Abstract
Renilla luciferase (RLUC) is a versatile tool for gene expression assays and in vivo biosensor applications, but its catalytic mechanism remains to be elucidated. RLUC is evolutionarily related to the α/β hydrolase family. Its closest known homologs are bacterial dehalogenases, raising the question of how a protein with a hydrolase fold can function as a decarboxylating oxygenase. Molecular docking simulations with the coelenterazine substrate against an RLUC homology model as well as a recently determined RLUC crystal structure were used to build hypotheses to identify functionally important residues, which were subsequently tested by site-directed mutagenesis, heterologous expression, and bioluminescence emission spectroscopy. The data highlighted two triads of residues that are critical for catalysis. The putative catalytic triad residues D120, E144, and H285 bear only limited resemblance to those found in the active site of aequorin, a coelenterazine-utilizing photoprotein, suggesting that the reaction scheme employed by RLUC differs substantially from the one established for aequorin. The role of H285 in catalysis was further supported by inhibition using diethylpyrocarbonate. Multiple substitutions of N53, W121, and P220—three other residues implicated in product binding in the homologous dehalogenase Sphingomonas LinB—also supported their involvement in catalysis. Together with luminescence spectra, our data lead us to propose that the conserved catalytic triad of RLUC is directly involved in the decarboxylation reaction of coelenterazine to produce bioluminescence, while the other active-site residues are used for binding of the substrate.
Keywords: coelenterazine, bioluminescence resonance energy transfer, α/β, hydrolase, dioxygenase
The blue-light emitting luciferase of the marine anthozoan Renilla reniformis (RLUC, E.C. number 1.13.12.5, luciferin-2-monooxygenase, decarboxylating) has become popular as a reporter enzyme for gene expression assays. RLUC also serves as an energy donor for protein interaction assays based on bioluminescence resonance energy transfer (BRET) (Xu et al. 1999; Subramanian et al. 2006). Attempts to improve the enzymatic properties by protein engineering are hampered by the fact that the enzymatic reaction mechanism of RLUC is not well understood, although mutagenesis approaches have nevertheless started to bear fruit (Loening et al. 2006; Hoshino et al. 2007). However, a better understanding of the enzymatic mechanism catalyzed by RLUC would be helpful. The substrate of RLUC, coelenterazine, is an aromatic imidazolo-pyrazinone, which is derivatized on three of its carbon ring atoms with p-hydroxy-phenyl (R1), benzyl (R2), and p-hydroxy-benzyl (R3) moieties. Coelenterazine is turned over in an oxidative decarboxylation reaction during which the imidazole ring is opened and carbon dioxide is released together with a photon of blue (470 nm) luminescence (Hori et al. 1973; Matthews et al. 1977a,b; Ohmiya and Hirano 1996).
Several high-resolution crystal structures in the presence or absence of the coelenterazine ligand have been solved for the calcium-stimulated photoproteins aequorin and obelin (Head et al. 2000; Liu et al. 2000; Vysotski et al. 2003; Deng et al. 2004). Here, coelenterazine reacts with molecular oxygen in a reaction coordinated by a catalytic triad of tyrosine (Y184 and Y190 in apo-aequorin and obelin, respectively), tryptophan (W173 and W179), and histidine (H169 and H175). The Y190 residue stabilizes the hydroperoxy group of the oxidized coelenterazine via a hydrogen bond. In the strict sense, the term “aequorin” refers to this complex between the apo-aequorin polypeptide and the reaction intermediate. Upon calcium binding, H175 shifts position, thus triggering a proton relay that deprotonates first Y190 and subsequently the hydroperoxy group of the reaction intermediate. The resulting peroxycoelenterazine anion then performs an intramolecular nucleophilic attack to form a dioxetanone ring. This highly unstable intermediate spontaneously decarboxylates, yielding one molecule of carbon dioxide and the electronically excited state of coelenteramide, which relaxes to the ground state by emission of a photon. Typically, the coelenteramide is thought to be deprotonated at the R1 hydroxyl group (phenolate anion), likely aided by a nearby histidine residue (H22). Under these conditions, photon emission will be in the blue range (470–490 nm) (Deng et al. 2004; Vysotski and Lee 2004), whereas if coelenteramide remains protonated, it emits a photon of purple light (405 nm) (Deng et al. 2004; Vysotski and Lee 2004; Liu et al. 2006).
Because RLUC uses the same substrate as apo-aequorin, yields the same products, and emits a photon with similar spectral characteristics, the reaction mechanisms are expected to be similar (Matthews et al. 1977a,b). Aequorin can also function as a calcium-dependent luciferase (Inouye and Sasaki 2007). However, RLUC itself is not calcium dependent. Therefore, RLUC may not possess a residue equivalent to Y190, the residue that stabilizes the hydroperoxy-coelenterazine intermediate in obelin. More importantly, RLUC and apo-aequorin are not homologous with each other. Instead, the RLUC oxygenase is clearly homologous to bacterial haloalkane dehalogenases of the LinB family (42% identity), thus joining the bacterial dioxygenases Hod and Qdo as an oxygenase derived from an α/β hydrolase ancestor (Frerichs-Deeken et al. 2004). Crystal structures of Sphingomonas paucimobilis LinB and related enzymes show the characteristic fold of the α/β hydrolase superfamily (Marek et al. 2000; Oakley et al. 2004). Dehalogenases hydrolyze the bond between carbon and halogen, thus producing inorganic halide ions and alcohols. In the catalytic triad of the haloalkane dehalogenase of Xanthobacter autotrophicus, whose mechanism has been well studied (for review, see Holmquist 2000), D124 functions as a nucleophile, H289 as a general base, and D260 as a catalytic acid. The substrate, 1,2-dichloroethane, binds to tryptophans 175 and 125 in the active site, followed by the attack of D124 on the carbon–halogen bond. The intermediate alkyl-enzyme complex is attacked by a nucleophilic water molecule that forms a hydrogen bond with H289 interacting with D260. The nucleophilic water attack results in a tetrahedral oxyanion intermediate, which decomposes, releasing a halogen and an alcohol molecule from the active site. The three equivalent catalytic triad residues in LinB are D108, H272, and E132 (Hynkova et al. 1999). They are conserved and functionally important in RLUC (Loening et al. 2006). Most recently, crystal structures of a stabilized form of RLUC carrying eight or 10 amino acid substitutions have been solved with and without the coelenteramide product (RLUC8) (Loening et al. 2007a). These structures confirm the overall α/β hydrolase fold and the arrangement of the putative catalytic triad residues at the bottom of the active-site cavity. Although the coelenteramide product is present in one of these structures (PDB 2PSJ), its position on the flank of the outer portion of the active site was interpreted as indicative of a nonproductive binding mode (Loening et al. 2007a).
