display vector fUSE55, which is derived from fUSE5, was used for the construction of peptide libraries (Scott and Smith 1990). Unless otherwise stated, E. coli cells were grown in NZY medium for phage infection and propagation containing the appropriate antibiotics at 37°C in a shaking incubator.
Protein purification
The mature recombinant KLK1 and KLK6 were expressed in a baculovirus/insect cell line system and purified as previous described (Laxmikanthan et al. 2005).
Construction of phage library
First, sequences encoding the Flag tag (DYKDDDDK) and XhoI restriction site were inserted into the 5′ of gene gIII by ligating a synthetic DNA fragment into the SfiI site of fUSE55, giving rise to fUSE55-Flag. Subsequently, the 8-mer library was synthesized by PCR using fUSE55-Flag DNA (100 pg) as the template, the 5′ primer, 5′-GATAAAGGACTCGAGGCTNNKNNKNNKNNKNNKNNKNNKNNKGGGGCCGAAACTGTTGAAAG-3′ (where N represents any nucleotide and K represents T/G), and the 3′ primer, 5′-CAAACGAATGGATCCTCATTAAAGCCAG-3′, followed by digestion with XhoI/BamHI and ligation into fUSE55-Flag (restriction sites are underlined). The ligated DNA (∼1 μg) was used to transform electrocompetent E. coli MC1061 (∼100 μL) via electroporation. The library was sequenced to confirm the randomization. No bias was found within the library.
Library screening and phage substrate purification
1011 cfu (colony forming unit) was first immobilized onto 3 mL of anti-Flag M2 Affinity Gel (Sigma) at 4°C for 4 h. The phage-bound resin was washed extensively with TBS buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5) to remove unbound phages then incubated with 100–200 nM of purified enzyme in TBS buffer containing 1 mM EDTA at 37°C for 45 min. The eluted phage particles were subsequently amplified in E. coli K91BK, purified with polyethylene glycol (PEG) 8000 (Sigma), and used as input in subsequent rounds of screening. From the third to the last round of screening, phage clones were randomly selected and plasmid DNA were isolated and sequenced to determine the displayed substrate peptide sequences. A total of six rounds of enrichment were performed for KLK1 and five rounds for KLK6.
Western blotting
Selected phage clones were purified and incubated with 0–1 μM of KLK6 or 4 μM of KLK1 in 50 mM Tris-HCl, 150 mM NaCl buffer (pH 7.5) containing 1 mM EDTA at 37°C for 45 min. The reaction mixture was separated with 4%–20% gradient SDS–polyacrylamide gel electrophoresis (PAGE) (Pierce), and then analyzed with anti-Flag antibody by Western blotting.
Kinetic measurements
Peptides were synthesized by solid-phase synthesis (EZBiolab, Inc.). Kinetic assays were carried out in 20 mM Tris-HCl buffer, containing 1 mM EDTA at pH 9.0 for KLK1 and 50 mM Tris-HCl, 150 mM NaCl containing 1 mM EDTA at pH 7.5 for KLK6, respectively. One to 200 μM substrate were incubated with 0.125–100 nM of purified enzyme at 37°C for 5–30 min. The reactions were quenched by liquid nitrogen and were analyzed on a C18 reverse-phase column (Phenomenex) by high-performance liquid chromatography (HPLC) using the following gradient: 5% acetonitrile/95% H2O for 1 min, increasing to 95% acetonitrile/5% H2O for 29 min, and returning to 5% acetonitrile/95% H2O for 5 min. The product amount was calculated upon the integration area at 280 nm and fitted to nonlinear regression of Michaelis-Menten equation by Prism software (GraphPad).
