Abstract
Enhanced long chain fatty acid synthesis may occur in breast cancer, where it is necessary for tumor growth and predicts a poor prognosis. “Spot 14” (S14) is a carbohydrate- and thyroid hormone-inducible nuclear protein specific to liver, adipose, and lactating mammary tissues that functions to activate genes encoding the enzymes of fatty acid synthesis. Amplification of chromosome region 11q13, where the S14 gene (THRSP) resides, also predicts a poor prognosis in breast tumors. We localized the S14 gene between markers D11S906 and D11S937, at the telomeric end of the amplified region at 11q13, and found that it was amplified and expressed in breast cancer-derived cell lines. Moreover, concordant expression of S14 and a key lipogenic enzyme (acetyl-CoA carboxylase) in a panel of primary breast cancer specimens strongly supported a role for S14 as a determinant of tumor lipid metabolism. S14 expression provides a pathophysiological link between two prognostic indicators in breast cancer: enhanced lipogenesis and 11q13 amplification.
“Spot 14” (S14) is a small (≈17 kDa), acidic (pI 4.9) protein with no similarity to other mammalian gene products. The S14 gene is specifically expressed in tissues that produce lipids for use as a metabolic fuel, including lactating mammary, white and brown adipose tissue, and liver (1). In liver, its expression is rapidly induced by stimuli for increased long chain fatty acid synthesis, including thyroid hormone, glucose, and insulin (1), and is inhibited by glucagon and other stimuli that reduce lipogenesis (2). These observations, coupled with immunohistochemical localization of S14 to the nucleus, prompted the hypothesis that S14 functioned in the tissue-specific regulation of genes encoding the enzymes of lipid synthesis (3). This concept was supported by experiments using rat hepatocytes in primary culture that were treated with a S14 antisense oligonucleotide during exposure to the lipogenic stimuli of glucose and thyroid hormone (4). Antisense-mediated disruption of S14 induction inhibited the induction of lipogenesis. This result was explained by impaired expression of the lipogenic enzymes and their respective mRNAs. Transfection studies showed that this effect was attributable to reduced transcription (5).
The role of S14 in the tissue-specific transduction of hormonal- and diet-induced signals for activation of genes involved in lipogenesis raised the hypothesis that aberrant S14 expression is important in conditions with abnormal lipid metabolism. One such condition is breast cancer. Overexpression of fatty acid synthase (FAS), a major enzyme of fatty acid biosynthesis, is a marker for poor prognosis in breast tumors (6, 7). FAS is also important for tumor metabolism and growth. Pharmacological inhibition of fatty acid synthesis in breast cancer cells with FAS overexpression caused reduced growth and programmed cell death, an effect that was reversed by addition of palmitic acid to the growth media (8, 9). Increased FAS expression also occurs in primary breast tumors (9). The mechanism underlying enhanced expression of lipogenic enzymes in the tumors is unknown.
We localized the human S14 gene (THRSP) to the long arm of chromosome 11 (11q13.5) (10), which has been confirmed independently (11). Amplification at 11q13 occurs in ≈20% of breast cancers, and this amplification predicts a poor prognosis (12, 13). It is believed that 11q13 amplification results in overexpression of genes that are critical for tumor growth. We therefore considered the possibility that the S14 gene is amplified and overexpressed in breast tumors and that this could result in increased tumor lipogenesis. Currently, cyclin D1 (CCND1) and EMS1 are the only genes known to be both amplified and expressed in breast tumors with 11q13 amplification (reviewed in ref. 14). Cyclin D1 is a nuclear protein that regulates the transition from the G1 to the S phase of the cell cycle and is an obvious candidate oncogene. Transgenic mice with constitutive CCND1 overexpression exhibited mammary neoplasia (15). EMS1 encodes a c-src substrate associated with adherence of tumor cells to the extracellular substratum (16, 17). It has been speculated that this protein could mediate interactions with the extracellular matrix that may be important for metastasis (14). Several features of the 11q13 amplicon have raised the possibility that other genes important to tumor growth must be expressed from it. First, the 11q13 amplicon is consistently large (reportedly up to 5 Mbp) (18, 19). Second, amplification at 11q13 is discontinuous (20, 21). Third, some breast cancer cell lines with 11q13 amplification do not exhibit overexpression of CCND1 (22). Fourth, overexpression of CCND1 in stably transfected human mammary epithelial cells (23) or diploid fibroblasts (24) inhibited rather than accelerated cell growth.
We now report a more precise localization of the human S14 gene and show that it was both amplified and expressed in several breast cancer-derived cell lines. Most importantly, S14 was expressed in the majority of primary breast cancer specimens, and its expression was highly concordant with that of acetyl-CoA carboxylase (ACC), a rate-determining enzyme of fatty acid biosynthesis. S14 therefore provides a critical metabolic link between two established prognostic indicators in human breast cancers: 11q13 amplification and enhanced fatty acid formation.
METHODS
PCR Screening of Yeast Artificial Chromosomes (YACs).
YAC 965-F-10 (1,170 kbp), 745-E-12 (990 kbp), and 796-E-11 (1,040 kbp) (Research Genetics, Huntsville, AL) were selected based on their localization to 11q13.5 and lack of chimerism. A cytogenetic map of the sequence-tagged sites and genes residing on these YACs was compiled by Kelley and coworkers (25) (Fig. 1A). YAC DNA was harvested as described (26). PCR was performed in a Perkin–Elmer Thermal Cycler (1 min at 94°C, then 30 cycles of 30 sec at 94°C, 30 sec at 65°C, 1 min at 72°C ). Primers (Oligos, Etc., Wilsonville, OR) are defined in Table 1.
