Abstract
Enoyl–acyl carrier protein (ACP) reductases are critical for bacterial type II fatty acid biosynthesis and thus are attractive targets for developing novel antibiotics. We determined the crystal structure of enoyl–ACP reductase (FabK) from Streptococcus pneumoniae at 1.7 Å resolution. There was one dimer per asymmetric unit. Each subunit formed a triose phosphate isomerase (TIM) barrel structure, and flavin mononucleotide (FMN) was bound as a cofactor in the active site. The overall structure was similar to the enoyl–ACP reductase (ER) of fungal fatty acid synthase and to 2-nitropropane dioxygenase (2-ND) from Pseudomonas aeruginosa, although there were some differences among these structures. We determined the crystal structure of FabK in complex with a phenylimidazole derivative inhibitor to envision the binding site interactions. The crystal structure reveals that the inhibitor binds to a hydrophobic pocket in the active site of FabK, and this is accompanied by induced-fit movements of two loop regions. The thiazole ring and part of the ureido moiety of the inhibitor are involved in a face-to-face π–π stacking interaction with the isoalloxazine ring of FMN. The side-chain conformation of the proposed catalytic residue, His144, changes upon complex formation. Lineweaver–Burk plots indicate that the inhibitor binds competitively with respect to NADH, and uncompetitively with respect to crotonoyl coenzyme A. We propose that the primary basis of the inhibitory activity is competition with NADH for binding to FabK, which is the first step of the two-step ping-pong catalytic mechanism.
Keywords: FabK, enoyl–acyl carrier protein reductase, fatty acid biosynthesis, antibiotics, inhibitor
Streptococcus pneumoniae causes community-acquired infections such as pneumonia, otitis media, and meningitis. The increase of penicillin- and/or macrolide-resistant S. pneumoniae is of great concern worldwide, and, moreover, the emergence of quinolone-resistant S. pneumoniae has been reported recently (Cohen 1992; Bartlett et al. 1998; Johnson et al. 2005). A key strategy to overcoming antibiotic resistance is the discovery of antibacterial agents with novel mechanisms of action that have no cross-resistance.
The bacterial type II fatty acid synthase complex comprises discrete enzyme activities encoded by discrete genes, in contrast to the multifunctional type I fatty acid synthase in mammals (Rock and Cronan 1996). These bacterial enzymes are attractive targets for the development of novel selective antibacterial agents (Heath et al. 2001). Enoyl–acyl carrier protein (ACP) reductase is responsible for catalyzing the final step in each elongation cycle of bacterial type II fatty acid biosynthesis (FAS II), and it plays a key role in regulation of the pathway (Heath and Rock 1995, 1996). Triclosan is known to inhibit FabI, the enoyl–ACP reductase from Escherichia coli (Heath et al. 1998) and Staphylococcus aureus (Heath et al. 2000; Slater-Radosti et al. 2001). The anti-tuberculosis agent isoniazid also targets the FabI homolog (InhA) of Mycobacterium tuberculosis (Quemard et al. 1995).
Recent genomic studies have demonstrated that an alternative triclosan-resistant enoyl–ACP reductase, FabK, is present in several clinical pathogens (Heath and Rock 2000). FabK is the sole enoyl–ACP reductase in S. pneumoniae, and both FabI and FabK have been found in pathogens such as Enterococcus faecalis and Pseudomonas aeruginosa (Heath and Rock 2000). Although novel FabI inhibitors targeting S. aureus have been reported by several groups (Heerding et al. 2001; Seefeld et al. 2001; Ling et al. 2004; Kitagawa et al. 2007a; Takahata et al. 2007; Yum et al. 2007), there are only a few reports of FabK inhibitors (Payne et al. 2002; Seefeld et al. 2003; Zheng et al. 2006). We have reported the inhibitor AG205 and more effective FabK inhibitors showing both FabK inhibitory activity and antibacterial activity against S. pneumoniae (Takahata et al. 2006; Kitagawa et al. 2007b,c). Inhibitors designed to target FabK are attractive as antibiotics against microorganisms that utilize the FabK pathway, in particular, S. pneumoniae.
S. pneumoniae FabK contains flavin mononucleotide (FMN) and requires NADH for its enzymatic activity (Marrakchi et al. 2003). Despite the importance of structural information about targeting proteins to guide drug design efforts, the crystal structure of FabK has not yet been determined. We report here the crystal structure of FabK and its complex with a phenylimidazole derivative inhibitor, compound 1 (Kitagawa et al. 2007c; Fig. 1). The structural information from this complex will facilitate the development of selective FabK and/or dual FabI/FabK inhibitors using structure-based drug design.
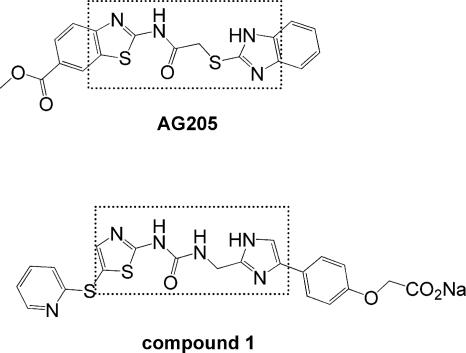
Figure 1.