Molecular modeling
Docking was performed using GOLD v3.1.1 (www.ccdc.cam.ac.uk) on TI-3D cluster with coordinates taken from human kallikrein 1 structure (PDB code 1SPJ) and human kallikrein 6 structure (PDB code 1L2E). For KLK6, the cavity was defined with a radius of 20 Å centered around the OH group of Ser195. Five peptides were tested, FRSA, FRSV, VRSA, VRSV, and FRFSQ, all capped with acetyl and N-methylamide groups. A total of 200 genetic algorithm (GA) simulations were performed on each peptide, with a population size of 150 and 1 × 106 operations. The following residues on target protein were defined with flexible side chains: His57, His99, Asp189, Ser190, Gln192, Ser195, Ser214, Trp215, Asn217, and Ile218, using standard rotamer libraries (Lovell et al. 2000). All figures were rendered in PyMOL (DeLano Scientific; http://www.pymol.org). For KLK1, seven peptides were tested: FYRR, LYSR, ARSA, SRSA, KRSY, FYSQ, and FYSR. The following residues on the protein were defined with flexible side chains: Gln41, His57, Tyr99, Asp102, Asp189, Thr190, Ser195, Ser214, Trp215, and Ser226. The rest of the docking/GA parameters were identical to KLK6 docking simulations.
Acknowledgments
This work was supported by National Institutes of Health grants (R01 GM065551, R01 GM073089, and 1R15NS057771-01). P.A.G. and P.R. acknowledge support from the National Institute of General Medical Sciences (grant R01GM079686).
Footnotes
Reprint requests to: George Georgiou, Department of Chemical Engineering, University of Texas, Campus Mail Code CO400, Austin, TX 78712-1062, USA; e-mail: gg@che.utexas.edu; fax: (512) 471-7963.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073333208.
References
- Angelo, P.F., Lima, A.R., Alves, F.M., Blaber, S.I., Scarisbrick, I.A., Blaber, M., Juliano, L., Juliano, M.A. Substrate specificity of human kallikrein 6: Salt and glycosaminoglycan activation effects. J. Biol. Chem. 2006;281:3116–3126. doi: 10.1074/jbc.M510096200. [DOI] [PubMed] [Google Scholar]
- Anisowicz, A., Sotiropoulou, G., Stenman, G., Mok, S.C., Sager, R. A novel protease homolog differentially expressed in breast and ovarian cancer. Mol. Med. 1996;2:624–636. [PMC free article] [PubMed] [Google Scholar]
- Bernett, M.J., Blaber, S.I., Scarisbrick, I.A., Dhanarajan, P., Thompson, S.M., Blaber, M. Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system. J. Biol. Chem. 2002;277:24562–24570. doi: 10.1074/jbc.M202392200. [DOI] [PubMed] [Google Scholar]
- Bhoola, K.D., Figueroa, C.D., Worthy, K. Bioregulation of kinins: Kallikreins, kininogens, and kininases. Pharmacol. Rev. 1992;44:1–80. [PubMed] [Google Scholar]
- Borgono, C.A., Michael, L.P., Diamandis, E.P. Human tissue kallikreins: Physiologic roles and applications in cancer. Mol. Cancer Res. 2004;2:257–280. [PubMed] [Google Scholar]
- Casabadan, M.J., Cohen, S.N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli . J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cloutier, S.M., Chagas, J.R., Mach, J.P., Gygi, C.M., Leisinger, H.J., Deperthes, D. Substrate specificity of human kallikrein 2 (hK2) as determined by phage display technology. Eur. J. Biochem. 2002;269:2747–2754. doi: 10.1046/j.1432-1033.2002.02960.x. [DOI] [PubMed] [Google Scholar]
- Debela, M., Magdolen, V., Schechter, N., Valachova, M., Lottspeich, F., Craik, C.S., Choe, Y., Bode, W., Goetting, P. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 2006;281:25678–25688. doi: 10.1074/jbc.M602372200. [DOI] [PubMed] [Google Scholar]
- Deperthes, D. Phage display substrate: A blind method for determining protease specificities. Biol. Chem. 2002;383:1107–1112. doi: 10.1515/BC.2002.119. [DOI] [PubMed] [Google Scholar]