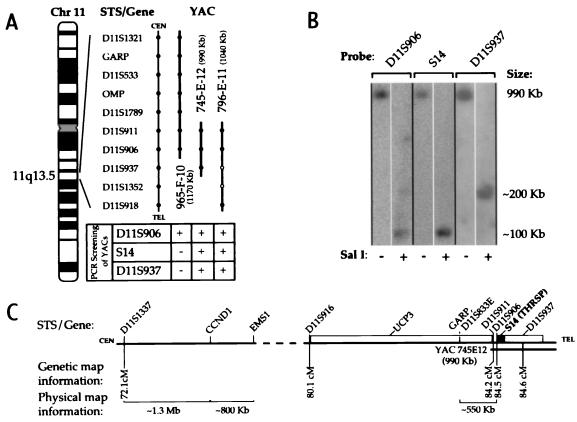
Figure 1.
Localization of the S14 gene to between D11S906 and D11S937. (A) A schematic representation of genes, sequence-tagged site markers, and YACs that span 11q13.5 without regard to genetic or physical distance, based on a cytogenetic map by Kelley and coworkers (25). From their study, the sequence-tagged site/gene content of YACs is represented with a solid circle; open circles indicate the markers not found on YAC 796-E-11 by those authors. In the current study, the S14 gene was amplified by PCR from YAC 745-E-12 and 796-E-11 but not 965-F-10. The presence of D11S906 was confirmed on all three YACs, and D11S937 was found on YACs 745-E-12 and 796-E-11. (B) Restriction and Southern blot analysis of YAC 745-E-12, resolved by pulsed-field gel electrophoresis. (C) An integrated map of the 11q13 amplicon, including centromeric and telomeric portions separated by a dashed line. Genetic map information (Généthon) can be found at: http://www.genethon.fr/genethon_en.html. Physical map information compiled from refs. 20 and 34–37. CEN, centromere; TEL, telomere; GARP, glycoprotein A repetitions predominant.
Table 1.
PCR primers, listed 5′ to 3′
| Gene/size, bp | 5′-Primer | 3′-Primer | Gene reference |
|---|---|---|---|
| S14 345* | CCAAGAACTGCCTGCTGACCGTCATGG | GGATGTGATGGAGGCTGGAGAAGTGC | 36-380, GDB:5446138 |
| S14 106† | GAGAATGGAACCGCAGAGACAGAGG | GGATGTGATGGAGGCTGGAGAAGTGC | 275-380, GDB:5446138 |
| IFNG 75† | CCCTCATCCAATGCTGGCAAACACC | CCAATCCAAGCCTTCTCCCTAGAGC | 5662-5736, GDB:119329 |
Primers used for YAC screening and RT-PCR. This PCR product was used to probe Southern blots.
Primers used for differential PCR. GDB, the Genome Database (http://gdbwww.gdb.org).
Southern Blot Analysis.
YAC 745-E-12 DNA in agarose plugs (27) was digested with NotI and SalI (Boehringer Mannheim) and resolved by pulsed-field gel electrophoresis in 0.8% agarose in 0.5X Tris/borate/EDTA buffer. Switching times [PPI-200 Programmable Electrophoresis Controller (MJ Research, Cambridge, MA)] were ramped from 40 sec to 6 min at 5-sec intervals for 96 hr at 100 V. Southern blots were probed with randomly primed (GIBCO/BRL), [α-32P]-labeled PCR products from markers D11S906 and D11S937 (Research Genetics) as well as a 345-bp product representing the S14 gene (Table 1).
Breast cancer cell line genomic DNA was isolated by using the Puregene DNA Isolation Kit (Gentra Systems). Control DNA was from human lymphocytes. DNA samples were digested with EcoRI (Boehringer Mannheim) and blotted as described. The probe for the uncoupling protein 3 (UCP3) gene was a 260-bp PCR product (28). The control probe targeted the T-cell antigen receptor constant β-chain gene (TCRBC), located on chromosome 7q (29). Signals were analyzed densitometrically [Personal Densitometer SI Model 375A (Molecular Dynamics)]; gene amplification was assessed by calculating a signal ratio of S14:TCRBC, normalized to that of control DNA.
Cell Culture.
Cell lines were from the American Type Culture Collection (ATCC). MDA-MB-134 cells were maintained in L-15 medium supplemented with 20% fetal calf serum (FCS) and 30 ng/ml epidermal growth factor (GIBCO/BRL); MDA-MB-435s cells were maintained in L-15 medium with 15% FCS, 2 mM l-glutamine, and 10 μg/ml insulin; MDA-MB-453 cells were maintained in L-15 medium with 10% FCS; MCF-7, SKBR3, and ZR-75-1 in DMEM/F12 were maintained with 10% FCS, and T47D cells were maintained in RPMI 1640 medium with 10% FCS, 2 mM l-glutamine, 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, 10 mM Hepes, 1.0 mM sodium pyruvate, and 10 μg/ml insulin. Penicillin and streptomycin were added to all culture media.
Differential PCR.
MDA-MB-134 genomic DNA was harvested by using the Trizol Reagent (GIBCO/BRL) and used as PCR template as described (30) (94°C for 25 sec and 64°C for 2.5 min × 29 cycles). Control DNA was from normal human lymphocytes. The interferon-γ (IFNG) gene (chromosome 12q) was used as a reference in this experiment. Primers are defined in Table 1.