In this work, docking simulations were performed with coelenterazine on the crystal structures of RLUC8 as well as a homology model of wild-type RLUC to make predictions about the positioning of the substrate in the active site and potential residues responsible for catalysis and spectral properties. These predictions were tested by site-directed mutagenesis and heterologous expression of recombinant RLUC, as well as using pharmacological inhibitors.
Results
In an attempt to build a hypothesis for how the native RLUC enzyme might position the coelenterazine substrate in preparation for catalysis, and prior to a crystal structure for RLUC, a homology model was generated using SwissModel ProModII (Schwede et al. 2003) based on the crystal structures of the bacterial haloalkane dehalogenase Sphingomonas paucimobilis LinB (PDB 1IZ8A, 1K6EA, 1IZ7A, 1MJ5A) (Oakley et al. 2004; Supplemental Fig. S1A). Protein sequence identity between LinB and RLUC is 42%. The root mean square deviation (RMSD) between the alpha carbon backbone, excluding loops, of the RLUC model and the LinB structure (262 out of 275 residues) was 0.34 Å, suggesting a highly parsimonious model. A very similar model was obtained using MOE software (Chemical Computing Group, Inc.). According to the homology model, the active site was made up of 26 residues, of which nine were aromatic, 17 hydrophobic, and 9 hydrophilic (Supplemental Fig. S1B). The outer portion of the active site was largely hydrophobic, while most of its more hydrophilic residues resided in the inner portion. The recently solved crystal structure of RLUC8 in the absence of product (Loening et al. 2007a) was also similar to our model (the RMSD with PDB 2PSD was 1.0 Å over 254 residues excluding several large surface loops), validating the strategy of using a homology model to guide the subsequent mutagenesis experiments. Our model, in line with the RLUC8 crystal structures, suggests that the putative catalytic triad residues—D120, E144, and H285—lie close together at the bottom of the active site in an almost identical spatial configuration as in LinB (Fig. 1A).
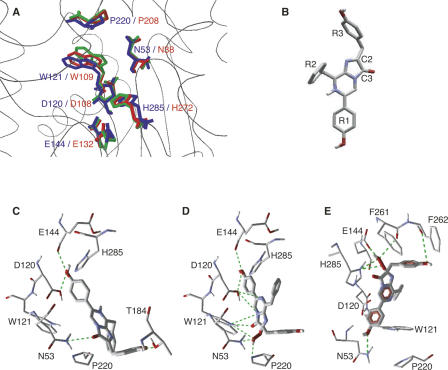
Figure 1.
RLUC protein structure displays and substrate docking simulations. (A) Overlay comparison between the catalytic triad and other active-site residues of Sphingomonas haloalkane dehalogenase, LinB (PDB IZ7A, red), the corresponding residues in the wild-type RLUC homology model (green), and the RLUC8 crystal structure (PDB 2PSD, blue) (Loening et al. 2007a). (B) Coelenterazine; oxygen and nitrogen are colored red and blue, respectively. Carbon and selected hydrogen are gray and white. (C–E) Docking simulations of coelenterazine to the lower portion of the RLUC active site including the putative catalytic triad. Hydrogen bonds are symbolized by green lines. (C,D) Docking of native coelenterazine (C) and the reaction intermediate, 2-hydroperoxy-coelenterazine (D) against the RLUC homology model. Note interactions between the hydroperoxy group and active-site residues N53, W121, and P220. (E) Docking of the reaction intermediate, 2-hydroperoxy-coelenterazine, was performed with the RLUC crystal structure obtained after exposure to substrate (PDB 2PSJ). In this alternative docking simulation, the reaction intermediate is suspended by hydrogen bonds between the R1 and R3 hydroxyls to N53 and the backbone of F262, respectively, while the reactive center is juxtaposed to the catalytic triad.
Docking simulations
The active site of apo-aequorin is highly hydrophobic and has three triads of tyrosine, histidine, and tryptophan. One of the triads functions as the catalytic triad, and two others are involved in substrate binding (Head et al. 2000). While the active site of RLUC is also substantially hydrophobic, it does not contain a triad of tyrosine, tryptophan, and histidine. To identify potential residues involved in the bioluminescence reaction, native coelenterazine (Fig. 1B) or the reaction intermediate, 2-hydroperoxy-coelenterazine, was docked against the homology model as well as the crystal structures. In the docking simulations with the homology model, the hydroxyl group on the R1 ring of coelenterazine interacted with both D120 and E144, while the hydroxyl group on the R3 ring was bound to T184 (Fig. 1C). When water was included in the docking simulation, the results were similar except that the R1 hydroxyl group bound to H285 via a water molecule (data not shown). Meanwhile, coelenterazine's reactive center, that is, the C2 carbon, which initially reacts with oxygen, and the C3 carbonyl, which is eliminated in the form of CO2, were docked in hydrogen-bonding distance to N53 (Fig. 1C). It is unclear whether the oxidation of coelenterazine occurs inside or outside of the active site. Likewise, the hydroperoxy group of the reaction intermediate had the potential to hydrogen bond with N53, W121, and P220 (Fig. 1D). Coincidentally, the same three residues are implicated in coordination of the eliminated halide anion in the LinB dehalogenase (Oakley et al. 2004). Similar results were obtained when 2-hydroperoxy-coelenterazine was docked to the crystal structure of RLUC8 that was obtained in the absence of substrate (PDB 2PSD and 2PSF) (data not shown). Crystal structures of RLUC8 differ substantially with respect to the width of the gateway (Supplemental Fig. S2; Loening et al. 2007a), a situation previously observed with the equivalent surface helix in the cap domain of LinB (Streltsov et al. 2003). When docking was performed with the crystal structure that was obtained after exposing RLUC8 to substrate (PDB 2PSJ), which has a wider gateway, most docking models showed the reactive center of coelenterazine juxtaposed to the putative catalytic triad residues, E144, and H285, as well as the backbone carbonyl of F261, while the R1 hydroxyl group interacted with N53 and the R3 hydroxyl group interacted with F262 (Fig. 1E). These different, yet highly reproducible docking models represent two distinct hypotheses concerning the roles of the putative catalytic triad consisting of D120, E144, and H285, on the one hand, and the triad consisting of N53, W121, and P220, on the other. In the following, these hypotheses were further tested by site-directed mutagenesis, drug inhibition, and scanning of emission spectra.