Chemical structures of FabK inhibitors, AG205 and compound 1. Dashed lines enclose similar components of the two compounds.
Results and Discussion
The structure of FabK
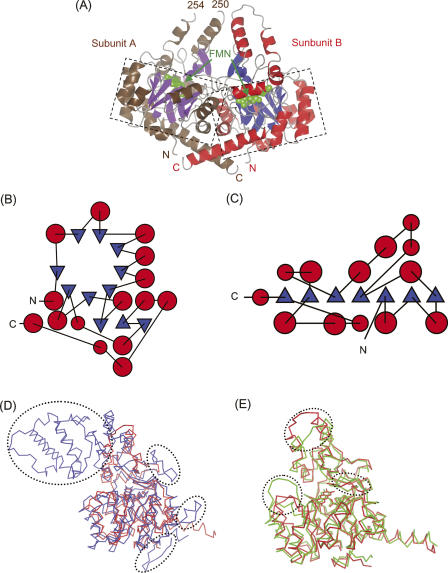
We determined the crystal structure of S. pneumoniae FabK by the multiple-wavelength anomalous dispersion (MAD) method using the selenomethionyl (SeMet)-substituted crystal (Table 1). The model of FabK consisted of one dimer in the asymmetric unit (Fig. 2A). The crystal structure revealed that each subunit formed a triose phosphate isomerase (TIM) barrel structure (Nagano et al. 2002) containing one FMN molecule bound as a cofactor. FabK was structurally unrelated to mammalian enoyl–ACP reductases (Maier et al. 2006) and most of the bacterial counterparts (FabI and InhA) that contain a Rossmann fold supporting a binding site for NADH or NADPH, but not for FMN (Dessen et al. 1995; Baldock et al. 1996; Fig. 2B,C). The subunits of FabK were highly similar to each other, with a root mean square deviation (r.m.s.d.) of 0.48 Å for 316 Cα atoms (amino acids 1–250 and 255–320). Because many more amino acid residues of subunit B (321 residues) were modeled into density than of subunit A (317 residues), subunit B was selected as the prototypical structure that is referred to in all further discussions. Recently, the crystal structure of fungal multifunctional fatty acid synthase (FAS) (Protein Data Bank [PDB] code 2UVA), which contains an enoyl–ACP reductase (abbreviated ER), has been reported (Jenni et al. 2007). The overall architecture of the fungal ER is similar to that of FabK, except in a few regions, with an r.m.s.d. of 2.2 Å for 291 equivalent Cα atoms (Fig. 2D). Several insertions, such as the α-helical insertion domain near the C terminus, are present in ER but not in FabK. In ER, this α-helical insertion domain interacts with the ketoacyl synthase and clamps the extended part of the linker region between the ketoacyl reductase and the ketoacyl synthase. This suggests that these insertions are essential for interactions with other functional domains of FAS, within multifunctional enzymes containing an ER moiety, and for proper assembly into complexes, but they are not essential for the enoyl–ACP reductase activity.
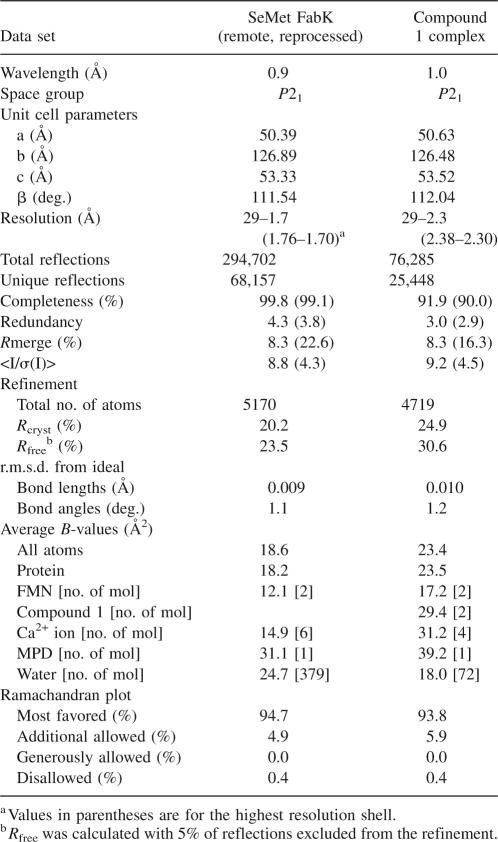
Table 1.
Data collection and refinement statistics
Figure 2.