Chromosome Analysis and Fluorescence in Situ Hybridization.
The probe for the S14 (THRSP) locus was an ≈80-kbp P1 clone (10) and biotin-labeled by nick translation (Bionick, GIBCO/BRL). Chromosomes prepared by standard techniques were conditioned by a 30 min, 37°C bath in 2X standard sodium citrate followed by dehydration in EtOH, at 22°C, and air dried. Slides were denatured in 70% formamide/2X standard sodium citrate followed by serial dehydration at 22°C. The probe was denatured (70°C × 5 min) and preannealed at 37°C for 2 h. Hybridization solution contained 0.2-μg probe, 10 μg of Cot-1 DNA (GIBCO/BRL), and 30 μg of herring sperm DNA (GIBCO/BRL) in 15 μl of Hybrisol VII (Oncor) per slide. The whole chromosome painting probe was from Oncor. Hybridization conditions were as described (31).
Reverse Transcriptase–PCR (RT-PCR).
Total RNA was extracted with Trizol Reagent (GIBCO/BRL). Contaminating DNA was digested with DNase I (GIBCO/BRL). RT-PCR was performed using the SuperScript One-Step System (GIBCO/BRL) using 100 ng of total RNA as template (55°C × 30 min and 94°C × 2 min, followed by 37 cycles of 94°C × 15 sec, 65°C × 30 sec, and 72°C × 1 min). Primers are listed in Table 1.
Western Blot.
Primary cultures of human hepatocytes (Clonetics, Walkersville, MD) were maintained for 3 days in modified William’s E medium (GIBCO/BRL) with 10% FCS, 50 nM thyroid hormone, and 27.5 μM glucose. Harvest of proteins, SDS-PAGE (hepatocytes, 10 μg/lane; MDA-MB-134 cells, 15 μg/lane) and Western blot with anti-S14 (residue 2–11 synthetic peptide) rabbit polyclonal antibody (1:1000 dilution) were as described (32).
Immunohistochemistry.
We obtained 21 paraffin-embedded primary breast cancer specimens (20 infiltrating ductal carcinomas and one lobular carcinoma) from the tissue archives of the Dartmouth-Hitchcock Medical Center. Tissue sections were steam-heated (20 min) and immunostained with anti-S14 (2–11) antibody (1:200) or anti-ACC rabbit polyclonal antibody (1:20) by using the avidin–biotin complex technique as described (3, 33). Counterstaining was with hematoxylin (1 min). Immunostaining was defined as positive for S14 or ACC if >20% of the tumor had discernible immunoreactivity at low power (×100) in the nucleus (S14) or the cytoplasm (ACC).
RESULTS
Localization of the S14 gene.
A band of the expected size for S14 was amplified by PCR from YAC 745-E-12 and 796-E-11 but not from 965-F-10 DNA. The presence of D11S906 was confirmed on all three YACs, and D11S937 was found on YACs 745-E-12 and 796-E-11 (Fig. 1A). Southern analysis of SalI-digested YAC 745-E-12 DNA localized S14 to the same ≈100-kbp band as D11S906 (Fig. 1B). Because S14 did not reside on the centromeric-most 100 kbp of 745-E-12, as shown by PCR analysis of overlapping YACs 965-F-10 (negative for S14) and 745-E-12 (positive for S14), its colocalization to within 100 kbp of D11S906 placed it on the telomeric side of this sequence-tagged site. D11S937 is telomeric to S14 because it localized to a different band (≈200 kbp) than did S14. Therefore, S14 is flanked by D11S906 (centromeric) and D11S937 (telomeric; Fig. 1C). This location is ≈550 kbp downstream from the telomeric-most marker shown thus far to be amplified in breast cancer [D11S833E (20, 21)]. Genetic map information for this region indicates that S14 is ≈11–12 Mbp telomeric to CCND1. Digestion of 745-E-12 with NotI revealed that D11S906, S14, and D11S937 were on the same 800-kbp DNA band (data not shown). Comparison with the results observed with the SalI digestion limit the location of D11S937 to a ≈600-kbp region telomeric to S14.
S14 Gene Amplification in Breast Cancer Cell Lines.
Southern analysis revealed an increased S14:TCRBC ratio in 2 of 7 breast cancer cell lines. The S14 gene was amplified 6.1-fold in MDA-MB-134 and 2.5-fold in T47D cells. The ratio of S14:TCRBC was between 1.5 and 1.7 for MDA-MB-453, SKBR3, and ZR-75-1 cells, a finding suggesting amplification. There was no amplification in MDA-MB-435s and MCF-7 cells. These results and the amplification status of CCND1 and EMS1 are summarized in Table 1. UCP3 recently was localized to a region of 11q13 lying between EMS1 and the S14 gene (28). Southern analysis with the UCP3 probe revealed no amplification in any of the cell lines examined, including MDA-MB-134 cells which exhibit amplification of CCND1, EMS1, and S14 (data not shown).
Differential PCR using a reference gene on chromosome 12q independently confirmed the Southern blot results for MDA-MB-134 cells, which used a reference gene from chromosome 7q. The intensity of the 106-bp band representing the S14 gene compared with the 76-bp product amplified from the IFNG control was markedly increased for MDA-MB-134 DNA compared with normal genomic DNA (Fig. 2B).