- Diamandis, E.P., Yousef, G.M., Clements, J., Ashworth, L.K., Yoshida, S., Egelrud, T., Nelson, P.S., Shiosaka, S., Little, S., Lilja, H., et al. New nomenclature for the human tissue kallikrein gene family. Clin. Chem. 2000;46:1855–1858. [PubMed] [Google Scholar]
- Diamandis, E.P., Scorilas, A., Fracchioli, S., Gramberen, M., Bruijin, H., Henrik, A., Soosaipillai, A., Grass, L., Yousef, G.M., Stenman, U.H., et al. Human kallikrein 6 (hK6): A new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J. Clin. Oncol. 2003;21:1035–1043. doi: 10.1200/JCO.2003.02.022. [DOI] [PubMed] [Google Scholar]
- Diamond, S.L. Methods for mapping protease specificities. Curr. Opin. Chem. Biol. 2007;11:46–51. doi: 10.1016/j.cbpa.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Engelender, S., Kaminsky, Z., Guo, X., Sharp, A.H., Amaravi, R.K., Kleiderlein, J.J., Margolis, R.L., Troncoso, J.C., Lanahan, A.A., Worley, P.F., et al. Synphilin-1 associates with α-synuclein and promotes the formation of cytosolic inclusions. Nat. Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- Felber, L.M., Borgono, C.A., Cloutier, S.M., Kundig, C., Kishi, T., Chagas, J.R., Jichlinski, P., Gygi, C.M., Leisinger, H.J., Diamandis, E.P., et al. Enzymatic profiling of human kallikrein 14 using phage-display substrate technology. Biol. Chem. 2005;386:291–298. doi: 10.1515/BC.2005.035. [DOI] [PubMed] [Google Scholar]
- Ferrieu-Weisbuch, C., Michel, S., Collomb-Clerc, E., Pothion, C., Deleage, G., Jolivet-Reynaud, C. Characterization of prostate-specific antigen binding peptides selected by phage display technology. J. Mol. Recognit. 2006;19:10–20. doi: 10.1002/jmr.762. [DOI] [PubMed] [Google Scholar]
- Fiedler, F., Leysath, G. Substrate specificity of porcine pancreatic kallikrein. Adv. Exp. Med. Biol. 1979;120A:261–271. doi: 10.1007/978-1-4757-0926-1_26. [DOI] [PubMed] [Google Scholar]
- Ghosh, M.C., Grass, L., Soosaipillai, A., Sotiropoulou, G., Diamandis, E.P. Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumor cells. Tumour Biol. 2004;25:193–199. doi: 10.1159/000081102. [DOI] [PubMed] [Google Scholar]
- Hwang, B.-Y., Varadarajan, N., Li, H., Rodriquez, S., Iverson, B.L., Georgiou, G. Substrate specificity of the Escherichia coli outer membrane protease OmpP. J. Bacteriol. 2007;189:522–530. doi: 10.1128/JB.01493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, A., Maruyama, M., Akagi, T., Hashikawa, T., Kanazawa, I., Tsuji, S., Nukima, N. α-Synuclein degradation by serine protease neurosin: Implication for pathogenesis of synucleinopathies. Hum. Mol. Genet. 2003;12:2625–2635. doi: 10.1093/hmg/ddg283. [DOI] [PubMed] [Google Scholar]
- Kerr, F.K., O'Brien, G., Quinsey, N.S., Whisstock, J.C., Boyd, S., de la Banda, M.G., Kaiserman, D., Matthews, A.Y., Bird, P.I., Pike, R.N. Elucidation of the substrate specificity of the C1s protease of the classical complement pathway. J. Biol. Chem. 2005;280:39510–39514. doi: 10.1074/jbc.M506131200. [DOI] [PubMed] [Google Scholar]
- Laxmikanthan, G., Blaber, S.I., Bernett, M.J., Scarisbrick, I.A., Juliano, M.A., Blaber, M. 1.70 Å X-ray structure of human apo kallikrein 1: Structural changes upon peptide inhibitor/substrate binding. Proteins. 2005;58:802–814. doi: 10.1002/prot.20368. [DOI] [PubMed] [Google Scholar]
- Little, S.P., Dixon, E.P., Norris, F., Buckley, W., Becker, G.W., Johnson, M., Dobbins, J.R., Wyrick, T., Miller, J.R., MacKellar, W., et al. Zyme, a novel and potentially amyloidogenic enzyme cDNA isolated from Alzheimer's disease brain. J. Biol. Chem. 1997;272:25135–25142. doi: 10.1074/jbc.272.40.25135. [DOI] [PubMed] [Google Scholar]