(A) Ribbon diagram of the dimer structure of S. pneumoniae FabK in the asymmetric unit. α-Helices in subunits A and B are colored in brown and red, respectively. β-Strands in subunits A and B are colored in purple and blue, respectively. Each FMN is indicated by arrows and shown as a sphere model colored green. TIM barrel structures are enclosed by dashed lines. (B) Topological diagram of S. pneumoniae FabK monomer. Red circles and blue triangles represent α-helices and β-strands, respectively. B and C were produced using the program TOPS (Michalopoulos et al. 2004) and then reproduced manually by tracing. (C) Topological diagram of E. coli FabI, which contains a Rossmann fold. Although FabK and FabI have distinct folds, they exhibit a similar enzymatic function. (D) Superposition of fungal enoyl–ACP reductase (ER, blue) on S. pneumoniae FabK (red) shown as a Cα trace. Each FMN molecule is shown as a stick model. Insertion regions observed only in ER are enclosed by dashed lines. (E) Superposition of P. aeruginosa 2-nitropropane dioxygenase (2-ND, light green) on S. pneumoniae FabK (red), shown as a Cα trace. Each FMN molecule is shown as a stick model. The loops in which the differences are observed between FabK and 2-ND are enclosed by dashed lines. Figures 2A,D,E, 3A,B, 4, and 6A,B were created using CCP4mg (Potterton et al. 2004).
A BLAST search (Altschul et al. 1990) of the fabK gene (GenBank accession no. AE008418) indicated that FabK belongs to the NAD(P)H-dependent flavin oxidoreductase family (Nagano et al. 2002). Among the family members that are in the PDB database, the 2-nitropropane dioxygenase from P. aeruginosa (abbreviated 2-ND, PDB code 2GJL) (Ha et al. 2006) showed the highest homology (sequence identity 28%) with FabK. The structural model of 2-ND was similar to that of FabK, with an r.m.s.d. of 1.4 Å for 303 equivalent Cα atoms (Fig. 2E). The most prominent differences between the structures were observed in three loops (amino acid numbers in FabK 71–76, 145–149, and 248–256) near FMN. This suggests that the differently shaped active sites of FabK and 2-ND correspond to distinct recognition sites for distinct reaction substrates for each enzyme. On the other hand, a putative catalytic role for the histidine at the active site (His144 in FabK) had been proposed previously for 2-ND (His152) (Ha et al. 2006) and for fungal ER (His751) (Jenni et al. 2007). Actually, His144 is located near FMN in FabK, similar to the positions of His152 and His751 in 2-ND and ER, respectively.
FabK–compound 1 complex structure
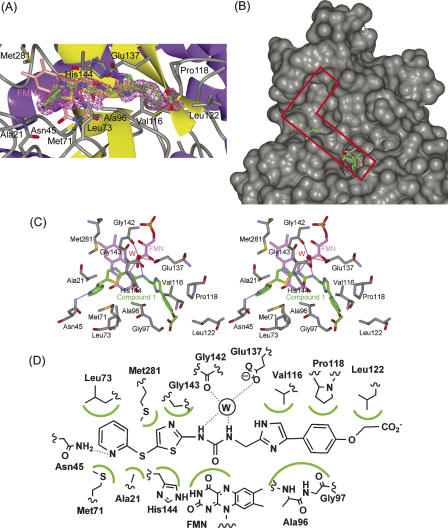
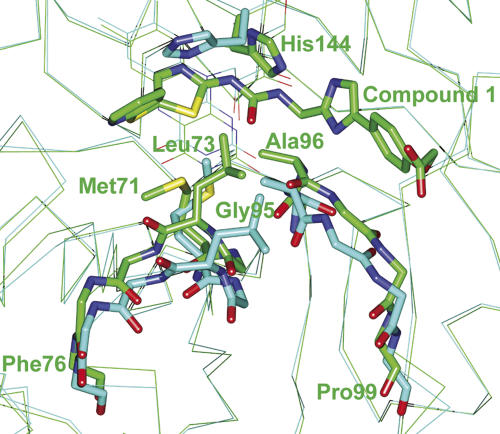
Crystals of FabK complexed with compound 1 were prepared by soaking native crystals in compound 1 solution, and the structure was determined using the SeMet-substituted FabK model for initial rigid-body refinement (Table 1). The model of FabK consists of one dimer per asymmetric unit. The compound 1 in each active site bound similarly; hence, subunit B was selected as the prototypical structure (Fig. 3A). Compound 1 is located on the right end of the L-shaped active pocket near FMN (Fig. 3B). The thiazole ring and part of the ureido moiety of the inhibitor are involved in a face-to-face π–π stacking interaction with the isoalloxazine ring of FMN (Fig. 3C). In addition, the thiazole ring forms hydrophobic interactions with Gly143, His144, and Met281. The ureido group forms hydrogen bonds to Gly142 and Glu137 by bridging an ordered solvent molecule. A previous study showed that after converting the ureido group to an acetamido or carbamate group, the resulting compounds exhibited lower FabK-inhibitory activity (Kitagawa et al. 2007b). Hence, the hydrogen bonds between the ureido group of compound 1 and the ordered water are considered to be important for binding. The phenylimidazole moiety is buried within a hydrophobic cleft, where it forms hydrophobic interactions with Val116, Pro118, Leu122, Ala96, and Gly97. The sulfur atom between the pyridine and thiazole rings forms hydrophobic interactions with Ala21 and Met71. The pyridine ring forms hydrophobic interactions with Met71 and Leu73, as well as hydrogen bonds to Asn45. The pyridine ring also faces an empty hydrophobic pocket in the upper part of the L-shaped active site. Therefore, additional chemical modifications of compound 1 to add moieties that will fit into the upper part of the active site may well enhance its inhibitory activity. A schematic representation of interactions between compound 1 and FabK is shown in Figure 3D. This structural information should be useful in the design of effective inhibitors.