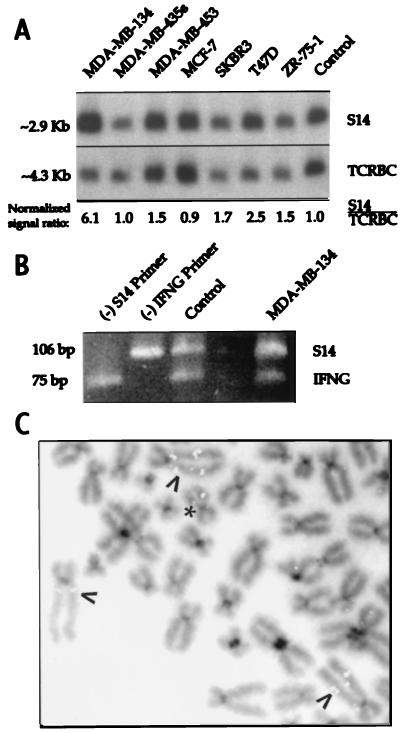
Figure 2.
Amplification of the S14 gene in a subset of breast cancer cell lines. (A) Southern analysis of DNA from seven breast cancer cell lines. (B) Differential PCR analysis of MDA-MB-134 cell DNA. Genomic DNA from normal lymphocytes (control) or from MDA-MB-134 cells was used as template for a multiplex PCR, using two primers for the S14 gene (yielding a 106-bp product) and two for IFNG (75 bp). In the first two lanes, omitting primers for S14 or IFNG [indicated by (−)] resulted in the disappearance of the bands representing those genes. In the lane marked MDA-MB-134, the increased S14:IFNG signal ratio compared with the control lane reflects the presence of S14 gene amplification in those cells. (C) Fluorescence in situ hybridization analysis of MDA-MB-134 chromosomes by using a S14 probe. Arrows point to the HSR in the aberrant chromosome 11s, previously observed on cytogenetic analysis. Intense fluorescent signals representing the S14 gene colocalize with the HSR. ∗ denotes a normal chromosome 11.
We undertook cytogenetic analysis of G-banded metaphases from MDA-MB-435s and MDA-MB-134 cells. MDA-MB-435s had a modal number of 53 chromosomes with numerous rearranged chromosomes, including a few that appeared to be partially derived from chromosome 11; no normal chromosome 11 was observed. The modal chromosome number of MDA-MB-134 cells was 85 and appeared to be a near tetraploid version of the karyotype initially established for this cell line (40). Of note were two apparently normal chromosome 11s in addition to three copies of a marker chromosome characterized by a large homogeneously staining region (HSR) on its long arm (data not shown). Paint analysis revealed that the HSR was composed of interspersed chromosome 11 sequences, whereas the short arm of the marker chromosome was not of chromosome 11 origin, consistent with a previous characterization of this marker chromosome (41, 42).
Fluorescence in situ hybridization with the S14 probe revealed hybridization to four chromosome-11-derived elements in MDA-MB-435s chromosomes, without any evidence of S14 gene amplification (data not shown). Line MDA-MB-134, however, showed the expected signal on the two normal chromosome 11s at q13.5 as well as intense signals interspersed in the HSR region of the marker chromosomes (Fig. 2C), indicating amplification of these sequences beyond that expected by chromosome copy number.
Expression of S14 in Breast Cancer Cell Lines and Primary Breast Cancers and Concordance with ACC Expression.
RT-PCR revealed a band of the expected size for S14 (345 bp) in MDA-MB-134, MDA-MB-453, MCF-7, SKBR3, T47D, and ZR-75-1 cell lines. The band representing MDA-MB-134 mRNA consistently demonstrated the strongest signal. The only cell line that did not express S14 was MDA-MB-435s, a line with no S14 gene amplification. The literature supports overexpression of CCND1 in MDA-MB-134, MDA-MB-453, and ZR-75-1 cells but not SKBR3 or T47D cells (22, 43, 44). Overexpression of EMS1 occurs in MDA-MB-134, MDA-MB-453, and ZR-75-1 but not MCF-7, SKBR3, or T47D cells. Excess lipogenesis and FAS content are reported in progesterone-treated MCF-7, SKBR3, T47D, and ZR-75-1 cells (45).