Inhibitor studies
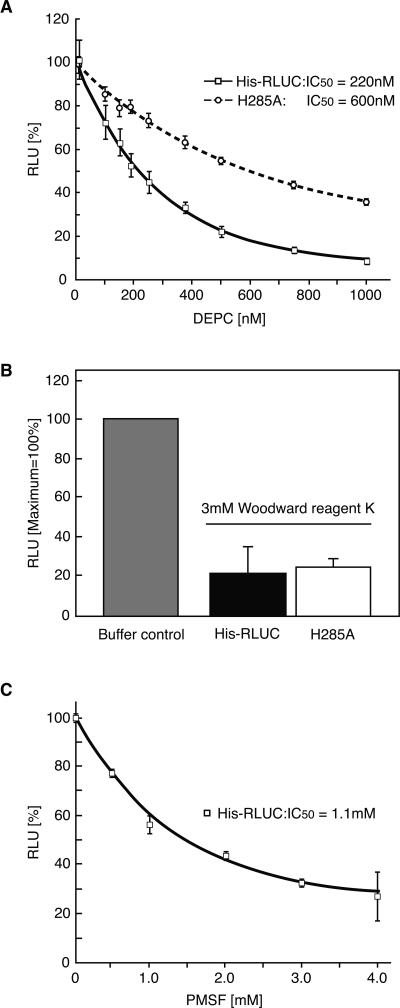
Several enzymes that rely on a catalytic histidine are sensitive to diethylpyrocarbonate treatment (DEPC), and H285 is the only histidine in the RLUC active site. RLUC enzyme purified from Escherichia coli was also inhibited by low concentrations of DEPC (IC50 at 2 μM substrate was 220 nM, kI was 160 nM), consistent with a role for H285 in RLUC catalysis (Fig. 2A). For comparison, RLUC was also sensitive to Woodward Reagent K (Fig. 2B). Because this reagent modifies acidic residues in a hydrophilic environment, the most likely target residue may be D162 (Supplemental Fig. S2). The serine/cysteine-reactive compound PMSF (phenylmethylsulfonylfluoride) also inhibited RLUC (Fig. 2C), perhaps by targeting S145 or S263, which lie near H285.
Figure 2.
Inhibitor studies of RLUC. (A) DEPC. (B) Woodward Reagent K. (C) PMSF. Error bars represent standard error from n = 3 repeats. Assays were performed with 10 nM RLUC, preincubated with inhibitor at the indicated concentration for 30 min. The substrate concentration was 2 μM. In A, the activity of each protein in the absence of inhibitor was normalized to the peak value for ease of comparison. The H285A mutant has 11% of wild-type activity (Table 1).
Site-directed mutagenesis of the putative catalytic triad
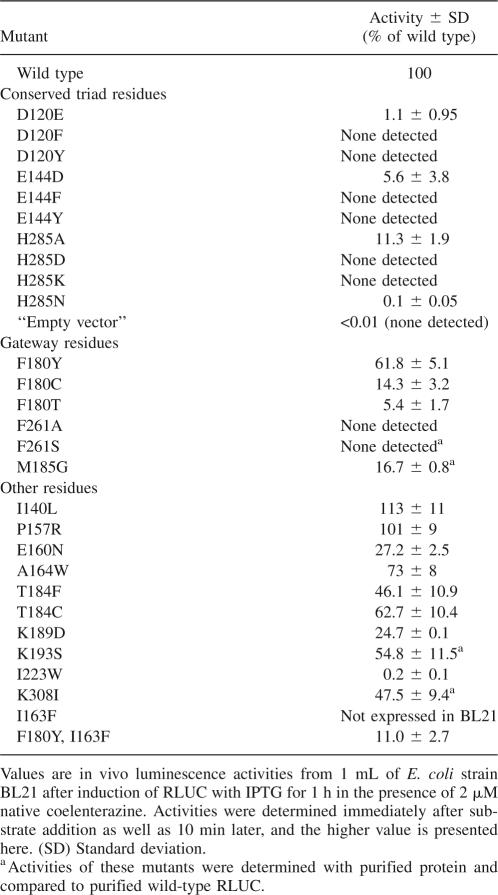
Based on the docking simulations, two sets of catalytic triads were postulated (Fig. 1D,E). One consists of D120, E144, and H285; the other of N53, W121, and P220. Residues D120, E144, and H285 were subjected to individual site-directed mutagenesis. Residue D120 represents the “nucleophile elbow” (Holmquist 2000) in the catalytic triad of the α/β hydrolase fold. When expressed in E. coli, D120E retained only 1% of wild-type activity, and more drastic changes to F or Y caused complete inactivation. Tyrosine was chosen because the catalytic triad of apo-aequorin contains a critical tyrosine residue. Likewise, for E144, the conservative change, E144D, had low activity, while changes to bulky aromatic side chains caused complete inactivation (Table 1). For comparison, mutation of another acidic residue near the active site, E160N, retained substantial activity. Taken together, and confirming similar mutagenesis results presented elsewhere (Loening et al. 2006), D120 and E144 must play important roles in catalysis.
Table 1.
RLUC enzyme activity of site-directed mutants
Mutations of H285 to D, K, or N caused complete or nearly complete inactivation of RLUC (Table 1). However, the H285A mutation of RLUC retained partial activity. The H285A mutant protein was also less sensitive to DEPC (Fig. 2A), underscoring that H285 is the residue most sensitive to DEPC inactivation and is important for efficient catalysis. In the hydrolases, the equivalent histidine often functions as an essential general base. For example, a similar H-to-A substitution in the LinB hydrolase is completely inactive (Hynkova et al. 1999). Aequorin, which is not related to hydrolases, also possesses a catalytic histidine (H169), whose mutation to alanine reduces activity to 1% (Ohmiya and Tsuji 1993).