Figure 3.
The structure of FabK complexed with compound 1. (A) The omit Fo–Fc difference density map observed for compound 1 bound in subunit B. The map, depicted as magenta mesh, is contoured at 2.5 σ. The inhibitor (green) and FMN (pink) are shown as stick models. Non-carbon atoms are colored according to atom type. The protein is shown as a ribbon model, and side chains of residues that bind to the inhibitor are shown as stick models. (B) The L-shaped active pocket in the complex of FabK with compound 1. FabK is shown as a surface model. The active pocket is enclosed by a red line. Compound 1 and FMN are shown as stick models colored green and pink, respectively. Non-carbon atoms are colored according to atom type. (C) Stereoview of the active site of the compound 1–FabK complex structure. Atoms are colored as described for Fig. 3A. Hydrogen bonds are represented by thin lines (magenta). The ordered solvent molecule is labeled W (red). (D) A schematic diagram of the inhibitor-binding mode. Hydrogen bonds are represented by broken lines (magenta), and hydrophobic contacts are shown as highlighted semicircular arcs (green); the ordered solvent molecule is labeled W.
Inhibitor-binding conformational changes
The overall structure of FabK complexed with compound 1 was similar to that of the uncomplexed structure, and the r.m.s.d. value is 0.49 Å for Cα atoms of the two superimposed structures (amino acids 1–223, 228–247, and 258–321). However, there were two noticeable differences between the structures. One was a conformational change of the His144 side chain, and the other was a conformational change in two loops (Fig. 4). In the uncomplexed structure, His144 is proximal to a water molecule on the si side of the isoalloxazine ring of FMN, but this side chain is flipped in the complexed structure, with compound 1 replacing the water molecule. In addition to this change, two loops present near the active site moved to create an induced fit with compound 1 upon formation of the complex. The most striking difference was a conformational change of the main chains of Gly95 and Ala96 in the loop (amino acids 95–99), which rotated to form hydrophobic interactions with the phenylimidazole moiety of compound 1. Moreover, the other loop (amino acids 71–76) moved toward compound 1 so as to introduce hydrophobic interactions between side chains (Met71 and Leu73) and compound 1 as described above (Fig. 3D).
Figure 4.
Comparison of the Cα traces for the complexed and uncomplexed structures. The uncomplexed structure is shown in cyan, and the structure of the complex with compound 1 is shown in green. The side chains of Ala96, Met71, Leu73, and His144 are shown in stick form. The main chains (amino acids 71–76 and 95–99) are shown as bold lines. Oxygen and nitrogen atoms are colored red and blue, respectively. Major differences between the two active-site loops can be observed between the uncomplexed and complexed forms (amino acids 71–76 and 95–99). Differences can also be observed in the side-chain conformation of His144.
Inhibition mechanism of compound 1
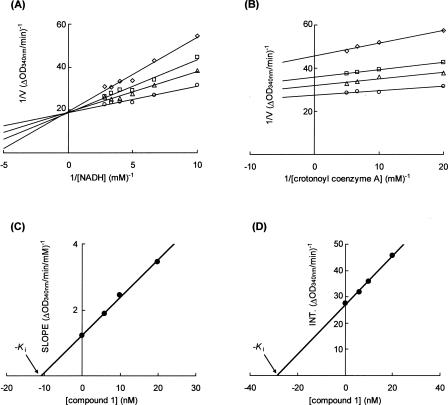
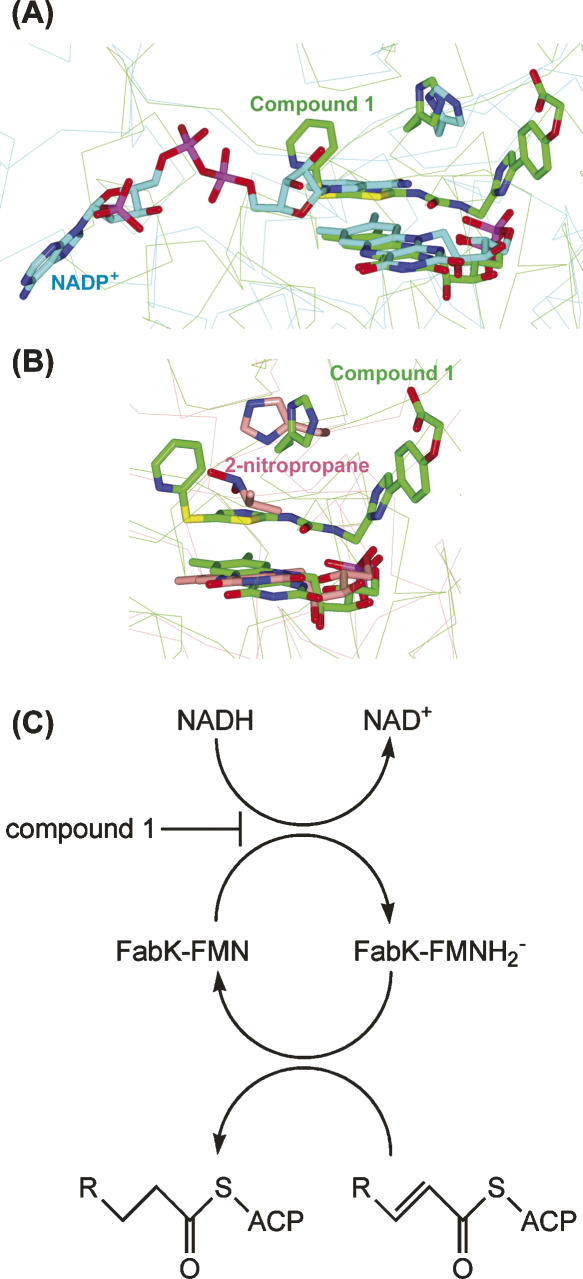