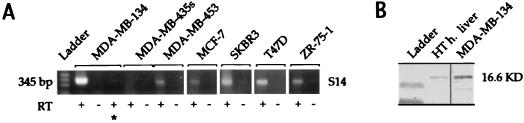
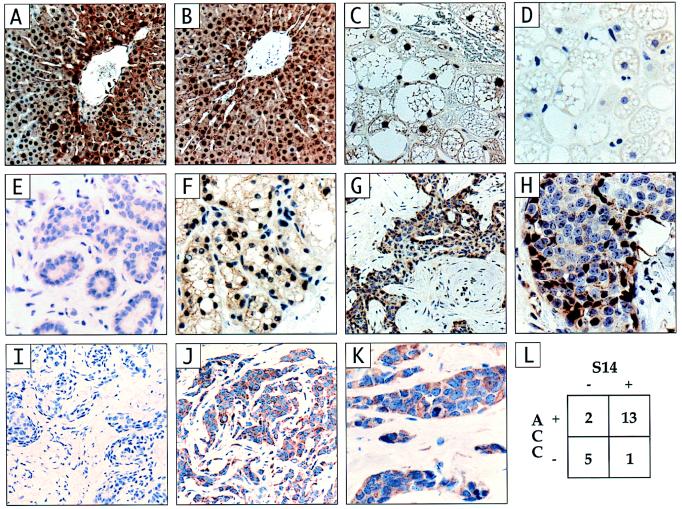
Conservation of residues 2–11 in the rat and human S14 polypeptides allowed the use of an antibody raised against the corresponding synthetic peptide for both Western and immunohistochemical analysis of S14 expression in human tissues. The utility of the antibody was validated by Western analysis of primary cultures of human hepatocytes treated with thyroid hormone and high glucose. This analysis revealed a band of the expected size for human S14 (16.6 kDa). Western analysis of MDA-MB-134 cells showed a band of the same size (Fig. 3B). We further validated the anti-S14 (2–11) antibody by comparing its pattern of immunohistochemical staining in hyperthyroid, carbohydrate-fed rat liver (Fig. 4A) with that of an antibody raised against a fusion protein containing the entire rat S14 protein as its carboxyl terminus (anti-GST-rS14 (3); Fig. 4B). Control experiments validating the specificity of both antibodies in Western analysis and the latter antibody in immunohistochemistry of rat tissues have been published (32, 33). The two antibodies yielded identical patterns of marked nuclear and weaker cytoplasmic immunoreactivity in a perivenous zonal distribution, as reported (3). Staining of a human, brown fat-derived tumor (hibernoma) with the anti-S14 (2–11) antibody yielded marked nuclear staining (Fig. 4C), as expected, that was attenuated by preincubation of the anti-S14 (2–11) antibody with the GST-rS14 fusion protein. This result was consistent with a recent report of S14 mRNA expression in a human lipoma (46). The weak cytoplasmic staining was unaffected by preincubation (Fig. 4D). Examination of five nonlactating human breast specimens yielded no nuclear staining and weak, variable cytoplasmic staining (for example, see Fig. 4E), whereas examination of four lactating mammary samples revealed nuclear staining (for example, see Fig. 4F). This result was consistent with the induction of S14 mRNA in lactating rat mammary gland (1). We analyzed 21 primary breast cancer specimens for S14 expression and found nuclear staining in 14 (67%) (see example in Fig. 4 G and H). The single lobular carcinoma was among the positive specimens (not shown). Staining of selected tumors that exhibited S14 immunoreactivity with antibody blocked with the GST-rS14 fusion protein showed a markedly attenuated signal (Fig. 4I). We also stained the panel of 21 tumors for ACC to investigate the metabolic relevance of S14 expression (example in Fig. 4 J and K). Fifteen (71%) of the tumors exhibited detectable ACC immunoreactivity, and this result was highly concordant (86%, P < 0.05) with S14 expression (Fig. 4L).
Figure 3.
Expression of S14 in the majority of breast cancer cell lines. (A) RT-PCR analysis of RNA extracted from seven breast cancer cell lines. ∗ indicates that the sample was exposed to RNase before RT-PCR. (B) Western analysis of hyperthyroid (HT) human hepatocytes exposed to high glucose, and MDA-MB-134 cells, using anti-S14 (2–11) IgG.
Figure 4.
Immunohistochemistry of hyperthyroid rat liver controls, nonlactating breast, lactating breast and 21 primary breast cancers for S14 and acetyl-CoA carboxylase. (A) Hyperthyroid, carbohydrate-fed rat liver probed with the anti-S14 (2–11) antibody; a region of the S14 peptide that is homologous in rats and humans. (B) The same liver probed with the anti-GST-rS14 fusion protein antibody. (C) Human hibernoma immunostained with the anti-S14 (2–11) antibody. (D) Hibernoma stained with the anti-S14 (2–11) antibody preincubated with the GST-rS14 fusion protein. (E) Normal breast epithelium. (F) Lactating breast epithelium. (G and H) Example of infiltrating ductal carcinoma that has positive immunoreactivity for S14 (I) The same tumor shown in G and H stained with the anti-S14 (2–11) antibody preincubated with the GST-rS14 fusion protein. (J and K) Example of infiltrating ductal carcinoma with positive immunostaining for ACC (L) Concordance analysis of immunostaining for S14 and ACC.
DISCUSSION
The major findings in the current study were that the S14 gene was both amplified and expressed in selected cell lines derived from human breast cancer and that the S14 protein also was readily detectable in two-thirds of a small sample of primary breast cancer specimens. CCND1, EMS1, and S14 are currently the only genes shown to be expressed from the consistently large 11q13 amplicon (14). The development of breast tumors in transgenic mice with mammary-specific overexpression established the CCND1 as a mammary oncogene (15). Our localization of the S14 gene between D11S906 and D11S937 placed it at the opposite, telomeric end of the amplicon from CCND1, raising the possibility that there was selective pressure for S14 expression in concert with that for CCND1. Published examples of breast cancers with amplification of distal 11q13 but not of the more centromeric CCND1 region (17, 20) further raise the speculation that S14 could provide an independent stimulus for tumor virulence. Indeed D11S833E, the closest marker to the S14 gene reported to be amplified in both breast cancer cell lines and primary breast tumors (21), was the only marker at 11q13 that was amplified in some tumors and in several tumors exhibited the greatest fold amplification of the genes and markers studied (20). Our finding that the UCP3 gene was not amplified in cell lines with amplification of S14 and with reported amplification of CCND1 and EMS1 confirmed the discontinuity of the 11q13 amplicon (17, 19–21). This finding raises the possibility of independent selection for multiple genes expressed from the amplicon.