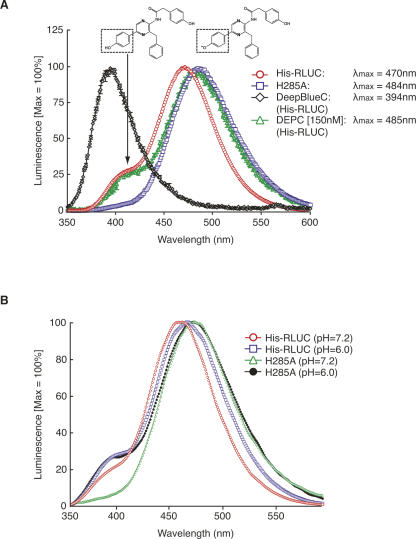
In the coelenterazine-utilizing luciferases as well as in the photoproteins obelin and aequorin, light emission at 470 nm is usually attributed to a negatively charged phenolate anion in which the p-hydroxyl on the R1 ring of coelenteramide is deprotonated, while emission at 400 nm is attributed to the neutral coelenteramide (Hart et al. 1979; Shimomura 1995; Ohmiya and Hirano 1996; Vysotski and Lee 2004). RLUC luminesces at 470 nm but also displays a weak shoulder near 400 nm (Matthews et al. 1977a). The substrate analog bisdehydroxycoelenterazine (tradename DeepBlueC) lacks the p-hydroxyl on the R1 ring and therefore cannot form a phenolate anion. Accordingly, bisdehydroxycoelenterazine luminesces with a peak near 400 nm (Fig. 3A; Hart et al. 1979). In the Ca2+-discharged aequorin, deprotonation on the R1 ring is mediated by one of the histidine/tryptophan/tyrosine triads (Vysotski et al. 2003). Aequorin generally lacks a 400-nm shoulder, but aequorin with a Y82F mutation does have a shoulder, which is attributed to less efficient deprotonation when F replaces Y82. Vice versa, obelin, which naturally carries F at the equivalent position, has a 400-nm shoulder, but the F88Y mutant does not (Stepanyuk et al. 2005). According to the hypothetical substrate configuration in Figure 1, C and D, H285 or other residues nearby might play a role as a proton acceptor for the R1 hydroxyl, thus ensuring formation of a phenolate that emits at 470 nm. This hypothesis predicts that nonpolar substitutions at these positions should enhance the 400-nm shoulder in RLUC. However, on the contrary, the H285A mutation displayed a loss of the shoulder (Fig. 3A). Similar results were obtained for the weak luminescence of E144D and D120E (data not shown). If H285 controlled the protonation state of the R1 hydroxyl, then its reaction with DEPC might enhance the 400-nm shoulder, but this was not observed (Fig. 3A). Moreover, one would predict that a pH below the pKa of histidine would favor protonation of the coelenteramide and thus increase the 400-nm shoulder; this trend was observed in the H285A mutant but not in the wild type, where it would have been expected (Fig. 3B). Taken together, although the model of coelenterazine with its R1 hydroxyl bound to the catalytic triad is typical in docking simulations with both our homology model as well as the crystal structure obtained in the absence of substrate (PDB 2PSD), the spectroluminescence data are difficult to reconcile with the docking simulation postulated in Figure 1C.
Figure 3.
Luminescence spectra. (A) Wild-type His-RLUC (10 nM, 2 μM native coelenterazine or 40 nM with 4 μM DeepBlueC) was compared with the H285A mutant (100 nM, 3.1 μM native coelenterazine). Samples were scanned in triplicate from 350 nm to 600 nm at 1 nm/sec and normalized to peak at 100%. Note that the spectra are distorted because of the loss of enzyme activity over the time of the scan (∼5 min), a necessary condition for highlighting the emission spectrum around 400 nm. The presumptive structures underlying emission at 400 nm (neutral coelenteramide) and 470 nm (phenolate anion) are shown. Control experiments were performed with wild-type His-RLUC by initiating the scan at different wavelengths to confirm that the shoulder at 400 nm is not specific to the early phase of the luminescence reaction (supplemental Fig. S3). (B) Wild-type RLUC (His-RLUC) and the H285A mutant RLUC with 2 μM native coelenterazine were scanned at pH 7.2 and pH 6.0.
Site-directed mutagenesis of the triad N53, W121, and P220
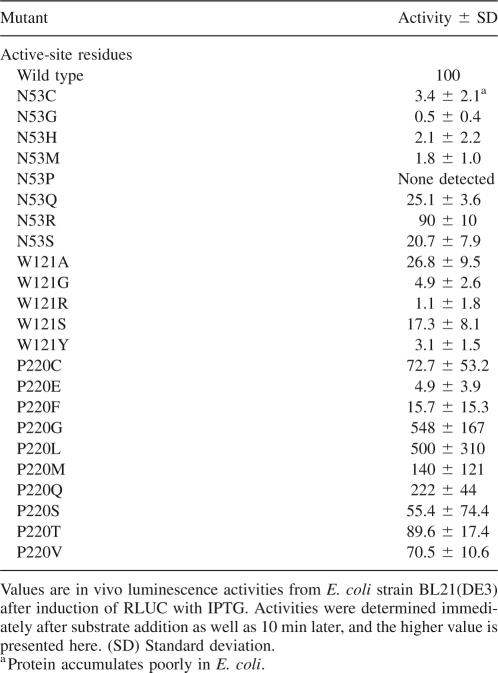
Site-directed mutagenesis was performed on residues N53, W121, and P220 with a randomized oligonucleotide with the goal of distinguishing whether these residues might interact with the reactive center of the substrate (Fig. 1C,D) or the p-hydroxylated R1 ring (Fig. 1E). For N53, the eight different substitutions tested caused varying loss of activity (R > Q,S > C,H,M > G > P) (Table 2) anywhere between 0% of wild type (N53P) and 90% (N53R). Here, we briefly mention that almost all RLUC mutants described in this study accumulated to the same level after induction in E. coli. However, N53C accumulated exceptionally poorly (Fig. 4A; data not shown). RLUC mutants were generally examined in E. coli in vivo, because the relative activities in vivo generally matched the activities obtained after purification of the His-tagged enzyme by nickel-affinity chromatography (Fig. 5). If N53 functioned as a hydrogen-bonding partner for the R1 hydroxyl as postulated in Figure 1E, then other hydrophilic residues, especially a basic one, might be able to substitute partially, as was, indeed, observed. Furthermore, when the emission spectra of N53M (poor activity) and N53R (high activity) were compared with the wild-type enzyme, N53M had an enhanced shoulder at <400 nm and a blueshifted emission maximum compared with N53R and His-RLUC. This result is consistent with the notion established with the Ca2+-discharged aequorin that a less polar environment around the R1 ring causes more of the coelenteramide product to adopt the neutral state, which emits at 400 nm. Specifically, in the Ca2+-discharged aequorin, a W86F mutation caused reduced luminescence, a strong shoulder at 400 nm, and blueshifted emission (Ohmiya et al. 1992), which was attributed to the lack of a hydrogen bond between the R1 hydroxyl and F86. No such spectral shift was observed when W129 or W179, which interact with the reactive imidazolopyrazinone ring, were changed to phenylalanine (Ohmiya et al. 1992; Head et al. 2000).