A Lineweaver–Burk plot analysis indicated that compound 1 acts as a competitive inhibitor with respect to NADH and as an uncompetitive inhibitor with respect to crotonoyl coenzyme A (Fig. 5A,B). The kinetic parameters indicated that the K m value for crotonoyl coenzyme A was 7 μM in the absence of compound 1, and the K m value for NADH was 67 μM in the absence of compound 1. The K i values of compound 1 were 11 nM and 29 nM for NADH and crotonoyl coenzyme A, respectively (Fig. 5C,D). Superposition of the crystal structure of the fungal ER complexed with NADP+ with another FMN-containing TIM barrel oxidoreductase, 2-ND, in complex with its substrate reveals that the substrate would sterically clash with the nicotinamide moiety of NADP+, suggesting that oxidized NADP is likely to be released before binding of enoyl–ACP (Jenni et al. 2007). In addition, superpositions of the FabK–compound 1 complex structure on ER and 2-ND also indicated that the thiazole moiety of compound 1 would sterically clash with the nicotinamide moiety of NADP+ in ER and 2-nitropropane, the substrate of 2-ND (Fig. 6A,B). It has been reported that competitive inhibitors for the first substrates are uncompetitive for the second substrates in human quinone oxidoreductase 2 and Saccharomyces cerevisiae NADH:Q6 oxidoreductase, which have only one substrate-binding site and follow the two-step ping-pong catalytic mechanism (Velazquez and Pardo 2001; Kwiek et al. 2004). Therefore, we propose that the main reason for the inhibitory activity of compound 1 is a competitive inhibition of NADH binding to FabK, which is the first step of the two-step ping-pong catalytic mechanism (Fig. 6C). According to this mechanism, compound 1 is considered to bind much more strongly to FabK–FMN (the oxidized form of the enzyme) than to FabK–FMNH2 − (the reduced form).
Figure 5.
The mechanism of FabK inhibition by compound 1. (A) Lineweaver–Burk plot showing competitive inhibition of FabK binding to NADH by compound 1. The enzyme was incubated in the presence of varying concentrations of compound 1 and NADH at a fixed concentration of crotonoyl coenzyme A (0.1 mM). The concentrations of compound 1 were 0 μg/mL (circle), 0.003 μg/mL (triangle), 0.005 μg/mL (square), and 0.01 μg/mL (diamond). (B) Lineweaver–Burk plot showing uncompetitive inhibition of FabK binding to crotonoyl coenzyme A by compound 1. The enzyme was incubated in the presence of varying concentrations of compound 1 and crotonoyl coenzyme A at a fixed concentration of NADH (0.1 mM). The concentrations of compound 1 were 0 μg/mL (circle), 0.003 μg/mL (triangle), 0.005 μg/mL (square), and 0.01 μg/mL (diamond). (C) The slope values of the lines from A are plotted versus compound 1 concentration. A K i of 11 nM for NADH was determined. (D) The intercept values of the lines from B are plotted versus compound 1 concentration. A K i of 29 nM for crotonoyl coenzyme A was determined.
Figure 6.
(A) Superposition of the active sites of S. pneumoniae FabK–compound 1 complex (green) with fungal ER–NADP+ complex (cyan). FMN, compound 1, NADP+, and side chains of the proposed catalytic histidine residues are shown as stick models. Non-carbon atoms are colored according to atom type. (B) Superposition of the active sites of S. pneumoniae FabK–compound 1 complex (green) with P. aeruginosa 2-ND in complex with the substrate, 2-nitropropane (pink). (C) Two-step ping-pong catalytic mechanism of FabK. FabK–FMN represents the oxidized form of the enzyme, and FabK–FMNH2 − represents the reduced form. R represents the CH3(CH2)n− (n = 1 − 12). Compound 1 is considered to be a competitive inhibitor for the first step of the reaction.