Increased synthesis of long chain fatty acids may occur in multiple common neoplasms, including those arising in the breast (8), prostate (47), ovary (48), colon (49), and endometrium (9), and has been shown to confer a poor prognosis. The mechanism underlying the overexpression of the lipogenic enzymes is unknown. Kuhajda and coworkers found that overexpression of FAS was not only a marker for unfavorable prognosis but also was critical for tumor growth. This was shown by treating breast cancer cells in tissue culture (9) or ovarian cancer xenografts in nude mice (50) with cerulenin, a pharmacological inhibitor of FAS. Cerulenin caused programmed cell death and slowed tumor growth. Cells were rescued from the cerulenin effect by addition of palmitic acid to the culture medium, indicating dependency on the fatty acid for growth. However, it is unclear which cellular function(s) crucial for growth depends on the ongoing supply of newly synthesized long chain fatty acids. The requirement of fatty acids for the synthesis of phospholipids used in plasma membranes is a possibility (9).
A single genetic abnormality involving genes coding for the enzymes cannot explain the enhanced lipid formation because the genes for the three major lipogenic enzymes (ACC, FAS, and ATP-citrate lyase) reside at different chromosomal locations (51–53). Previous work from our laboratory established a role for S14 in the integrated switching of metabolism from the fasted to the fed state, including induction of the lipogenic enzymes, at the level of transcription (5). S14 protein exists both as a homodimer and as a heterodimer with a currently unidentified 36-kDa protein in vivo (54). Because S14 regulates expression of the lipogenic enzymes, an increase in its expression provides a unitary explanation for the increased lipid synthesis in tumors. Breast cancer patients with a poor prognosis associated with either 11q13 amplification or increased FAS expression may therefore represent overlapping clinical groups.
Under physiological circumstances, S14 expression is induced in rat liver by dietary carbohydrate and thyroid hormone (55) and disappears in response to glucagon administration (2), dietary polyunsaturated fatty acids (56), fasting, or diabetes mellitus (57). These effects are mediated at the transcriptional level and reflect the array of response elements present in the S14 gene promoter that allow it to integrate multiple input signals initiated by fuel-related hormones and dietary substrates. Little is known about the regulation of S14 expression in mammary tissue, except that it is induced during the lipogenic demands of lactation in rat (1) and in human (Fig. 4F). In breast cancer-derived cell lines that overexpress FAS, the enzyme is inducible by progestin, indicating the presence of an intact regulatory mechanism (43). It remains to be seen whether this induction requires S14, analogous to its function in the transduction of thyroid hormone-induced signals for lipogenesis in liver. If so, hormonal or other means of manipulating S14 expression could affect metabolism and growth of selected tumors in a manner similar to that observed upon pharmacological inhibition of lipogenesis.
Table 2.
Amplification status of genes at 11q13
| Cell line (ref.) | CCND1 | EMS1 | S14 |
|---|---|---|---|
| MDA-MB-134 (17, 21, 22) | + | + | + |
| MDA-MB-435s (39) | − | − | − |
| MDA-MB-453 (17, 22) | + | + | −* |
| MCF-7 (17) | − | + | − |
| SKBR3 (17, 22) | − | − | −* |
| T47D (17, 22, 38) | +/− | − | + |
| ZR-75-1 (17, 22) | + | + | −* |
The S14:TCRBC signal ratio was between 1.5 and 1.7.
Acknowledgments
Help was kindly supplied by J. Dunlap, L. Witters, and W. Noll (all of Dartmouth Medical School). The hibernoma specimen was kindly supplied by M. Miettinen, Armed Forces Institute of Pathology, and primers for UCP3 were kindly supplied by B. Lowell, Harvard Medical School. We thank Christine Hodorowski and Maudine Waterman for their expert advice on immunohistochemistry, Reed Lowrie and Peggy Sleeth for their editorial assistance, and Bernard Cole for the concordance analysis. This work was supported by National Institutes of Health Grant RO1 DK43142 (to W.B.K.), National Institutes of Health Training Grant DK07508, and U.S. Army Health Professions Scholarship Program support (to J.T.M.).
ABBREVIATIONS
- S14
Spot 14
- CCND1
cyclin D1
- IFNG
interferon-γ
- FAS
fatty acid synthase
- RT
reverse transcription
- HSR
homogeneously staining region
- YAC
yeast artificial chromosome
- ACC
acetyl-CoA carboxylase
- FCS
fetal calf serum
- UCP3
uncoupling protein 3
- TCRBC
T-cell antigen receptor constant β-chain
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. 5446138 and 119329).