Table 2.
Activities of active-site mutants N53, W121, and P220
Figure 4.
Expression levels and luminescence spectra of representative mutant RLUC proteins. (A) Coomassie blue-stained polyacrylamide gel demonstrating equal accumulation of wild-type RLUC and several representative RLUC mutant proteins after 1 h of induction of expression with IPTG. The N53C mutant is a rare exception. (Right) A dilution series of RLUC extract in empty vector extract (EV). The arrow points to recombinant RLUC. (B) Luminescence spectra of two mutations affecting residue N53. The conditions are as for Figure 3.
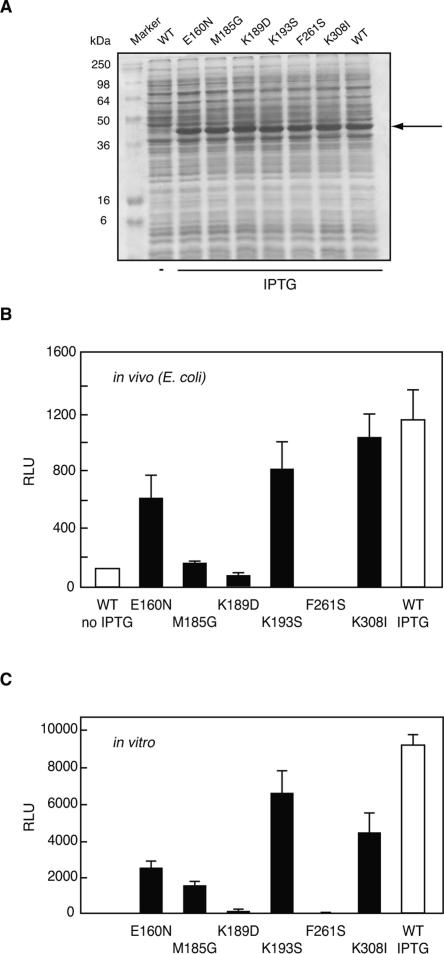
Figure 5.
Enzyme activities (relative light units) for wild-type RLUC and selected mutants. (A) Polyacrylamide gel for the RLUC expression strains shown in B. (B) Values are in vivo luminescence activities from E. coli strain BL21 after induction of RLUC with IPTG. (C) Values are in vitro luminescence activities of purified proteins in the presence of coelenterazine substrate. Note the similarity between the activity profiles (B) in vivo and (C) in vitro.
Each of five substitutions at W121 (A, G, R, S, and Y) caused between 80% (W121S) and 99% (W121R) loss of activity. The loss of activity with W121R compared to N53R might point to a role for W121 in guiding the R1 hydroxyl to interact with the N53 residue. The W121R mutation may hamper the proper positioning of the substrate in the active site. For comparison, 10 different substitutions for P220 showed a wide range of activities (Table 2), generally indicating that other small or medium-sized residues were tolerated or even beneficial at this position, but glutamic acid and phenylalanine were detrimental. Other properties of the P220 mutants will be described elsewhere (J.C. Woo and A.G. von Arnim, in prep.).
Aside from mutations affecting the lower portion of the active site, we also examined several residues thought to compose the entrance to the active site (gateway) (listed in Table 1). Among these, F180, which resides at the rim of the active site in all structures including our homology model, was sensitive to mutation, suggesting that its strongly hydrophobic character might play a role in initial binding of the substrate. Alteration of F261 to serine or alanine completely disrupted RLUC activity, consistent with similar experiments presented elsewhere (Loening et al. 2006, 2007b). Several additional mutations are presented for the record. Taken together, the active-site mutagenesis data support the view that the oxygenase activity of RLUC relies on the catalytic triad inherited from a hydrolase ancestor and suggest that a pocket of N53 and W121 has been co-opted to function in substrate binding.
Discussion
In conclusion, the combined data from molecular docking simulations, pharmacological inhibitors, site-directed mutagenesis, and luminescence spectroscopy extend previously published structure–function results (Loening et al. 2006) and suggest that the catalytic mechanism used by RLUC must differ substantially from the one established for the aequorin family. Although both of their active sites are rich in aromatic residues, RLUC does not possess a spatial arrangement of catalytic triad residues analogous to that in aequorin. Instead, several active-site residues in the evolutionarily related dehalogenase LinB, including those forming the catalytic triad (D120, E144, and H285 in RLUC) and also N53, W121, and P220, are completely conserved, including in their spatial arrangement. Meanwhile, the great majority of RLUC's gateway residues (Supplemental Fig. S2) are different in LinB (data not shown) and show considerable flexibility between the homology model and the various crystal structures.