- Lovell, J.M., Richardson, J.S., Richardson, D.C. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- Magklara, A., Mellati, A.A., Wasney, G.A., Little, S.P., Sotiropoulou, G., Becker, G.W., Diamandis, E.P. Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and regulation by inhibitors. Biochem. Biophys. Res. Commun. 2003;307:948–955. doi: 10.1016/s0006-291x(03)01271-3. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou, K., Hansen, K.K., Saifeddine, M., Tea, I., Blaber, M., Blaber, S.I., Scarisbrick, I., Andrade-Gordou, P., Cottrell, G.S., Bunnett, N.W., et al. Proteinase-activated receptors, targets for kallikrein signaling. J. Biol. Chem. 2006;281:32095–32112. doi: 10.1074/jbc.M513138200. [DOI] [PubMed] [Google Scholar]
- Paliouras, M., Diamandis, E.P. The kallikrein world: An update on the human tissue kallikreins. Biol. Chem. 2006;387:643–652. doi: 10.1515/BC.2006.083. [DOI] [PubMed] [Google Scholar]
- Pimenta, D.C., Juliano, M.A., Juliano, L. Hydrolysis of somatostatin by human tissue kallikrein after the amino acid pair Phe–Phe. Biochem. J. 1997;327:27–30. doi: 10.1042/bj3270027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Towner, M.D., Isackson, P.J. Nervous system-specific expression of a novel serine protease: Regulation in the adult rat spinal cord by excitotoxic injury. J. Neurosci. 1997;17:8156–8168. doi: 10.1523/JNEUROSCI.17-21-08156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J.K., Smith, G.P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Sedlacek, R., Chen, E. Screening for protease substrate by polyvalent phage display. Comb. Chem. High Throughput Screen. 2005;8:197–203. doi: 10.2174/1386207053258541. [DOI] [PubMed] [Google Scholar]
- Spillantini, M.G., Schmidt, M.L., Lee, V.M., Trojanowski, J.Q., Jakes, R., Goedert, M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Sueiras-Diaz, J., Jones, D.M., Ashworth, D., Horton, J., Evans, D.M., Szelke, M. Cleavage of human kininogen fragments at Met–Lys by human tissue kallikrein. Braz. J. Med. Biol. Res. 1994;27:1935–1942. [PubMed] [Google Scholar]
- Yamashiro, K., Tsuruoka, N., Kodama, S., Tsujimoto, M., Yamamura, Y., Tanaka, T., Nakazato, H., Yamaguchi, N. Molecular cloning of a novel trypsin-like serine protease (neurosin) preferentially expressed in brain. Biochim. Biophys. Acta. 1997;1350:11–14. doi: 10.1016/s0167-4781(96)00187-x. [DOI] [PubMed] [Google Scholar]
- Yoon, H., Laxmikanthan, G., Lee, J., Blaber, S.I., Rodriguez, A., Kogot, J.M., Scarisbrick, I.A., Blaber, M. Activation profiles and regulatory cascades of the human kallikrein-related peptidases. Biochemistry. 2007;46:5209–5217. doi: 10.1074/jbc.M705190200. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Diamandis, E.P. The new human tissue kallikrein gene family: Structure, function, and association to disease. Endocr. Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Luo, L.Y., Scherer, S.W., Sotiropoulou, G., Diamandis, E.P. Molecular characterization of zyme/protease M/neurosin (PRSS9), a hormonally regulated kallikrein-like serine protease. Genomics. 1999;62:251–259. doi: 10.1006/geno.1999.6012. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Plymeris, M.E., Yacoub, G.M., Scorilas, A., Soosaipillai, A., Popalis, C., Fracchioli, S., Katsaros, D., Diamandis, E.P. Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res. 2003;63:2223–2227. [PubMed] [Google Scholar]
- Zhou, G.X., Chao, L., Chao, J. Kallistatin: A novel human tissue kallikrein inhibitor. Purification, characterization, and reactive center sequence. J. Biol. Chem. 1992;267:25873–25880. [PubMed] [Google Scholar]