References
- 1.Jump D, Oppenheimer J. Endocrinology. 1985;117:2259–2266. doi: 10.1210/endo-117-6-2259. [DOI] [PubMed] [Google Scholar]
- 2.Kinlaw W B, Schwartz H L, Towle H C, Oppenheimer J H. J Clin Invest. 1986;78:1091–1096. doi: 10.1172/JCI112665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinlaw W B, Tron P, Friedmann A S. Endocrinology. 1992;131:3120–3122. doi: 10.1210/endo.131.6.1446647. [DOI] [PubMed] [Google Scholar]
- 4.Kinlaw W, Church J, Harmon J, Mariash C. J Biol Chem. 1995;270:16615–16618. doi: 10.1074/jbc.270.28.16615. [DOI] [PubMed] [Google Scholar]
- 5.Brown S B, Maloney M, Kinlaw W B. J Biol Chem. 1997;272:2163–2166. [PubMed] [Google Scholar]
- 6.Alo P, Visca P, Marci A, Mangoni A, Botti C, Di Tondo U. Cancer. 1995;77:474–482. doi: 10.1002/(SICI)1097-0142(19960201)77:3<474::AID-CNCR8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Kuhajda F, Piantadosi S, Pasternack G. N Engl J Med. 1989;321:636–641. doi: 10.1056/NEJM198909073211003. [DOI] [PubMed] [Google Scholar]
- 8.Kuhajda F, Jenner K, Wood F, Hennigar R, Jacobs L, Dick J, Pasternak G. Proc Natl Acad Sci USA. 1994;91:6279–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pizer E, Jackisch C, Wood F, Pasternak G, Davidson N, Kuhajda F. Cancer Res. 1996;56:2745–2747. [PubMed] [Google Scholar]
- 10.Moncur J, Park J, Maloney M, Mohandas T K, Kinlaw W B. Cytogenet Cell Genet. 1997;78:131–132. doi: 10.1159/000134644. [DOI] [PubMed] [Google Scholar]
- 11.Taviaux S, Planells R, Gastaldi M, Torresani J, Grillasca J. Cytogenet Cell Genet. 1997;76:219–220. doi: 10.1159/000134553. [DOI] [PubMed] [Google Scholar]
- 12.Champeme M, Bieche I, Lizard S, Lidereau R. Genes Chromosomes Cancer. 1995;12:128–133. doi: 10.1002/gcc.2870120207. [DOI] [PubMed] [Google Scholar]
- 13.Schuuring E, Verhoeven E, van Tintern H, Peterse J, Nunnink B, Thunnisen F, Devilee P, Cornelisse C, van de Vijver M, Mooi W, et al. Cancer Res. 1992;52:5229–5234. [PubMed] [Google Scholar]
- 14.Schuuring E. Gene. 1995;159:83–96. doi: 10.1016/0378-1119(94)00562-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Cardiff R, Zukerberg L, Lees E, Arnold A, Schmidt E. Nature (London) 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 16.Schuuring E, Verhoeven E, Litvinov S, Michalides R. Mol Cell Biol. 1993;13:2891–2898. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell D, deFazio A, Sutherland R, Daly R. Int J Cancer. 1996;68:485–492. doi: 10.1002/(SICI)1097-0215(19961115)68:4<485::AID-IJC14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Hui R, Campbell D, Lee C, McCaul K, Horsfall D, Musgrove E, Daly R, Seshadri R, Sutherland R. Oncogene. 1997;15:1517–1623. doi: 10.1038/sj.onc.1201311. [DOI] [PubMed] [Google Scholar]
- 19.Tanigami A, Tokino T, Takita K, Ueda M, Kasumi F, Nakamura Y. Genomics. 1992;13:21–24. doi: 10.1016/0888-7543(92)90196-y. [DOI] [PubMed] [Google Scholar]
- 20.Karlseder J, Zeillinger R, Schneeberger C, Czerwenka K, Speiser P, Kubista E, Birnbaum D, Gaudray P, Theillet C. Genes Chromosomes Cancer. 1994;9:42–48. doi: 10.1002/gcc.2870090108. [DOI] [PubMed] [Google Scholar]
- 21.Szeptowski P, Ollendorff V, Grosgeorge J, Courseuax A, Birnbaum D, Theillet C, Gaudray P. Oncogene. 1992;7:2513–1517. [PubMed] [Google Scholar]
- 22.Buckley M, Sweeney K, Hamilton J, Sini R, Manning D, Nicholson R, deFazio A, Watts C, Musgrove E, Sutherland R. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 23.Han E, Sgambato A, Jiang W, Santella R, Doki Y, Cacace A. Oncogene. 1995;10:953–961. [PubMed] [Google Scholar]
- 24.Atadja P, Wong H, Veillette C, Riabowol K. Exp Cell Res. 1995;217:205–216. doi: 10.1006/excr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 25.Kelley P M, Weston M D, Chen Z, Orten D J, Hasson T, Overbeck L D, Pinnt J, Talmadge C B, Ing P, Mooseker M S, et al. Genomics. 1997;40:73–79. doi: 10.1006/geno.1996.4545. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 27.Ragoussis J. YAC Protocols. Totowa, NJ: Humana; 1996. [Google Scholar]
- 28.Solanes G, Vidal-Puig A, Grujic D, Flier J, Lowell B. J Biol Chem. 1997;272:25433–25436. doi: 10.1074/jbc.272.41.25433. [DOI] [PubMed] [Google Scholar]