In the calcium-stimulated photoproteins, the oxidative decarboxylation reaction uses histidine as a general base. We propose that H285 functions in a similar capacity in RLUC, for the following reasons. First, RLUC was inactivated by the histidine-reactive compound, DEPC (Fig. 2A), and H285A was more resistant to DEPC compared to wild type. The residual sensitivity of the H285A mutant to DEPC clearly shows that other functionally important histidines are also being modified. Interestingly, four out of eight histidines in RLUC (H119, H128, H133, and H142) lie near the two acidic catalytic residues (D120 and E144), and although neither is exposed to the active-site cavity, their reaction with DEPC might contribute to the residual inhibition of the H285A mutant by DEPC. Second, the drop in RLUC activity below the pH optimum at 7.2 (Matthews et al. 1977b) is consistent with a titratable histidine. Third, mutation of H285 to most other residues caused loss of activity, as did mutation of D120 and E144. And finally, molecular docking simulations suggested that RLUC can suspend the substrate on its R1 and R3 rings using hydrogen bonds to defined residues, possibly N53 and F262, in such a fashion that the reactive center of coelenterazine with its C2 and C3 ring atoms become juxtaposed to H285 and the remaining catalytic triad residues. Such a docking model was also plausible for the reaction intermediate, 2-hydroperoxy-coelenterazine (Fig. 1E). Ancillary evidence is that RLUC is expected to bind the R1 hydroxyl group in such a way as to facilitate its deprotonation into a phenolate anion, which gives rise to the emission peak at around 470 nm. While H285 could potentially perform this role, the presence of two acidic residues nearby does not make this scenario very likely, and the luminescence spectrum of the H285A mutant was inconsistent with such a role (Fig. 3). If, as seems more likely, the R1 hydroxyl of coelenterazine is bound by N53, it is unclear which residue causes its deprotonation in the majority of cases or whether coelenteramide may luminesce in its amide anion form (Shimomura 1995) rather than as a phenolate anion, a hypothesis that does not require a proton acceptor near the R1.
That said, certain considerations potentially support the argument against the notion that H285 forms a component of the catalytic triad. For one, the H285A mutant retains ∼11% of activity, suggesting that another residue can substitute as a base, if poorly. However, the Ca2+-discharged aequorin with an H-to-A mutation also retains low but detectable activity, around 1% of wild type (Ohmiya and Tsuji 1993). Another consideration is that one X-ray structure in the presence of the reaction product, coelenteramide, shows the reaction product near the surface of the active site rather than engaged with the proposed catalytic triad (PDB 2PSJ). However, Loening and coworkers already pointed out that this position may represent a nonproductive binding mode. For example, it does not explain why active-site residues such as D120, E144, W121, and N53 are important for function and, moreover, why mutations that ought to disrupt the hydrogen bonds with coelenteramide in the surface position had only mild effects on enzymatic activity (Loening et al. 2007a). Moreover, our spectroscopic results (Fig. 3) were difficult to reconcile with a hydrogen bond between the R1 hydroxyl and H285, a feature that the reaction product in 2PSJ shares with our disfavored docking model (Fig. 1C).
Finally, the only crystal structure that allowed coelenterazine's reactive center to be docked to the catalytic triad was PDB 2PSJ (with coelenteramide removed) (Fig. 1E; Loening et al. 2007a). This conformation has a wide gateway and a bowl-shaped active site, with the side chain of F262 flipped out of the active site, thus exposing H285 and E144 (Supplemental Fig. S2), while another conformation (PDB 2PSD) possesses a narrow gateway and a vase-shaped active site, in which the side chains of both F262 and W156 are packed into the active-site cavity to cover E144 and H285. Although the structure with the bowl-shaped active site appears consistent with our model, whether coelenterazine encounters RLUC with a vase-shaped or a bowl-shaped active site is unknown. However, it would be premature to argue that the only conformation of RLUC able to engage the substrate as predicted by our model is a conformation that can only be seen once RLUC has reacted with the substrate. Instead, we point out that the structure in 2PSF, which was obtained without substrate, does have a fully open gateway, although F262 is still flipped into the active site. These results point to considerable flexibility in the conformation of RLUC, some of which may be driven by substrate binding. In summary, while additional work is needed to clarify the mechanism of action of Renilla luciferase, there is now a framework on which additional mutagenesis of the enzyme with the goal of enhancing its activity in heterologous expression systems can be conducted. BRET applications, in particular, would benefit from such improvements, in the interest of reducing protein expression levels of RLUC-tagged proteins, shortening measurement times, and improving spatial resolution, especially for BRET imaging (Xu et al. 2007; Coulon et al. 2008). In this regard, although most mutations studied here compromised enzymatic activity (Table 1), both P220G and P220L are of interest because their in vivo activities were elevated fivefold over wild type after 10 min of incubation with the substrate (Table 2). The utility of these and other mutations for enhancing RLUC activity is currently under investigation.
Materials and Methods
Site-directed mutagenesis and other recombinant DNA techniques
The wild-type Renilla reniformis luciferase cDNA obtained from plasmid pBS-35S:RLUC-attR (AY995136) (Subramanian et al. 2006) was subcloned into the expression vector pET30(a) as an NcoI–BamHI fragment, thus adding an N-terminal histidine tag and linker sequence (His-RLUC). Site-directed mutagenesis was performed using the QuikChange procedure (Stratagene). The E. coli strain was BL21(DE3). Mutations were confirmed by DNA sequencing, including resequencing of the entire RLUC coding region to guard against unintended secondary mutations. For several codon positions, mutagenesis was performed using a mutagenic oligonucleotide pool in which all four bases were allowed at all three codon positions (site-directed random mutagenesis). In this case, approximately 90 E. coli transformants were picked from the library and grown in LB in a white 96-well microtiter plate (Packard) to an optical density of ∼1. Colonies were pre-screened for RLUC activity in vivo by adding 2 μM coelenterazine to the culture medium and mixing; luminescence was recorded in a PolarStar luminescence microplate reader for 3 sec per well (BMG Labtech) alongside the original wild-type RLUC strain and non-transformed E. coli. Colonies representing high, medium, and low RLUC levels were saved and the mutations identified by DNA sequencing.