Conclusions
S. pneumoniae FabK, which catalyzes the last step in each cycle of fatty acid elongation, is a promising drug target because it plays a key role in regulation of the pathway. We have determined the crystal structure of FabK and its complex with a phenylimidazole derivative inhibitor. The resulting structural information regarding the active site pocket, and details of the inhibitor-binding mode, should be useful in the rational design of more effective FabK inhibitors that can be used against S. pneumoniae that shows resistance to conventional antibiotics, and/or dual FabI/FabK inhibitors that have a broader spectrum of antibacterial activity.
Materials and Methods
Crystallization and data collection
SeMet-substituted FabK was crystallized as described previously (Saito et al. 2006). The native form of FabK was expressed and purified by the same procedure described previously (Saito et al. 2006). Native crystals were obtained by the hanging-drop vapor-diffusion method at 277 K using a reservoir solution containing 0.1 M MES, pH 5.5–7.0, 0.1 M NH4Cl, 0.2 M CaCl2, 8%–15% 2-methyl-2,4-pentanediol (MPD), 8%–15% polyethylene glycol 1000, and 5 mM dithiothreitol. Crystals of FabK complexed with compound 1 were prepared by soaking native crystals in a reservoir solution containing 50 mM compound 1 (Kitagawa et al. 2007c) and 5% dimethylsulfoxide overnight. The resulting crystals were cryoprotected by using 15% glycerol. A set of MAD data has been collected to 2.0 Å resolution using the SeMet-substituted crystals, as reported previously (Saito et al. 2006). The previously reported remote data were reprocessed using the CrystalClear package (Rigaku) without distinguishing a Bijvoet pair of reflections, thus improving the resolution to 1.7 Å. The diffraction data for the FabK–compound 1 complex crystals were collected at 100 K at a wavelength of 1.0 Å on the BL32B2 beamline at the SPring-8 synchrotron facility. The diffraction data were integrated and scaled using the CrystalClear package (Rigaku).
Structure determination and refinement
Initial phases were obtained from the MAD data at 2.2 Å using the program SOLVE (Terwilliger and Berendzen 1999). The mean figure of merit was 0.40. After density modification with the program RESOLVE (Terwilliger 2000), the mean figure of merit was 0.65. The resulting experimental electron-density map was readily interpretable and allowed RESOLVE (Terwilliger 2003a,b) to build an initial model. Manual model building was carried out using the programs Coot (Emsley and Cowtan 2004), DS Modeling (Accelrys, Inc.), and TurboFrodo (AFMB–CNRS). The model was refined with the programs CNX (Accelrys, Inc.) and Refmac5 (Murshudov et al. 1997). Subsequently, this model was used to refine the high-resolution structure from the reprocessed remote data to 1.7 Å resolution. One FabK dimer is present in the asymmetric unit. The final model of the SeMet-substituted FabK consists of 317 residues in subunit A, 321 residues in subunit B, two FMNs, one MPD, six calcium ions, and 379 water molecules in the asymmetric unit. Crystals of the FabK–compound 1 complex were isomorphous with those of the SeMet-substituted FabK. The structure of the SeMet-substituted FabK was used for initial rigid-body refinement of the structure complexed with compound 1. Model building and refinement were carried out using the programs as described above. The occupancy of compound 1 in each subunit was set to 1.0, and this was consistent with the electron density levels. The final model of the compound 1 complex consists of 307 residues in subunit A, 307 residues in subunit B, two FMNs, two compound 1 molecules, one MPD, four calcium ions, and 72 water molecules in the asymmetric unit. All structures were validated using PROCHECK (Laskowski et al. 1993). A summary of statistics from data collection and refinement is given in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code 2Z6I for the SeMet-substituted structure and 2Z6J for the compound 1 complex.
Enzyme inhibition assays
FabK catalyzes the reduction of enoyl–ACPs with the concomitant oxidation of NADH (Marrakchi et al. 2003). The inhibitory activity of compound 1 against FabK was assayed using crotonoyl coenzyme A as a substrate, by monitoring the decrease in absorbance at 340 nm (Takahata et al. 2006; Kitagawa et al. 2007b). To evaluate the inhibitory effect of compound 1 on the activity of FabK, two sets of experiments were performed. In the first, the crotonoyl coenzyme A concentration was held constant while the concentrations of NADH and compound 1 were varied systematically. Assays (100 μL) were performed for 5 min in 0.3 μg/mL FabK, 0.1 M MES, pH 7.0, 0.1 mM crotonoyl coenzyme A, 0.1 M NH4Cl for NADH in a concentration range from 0.1 to 0.35 mM and in the absence or presence of 0.003–0.01 μg/mL compound 1. In the second set of experiments, the NADH concentration was held constant while the concentrations of crotonoyl coenzyme A and compound 1 were varied systematically. Assays (100 μL) were performed for 5 min in 1.5 μg/mL FabK, 0.1 M MES, pH 7.0, 0.1 mM NADH, 0.1 M NH4Cl for the crotonoyl coenzyme A concentration range from 0.05 to 0.2 mM and in the absence or presence of 0.003–0.01 μg/mL compound 1. Michaelis constants (K m) for crotonoyl coenzyme A and NADH were determined by means of Lineweaver–Burk plots, using initial velocities obtained by varying the concentration of one substrate while maintaining the other substrate at a fixed concentration in the absence of compound 1.
Acknowledgment
We thank Dr. Y. Katsuya at the SPring-8 synchrotron for help with data collection.
Footnotes
Reprint requests to: Jun Saito, Pharmaceutical Research Center, Meiji Seika Kaisha, Ltd., 760 Morooka-cho, Kohoku-ku, Yokohama 222-8567, Japan; e-mail: jun_saito@meiji.co.jp; fax: 81-45-545-3152.
Abbreviations: ACP, acyl carrier protein; FAD, flavin adenine dinucleotide; FAS II, bacterial type II fatty acid biosynthesis; FMN, flavin mononucleotide; MAD, multiple-wavelength anomalous dispersion; NADH, nicotinamide adenine dinucleotide; ER, enoyl–ACP reductase; 2-ND, 2-nitropropane dioxygenase; TIM, triose phosphate isomerase.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073288808.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baldock, C., Rafferty, J.B., Sedelnikova, S.E., Baker, P.J., Stuitje, A.R., Slabas, A.R., Hawkes, T.R., Rice, D.W. A mechanism of drug action revealed by structural studies of enoyl reductase. Science. 1996;274:2107–2110. doi: 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- Bartlett, J.G., Breiman, R.F., Mandell, L.A., File T.M., Jr Community-acquired pneumonia in adults: Guidelines for management. The Infectious Diseases Society of America. Clin. Infect. Dis. 1998;26:811–838. doi: 10.1086/513953. [DOI] [PubMed] [Google Scholar]
- Cohen, M.L. Epidemiology of drug resistance: Implications for a post-antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- Dessen, A., Quemard, A., Blanchard, J.S., Jacobs W.R., Jr, Sacchettini, J.C. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis . Science. 1995;267:1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ha, J.Y., Min, J.Y., Lee, S.K., Kim, H.S., Kim do, J., Kim, K.H., Lee, H.H., Kim, H.K., Yoon, H.J., Suh, S.W. Crystal structure of 2-nitropropane dioxygenase complexed with FMN and substrate. Identification of the catalytic base. J. Biol. Chem. 2006;281:18660–18667. doi: 10.1074/jbc.M601658200. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., Rock, C.O. Enoyl–acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli . J. Biol. Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., Rock, C.O. Regulation of fatty acid elongation and initiation by acyl–acyl carrier protein in Escherichia coli . J. Biol. Chem. 1996;271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., Rock, C.O. A triclosan-resistant bacterial enzyme. Nature. 2000;406:145–146. doi: 10.1038/35018162. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., Yu, Y.T., Shapiro, M.A., Olson, E., Rock, C.O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 1998;273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., Li, J., Roland, G.E., Rock, C.O. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl–acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 2000;275:4654–4659. doi: 10.1074/jbc.275.7.4654. [DOI] [PubMed] [Google Scholar]
- Heath, R.J., White, S.W., Rock, C.O. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- Heerding, D.A., Chan, G., DeWolf, W.E., Fosberry, A.P., Janson, C.A., Jaworski, D.D., McManus, E., Miller, W.H., Moore, T.D., Payne, D.J., et al. 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl–acyl carrier protein reductase (FabI) Bioorg. Med. Chem. Lett. 2001;11:2061–2065. doi: 10.1016/s0960-894x(01)00404-8. [DOI] [PubMed] [Google Scholar]
- Jenni, S., Leibundgut, M., Boehringer, D., Frick, C., Mikolasek, B., Ban, N. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- Johnson, C.N., Briles, D.E., Benjamin W.H., Jr, Hollingshead, S.K., Waites, K.B. Relative fitness of fluoroquinolone-resistant Streptococcus pneumoniae . Emerg. Infect. Dis. 2005;11:814–820. doi: 10.3201/eid1106.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, H., Kumura, K., Takahata, S., Iida, M., Atsumi, K. 4-Pyridone derivatives as new inhibitors of bacterial enoyl–ACP reductase FabI. Bioorg. Med. Chem. Lett. 2007a;15:1106–1116. doi: 10.1016/j.bmc.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Kitagawa, H., Ozawa, T., Takahata, S., Iida, M. Phenylimidazole derivatives as new inhibitors of bacterial enoyl–ACP reductase FabK. Bioorg. Med. Chem. Lett. 2007b;17:4982–4986. doi: 10.1016/j.bmcl.2007.06.040. [DOI] [PubMed] [Google Scholar]
- Kitagawa, H., Ozawa, T., Takahata, S., Iida, M., Saito, J., Yamada, M. Phenylimidazole derivatives of 4-pyridone as dual inhibitors of bacterial enoyl–acyl carrier protein reductases FabI and FabK. J. Med. Chem. 2007c;50:4710–4720. doi: 10.1021/jm0705354. [DOI] [PubMed] [Google Scholar]