Docking simulations
For the docking simulations, molecular structures of native coelenterazine and 2-hydroperoxy-coelenterazine were drawn by the MOE program (Chemical Computing Group, Inc.). The homology model of RLUC and the crystallographic structures (PDB 2PSD, 2PSJ) were used for multiple docking simulations using the MOLEGRO/MolDock program (Thomsen and Christensen 2006). The volume of the active site was estimated by this program with the 1.6 Å probe. Generally, between 10 and 100 individual docking simulations were performed, and every possible docking model was analyzed by MolDock scores and hydrogen bonds. After multiple docking simulations, consistent docking models among the best-fitted models were used for the development of the hypotheses.
Expression and purification of RLUC
RLUC expression in E. coli strain BL21(DE3)pLysS was induced with 1 mM IPTG for 3 h at 30°C. The accumulation of RLUC in E. coli was routinely checked by cell lysis and gel electrophoresis and Coomassie blue staining. With just two exceptions (N53C and I163F), all site-directed mutants tested accumulated to similar levels. RLUC was purified from the soluble cytosolic fraction over a nickel column (His-Bind Kit; Novagen) following standard procedures that included sonication, centrifugation of cell debris at 12,000 rpm for 10 min at 4°C, and filtration of the supernatant through a 0.45-micron filter to prevent clogging of resin. Protein was affinity-purified according to the manufacturer's protocol and eluted with 1 M imidazole (1 M imidazole, 0.5 M NaCl, 20 mM Tris-Hcl, pH 7.9). After elution, RLUC was dialyzed overnight against 2 L of phosphate-buffered saline (PBS, pH 7.2) in order to remove inhibitory imidazole (Inouye and Sasaki 2007). Protein concentration was determined using the BCA assay (Pierce) with BSA as a standard. Alternatively, the protein concentration of preparations that were free of imidazole was measured by UV absorbance using an extinction coefficient of 65,040 M−1 cm−1 (Mach et al. 1992). Purified RLUC protein was stored in PBS buffer with 50% glycerol at −70°C in small aliquots or stored at 4°C for up to 2 wk.
Kinetics of RLUC enzyme activity
Enzyme assays were conducted using freshly purified RLUC enzyme at a concentration of 1 nM or as otherwise indicated in 1 mL of PBS (pH 7.2). Native coelenterazine substrate was added from a 250× stock solution of the indicated concentration in ethanol (final ethanol concentration, 0.8%), the solution was mixed by tapping to ensure a maximal supply of oxygen, and the luminescence activity was recorded in the TD20/20 luminometer (Turnerbiosystems). Because luciferase activity drops sharply over time, the first 5-sec luminescence reading was taken as a measure of enzyme activity.
Drug inhibition
Diethyl pyrocarbonate (DEPC) (Sigma) and phenylmethanesulfonyl fluoride (PMSF) (Sigma) were dissolved in ethanol and isopropyl alcohol, respectively. N-ethyl-5-phenylisoxazolium-3′-sulphonate (Woodward's reagent K) (Sigma) was added in 50 mM HEPES (pH 6.0). All inhibitors were prepared freshly before use. Ten nM purified protein was incubated with the indicated inhibitor for 30 min at room temperature, and then the LUC activity was measured in the TD20/20. The indicated substrate was added just before the measurement.
Emission spectra
Every bioluminescence spectrum was recorded under the same condition as the enzyme assay using a spectroluminometer (Photon Technology International, Inc.), except that the assay volume was 2 mL. The native coelenterazine substrate was 2 μM (ethanol concentration, 0.8%). The protein concentration was 10 nM purified enzyme or as otherwise indicated. Generally, the emission spectrum was analyzed with the Felix32 software (Photon Technology International, Inc.). All spectra were recorded at 1 nm/sec.
Acknowledgments
This work was supported by NSF grants MCB-0114653 and DBI-0619631 and by University of Tennessee Research Incentive Funds in support of the purchase of a spectroluminometer and a microplate reader. We are grateful for enzymological advice from Liz Howell and additional comments on the manuscript by Byung-Hoon Kim. We thank Tim Sparer for access to the spectroluminometer, the MolDock staff for access to software, and A. Loening and S.S. Gambhir for communicating results prior to publication.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Albrecht G. von Arnim, Department of Biochemistry and Cellular and Molecular Biology, M407 Walters Life Sciences, University of Tennessee, Knoxville, TN 37996-0840, USA; e-mail: vonarnim@utk.edu; fax: (865) 974-6306.
Abbreviations: RLU, relative light units; RLUC, Renilla luciferase.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073355508.