- Kwiek, J.J., Haystead, T.A., Rudolph, J. Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry (Mosc.) 2004;43:4538–4547. doi: 10.1021/bi035923w. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Ling, L.L., Xian, J., Ali, S., Geng, B., Fan, J., Mills, D.M., Arvanites, A.C., Orgueira, H., Ashwell, M.A., Carmel, G., et al. Identification and characterization of inhibitors of bacterial enoyl–acyl carrier protein reductase. Antimicrob. Agents Chemother. 2004;48:1541–1547. doi: 10.1128/AAC.48.5.1541-1547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T., Jenni, S., Ban, N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- Marrakchi, H., Dewolf W.E., Jr, Quinn, C., West, J., Polizzi, B.J., So, C.Y., Holmes, D.J., Reed, S.L., Heath, R.J., Payne, D.J., et al. Characterization of Streptococcus pneumoniae enoyl–(acyl-carrier protein) reductase (FabK) Biochem. J. 2003;370:1055–1062. doi: 10.1042/BJ20021699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos, I., Torrance, G.M., Gilbert, D.R., Westhead, D.R. TOPS: An enhanced database of protein structural topology. Nucleic Acids Res. 2004;32:D251–D254. doi: 10.1093/nar/gkh060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagano, N., Orengo, C.A., Thornton, J.M. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Payne, D.J., Miller, W.H., Berry, V., Brosky, J., Burgess, W.J., Chen, E., DeWolf W.E., Jr, Fosberry, A.P., Greenwood, R., Head, M.S., et al. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob. Agents Chemother. 2002;46:3118–3124. doi: 10.1128/AAC.46.10.3118-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G.N., Cohen, S., Perrakis, A., Noble, M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Quemard, A., Sacchettini, J.C., Dessen, A., Vilcheze, C., Bittman, R., Jacobs W.R., Jr, Blanchard, J.S. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis . Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- Rock, C.O., Cronan, J.E. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- Saito, J., Yamada, M., Watanabe, T., Kitagawa, H., Takeuchi, Y. Crystallization and preliminary X-ray analysis of enoyl–acyl carrier protein reductase (FabK) from Streptococcus pneumoniae . Acta Crystallograph. F Struct. Biol. Cryst. Commun. 2006;62:576–578. doi: 10.1107/S1744309106017039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeld, M.A., Miller, W.H., Newlander, K.A., Burgess, W.J., Payne, D.J., Rittenhouse, S.F., Moore, T.D., DeWolf W.E., Jr, Keller, P.M., Qiu, X., et al. Inhibitors of bacterial enoyl–acyl carrier protein reductase (FabI): 2,9-Disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents. Bioorg. Med. Chem. Lett. 2001;11:2241–2244. doi: 10.1016/s0960-894x(01)00405-x. [DOI] [PubMed] [Google Scholar]
- Seefeld, M.A., Miller, W.H., Newlander, K.A., Burgess, W.J., DeWolf W.E., Jr, Elkins, P.A., Head, M.S., Jakas, D.R., Janson, C.A., Keller, P.M., et al. Indole naphthyridinones as inhibitors of bacterial enoyl–ACP reductases FabI and FabK. J. Med. Chem. 2003;46:1627–1635. doi: 10.1021/jm0204035. [DOI] [PubMed] [Google Scholar]
- Slater-Radosti, C., Van Aller, G., Greenwood, R., Nicholas, R., Keller, P.M., DeWolf W.E., Jr, Fan, F., Payne, D.J., Jaworski, D.D. Biochemical and genetic characterization of the action of triclosan on Staphylococcus aureus . J. Antimicrob. Chemother. 2001;48:1–6. doi: 10.1093/jac/48.1.1. [DOI] [PubMed] [Google Scholar]
- Takahata, S., Iida, M., Osaki, Y., Saito, J., Kitagawa, H., Ozawa, T., Yoshida, T., Hoshiko, S. AG205, a novel agent directed against FabK of Streptococcus pneumoniae . Antimicrob. Agents Chemother. 2006;50:2869–2871. doi: 10.1128/AAC.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, S., Iida, M., Yoshida, T., Kumura, K., Kitagawa, H., Hoshiko, S. Discovery of 4-pyridone derivatives as specific inhibitors of enoyl–acyl carrier protein reductase (FabI) with antibacterial activity against Staphylococcus aureus . J. Antibiot. (Tokyo) 2007;60:123–128. doi: 10.1038/ja.2007.11. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2003a;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 2003b;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez, I., Pardo, J.P. Kinetic characterization of the rotenone-insensitive internal NADH: Ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae . Arch. Biochem. Biophys. 2001;389:7–14. doi: 10.1006/abbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- Yum, J.H., Kim, C.K., Yong, D., Lee, K., Chong, Y., Kim, C.M., Kim, J.M., Ro, S., Cho, J.M. In vitro activities of CG400549, a novel FabI inhibitor, against recent clinical staphylococcal isolates in Korea. Antimicrob. Agents Chemother. 2007;51:2591–2593. doi: 10.1128/AAC.01562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C.J., Sohn, M.J., Kim, W.G. Atromentin and leucomelone, the first inhibitors specific to enoyl–ACP reductase (FabK) of Streptococcus pneumoniae . J. Antibiot. (Tokyo) 2006;59:808–812. doi: 10.1038/ja.2006.108. [DOI] [PubMed] [Google Scholar]