References
- Coulon, V., Audet, M., Homburger, V., Bockaert, J., Fagni, L., Bouvier, M., Perroy, J. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys. J. 2008;94:1001–1009. doi: 10.1529/biophysj.107.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., Markova, S.V., Vysotski, E.S., Liu, Z.J., Lee, J., Rose, J., Wang, B.C. Crystal structure of a Ca2+-discharged photoprotein: Implications for mechanisms of the calcium trigger and bioluminescence. J. Biol. Chem. 2004;279:33647–33652. doi: 10.1074/jbc.M402427200. [DOI] [PubMed] [Google Scholar]
- Frerichs-Deeken, U., Ranguelova, K., Kappl, R., Huttermann, J., Fetzner, S. Dioxygenases without requirement for cofactors and their chemical model reaction: Compulsory order ternary complex mechanism of 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase involving general base catalysis by histidine 251 and single-electron oxidation of the substrate dianion. Biochemistry. 2004;43:14485–14499. doi: 10.1021/bi048735u. [DOI] [PubMed] [Google Scholar]
- Hart, R.C., Matthews, J.C., Hori, K., Cormier, M.J. Renilla reniformis bioluminescence: Luciferase-catalyzed production of non-radiating excited states from luciferin analogues and elucidation of the excited state species involved in energy transfer to Renilla green fluorescent protein. Biochemistry. 1979;18:2204–2210. doi: 10.1021/bi00578a011. [DOI] [PubMed] [Google Scholar]
- Head, J.F., Inouye, S., Teranishi, K., Shimomura, O. The crystal structure of the photoprotein aequorin at 2.3 Å resolution. Nature. 2000;405:372–376. doi: 10.1038/35012659. [DOI] [PubMed] [Google Scholar]
- Holmquist, M. α/β-Hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Hori, K., Wampler, J.E., Matthews, J.C., Cormier, M.J. Identification of the product excited states during the chemiluminescent and bioluminescent oxidation of Renilla (sea pansy) luciferin and certain of its analogs. Biochemistry. 1973;12:4463–4468. doi: 10.1021/bi00746a025. [DOI] [PubMed] [Google Scholar]
- Hoshino, H., Nakajima, Y., Ohmiya, Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nat. Methods. 2007;4:637–639. doi: 10.1038/nmeth1069. [DOI] [PubMed] [Google Scholar]
- Hynkova, K., Nagata, Y., Takagi, M., Damborsky, J. Identification of the catalytic triad in the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. FEBS Lett. 1999;446:177–181. doi: 10.1016/s0014-5793(99)00199-4. [DOI] [PubMed] [Google Scholar]
- Inouye, S., Sasaki, S. Imidazole-assisted catalysis of luminescence reaction in blue fluorescent protein from the photoprotein aequorin. Biochem. Biophys. Res. Commun. 2007;354:650–655. doi: 10.1016/j.bbrc.2006.12.233. [DOI] [PubMed] [Google Scholar]
- Liu, Z.J., Vysotski, E.S., Chen, C.J., Rose, J.P., Lee, J., Wang, B.C. Structure of the Ca2+-regulated photoprotein obelin at 1.7 Å resolution determined directly from its sulfur substructure. Protein Sci. 2000;9:2085–2093. doi: 10.1110/ps.9.11.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.J., Stepanyuk, G.A., Vysotski, E.S., Lee, J., Markova, S.V., Malikova, N.P., Wang, B.C. Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state. Proc. Natl. Acad. Sci. 2006;103:2570–2575. doi: 10.1073/pnas.0511142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening, A.M., Fenn, T.D., Wu, A.M., Gambhir, S.S. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- Loening, A.M., Fenn, T.D., Gambhir, S.S. Crystal structures of the luciferase and green fluorescent protein from Renilla reniformis . J. Mol. Biol. 2007a;374:1017–1028. doi: 10.1016/j.jmb.2007.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening, A.M., Wu, A.M., Gambhir, S.S. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods. 2007b;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- Mach, H., Middaugh, C.R., Lewis, R.V. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal. Biochem. 1992;200:74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]
- Marek, J., Vevodova, J., Smatanova, I.K., Nagata, Y., Svensson, L.A., Newman, J., Takagi, M., Damborsky, J. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry. 2000;39:14082–14086. doi: 10.1021/bi001539c. [DOI] [PubMed] [Google Scholar]
- Matthews, J.C., Hori, K., Cormier, M.J. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977a;16:85–91. doi: 10.1021/bi00620a014. [DOI] [PubMed] [Google Scholar]
- Matthews, J.C., Hori, K., Cormier, M.J. Substrate and substrate analogue binding properties of Renilla luciferase. Biochemistry. 1977b;16:5217–5220. doi: 10.1021/bi00643a009. [DOI] [PubMed] [Google Scholar]
- Oakley, A.J., Klvana, M., Otyepka, M., Nagata, Y., Wilce, M.C., Damborsky, J. Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 Å resolution: Dynamics of catalytic residues. Biochemistry. 2004;43:870–878. doi: 10.1021/bi034748g. [DOI] [PubMed] [Google Scholar]
- Ohmiya, Y., Tsuji, F.I. Bioluminescence of the Ca2+-binding photoprotein, aequorin, after histidine modification. FEBS Lett. 1993;320:267–270. doi: 10.1016/0014-5793(93)80600-y. [DOI] [PubMed] [Google Scholar]
- Ohmiya, Y., Hirano, T. Shining the light: The mechanism of the bioluminescence reaction of calcium-binding photoproteins. Chem. Biol. 1996;3:337–347. doi: 10.1016/s1074-5521(96)90116-7. [DOI] [PubMed] [Google Scholar]
- Ohmiya, Y., Ohashi, M., Tsuji, F.I. Two excited states in aequorin bioluminescence induced by tryptophan modification. FEBS Lett. 1992;301:197–201. doi: 10.1016/0014-5793(92)81247-j. [DOI] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura, O. Cause of spectral variation in the luminescence of semisynthetic aequorins. Biochem. J. 1995;306:537–543. doi: 10.1042/bj3060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyuk, G.A., Golz, S., Markova, S.V., Frank, L.A., Lee, J., Vysotski, E.S. Interchange of aequorin and obelin bioluminescence color is determined by substitution of one active site residue of each photoprotein. FEBS Lett. 2005;579:1008–1014. doi: 10.1016/j.febslet.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Streltsov, V.A., Prokop, Z., Damborsky, J., Nagata, Y., Oakley, A., Wilce, M.C. Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates. Biochemistry. 2003;42:10104–10112. doi: 10.1021/bi027280a. [DOI] [PubMed] [Google Scholar]
- Subramanian, C., Woo, J., Cai, X., Xu, X., Servick, S., Johnson, C.H., Nebenfuhr, A., von Arnim, A.G. A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET) Plant J. 2006;48:138–152. doi: 10.1111/j.1365-313X.2006.02851.x. [DOI] [PubMed] [Google Scholar]
- Thomsen, R., Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- Vysotski, E.S., Lee, J. Ca2+-regulated photoproteins: Structural insight into the bioluminescence mechanism. Acc. Chem. Res. 2004;37:405–415. doi: 10.1021/ar0400037. [DOI] [PubMed] [Google Scholar]
- Vysotski, E.S., Liu, Z.J., Markova, S.V., Blinks, J.R., Deng, L., Frank, L.A., Herko, M., Malikova, N.P., Rose, J.P., Wang, B.C., et al. Violet bioluminescence and fast kinetics from W92F obelin: Structure-based proposals for the bioluminescence triggering and the identification of the emitting species. Biochemistry. 2003;42:6013–6024. doi: 10.1021/bi027258h. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Piston, D.W., Johnson, C.H. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Soutto, M., Xie, Q., Servick, S., Subramanian, C., von Arnim, A.G., Johnson, C.H. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc. Natl. Acad. Sci. 2007;104:10264–10269. doi: 10.1073/pnas.0701987104. [DOI] [PMC free article] [PubMed] [Google Scholar]