- 29.Yoshikai Y, Anatoniou D, Clark S, Yanagi Y, Sangster R, Van den Elsen P, Terhorst C, Mak T. Nature (London) 1984;312:521–524. doi: 10.1038/312521a0. [DOI] [PubMed] [Google Scholar]
- 30.Neubauer A, Neubauer B, He M, Effert P, Iglehart D, Frye R, Liu E. Oncogene. 1991;7:1019–1025. [PubMed] [Google Scholar]
- 31.Park J P. Cytogenet Cell Genet. 1996;74:133–137. doi: 10.1159/000134400. [DOI] [PubMed] [Google Scholar]
- 32.Kinlaw W B, Ling N C, Oppenheimer J H. J Biol Chem. 1989;264:19779–19783. [PubMed] [Google Scholar]
- 33.Kinlaw W B, Tron P, Witters L A. Endocrinology. 1993;133:645–650. doi: 10.1210/endo.133.2.8102096. [DOI] [PubMed] [Google Scholar]
- 34.Peters G, Fantl V, Smith R, Brookes S, Dickson C. Breast Cancer Res Treat. 1995;3:125–135. doi: 10.1007/BF00682720. [DOI] [PubMed] [Google Scholar]
- 35.Brookes S, Lammie G, Schuuring E, Boer C, Michalides R, Dickson C, Peters G. Genes Chromosomes Cancer. 1993;6:221–231. doi: 10.1002/gcc.2870060406. [DOI] [PubMed] [Google Scholar]
- 36.Merscher S, Bekri S, Leeuw B, Pedeutour F, Grosgeorge J, Shows T, Mullenbach R, Paslier D, Nowak N, Gaudray P. Genomics. 1997;39:340–347. doi: 10.1006/geno.1996.4460. [DOI] [PubMed] [Google Scholar]
- 37.Courseaux A, Szepetowski P, Fernandes M, Serizet C, Kawaguchi Y, Grosgeorge J, Perucca-Lostanlein D, Shows T, Todd J, Nowak N, et al. Genomics. 1997;40:13–23. doi: 10.1006/geno.1996.4527. [DOI] [PubMed] [Google Scholar]
- 38.Faust J, Meeker T. Cancer Res. 1992;52:2460–2463. [PubMed] [Google Scholar]
- 39.Kallioneimi A, Kallioneimi O, Piper J, Tanner M, Stokke T, Chen L, Smith H, Pinkel D, Gray J, Waldman F. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafage M, Nguyen C, Szepetowski P, Pebusque M, Simonetti J, Courtois G, Gaudray P, DeLapeyriere O, Jordan B, Birnbaum D. Genes Chromosomes Cancer. 1990;2:171–181. doi: 10.1002/gcc.2870020302. [DOI] [PubMed] [Google Scholar]
- 41.Lafage M, Peudeutour F, Sylvie M, Simonetti J, Prosperi M, Gaudray P, Birnbaum D. Genes Chromosomes Cancer. 1992;5:40–49. doi: 10.1002/gcc.2870050107. [DOI] [PubMed] [Google Scholar]
- 42.Lemieux N, Apiou F, Vogt N, Malfoy B, Dutrillaux B. Cancer Genet Cytogenet. 1996;90:75–79. doi: 10.1016/0165-4608(96)00061-1. [DOI] [PubMed] [Google Scholar]
- 43.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 44.Hosokawa Y, Gadd M, Smith A, Koerner F, Schmidt E, Arnold A. Cancer Lett. 1997;113:123–130. doi: 10.1016/s0304-3835(97)04605-3. [DOI] [PubMed] [Google Scholar]
- 45.Chalbos D, Joyeux C, Galthier F, Rochefort H. J J Steroid Biochem Mol Biol. 1992;43:223–228. doi: 10.1016/0960-0760(92)90211-z. [DOI] [PubMed] [Google Scholar]
- 46.Grillasca J, Gastaldi M, Khiri H, Dace A, Peyrol N, Reynier P, Torresani J, Planells R. FEBS Lett. 1997;401:38–42. doi: 10.1016/s0014-5793(96)01433-0. [DOI] [PubMed] [Google Scholar]
- 47.Shurbaji M, Kalbfleisch J, Thurmond T. Hum Pathol. 1996;27:917–921. doi: 10.1016/s0046-8177(96)90218-x. [DOI] [PubMed] [Google Scholar]
- 48.Gansler T, Hardman W, Hunt D, Schaffel S, Hennigar R. Hum Pathol. 1997;28:686–692. doi: 10.1016/s0046-8177(97)90177-5. [DOI] [PubMed] [Google Scholar]
- 49.Reston M, Kern S, Vogelstein B. Lab Invest. 1992;66:47. (abstr.). [Google Scholar]
- 50.Pizer E, Wood F, Heine H, Romanstev F, Pasternak G, Kuhajda F. Cancer Res. 1996;56:1189–1193. [PubMed] [Google Scholar]
- 51.Ullrich C, Widmer J, Park J, Mohandas T, Witters L. Cytogenet Cell Genet. 1997;77:176–177. doi: 10.1159/000134568. [DOI] [PubMed] [Google Scholar]
- 52.Jayakumar A, Chirala S, Chinault A, Baldini A, Abu-Elhegia L, Wakil S. Genomics. 1994;23:420–424. doi: 10.1006/geno.1994.1518. [DOI] [PubMed] [Google Scholar]
- 53.Couch F, Abel K, Brody L, Boehnke M, Collins F, Weber B. Genomics. 1994;21:444–446. doi: 10.1006/geno.1994.1293. [DOI] [PubMed] [Google Scholar]
- 54.Cunningham B, Maloney M, Kinlaw W. Endocrinology. 1997;138:5184–5188. doi: 10.1210/endo.138.12.5597. [DOI] [PubMed] [Google Scholar]
- 55.Jump D B, Narayan P, Towle H, Oppenheimer J H. J Biol Chem. 1984;259:2789–2797. [PubMed] [Google Scholar]
- 56.Jump D B, Clarke S D, MacDougald O, Thelen A. Proc Natl Acad Sci USA. 1993;90:8454–8458. doi: 10.1073/pnas.90.18.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carr F E, Bingham C, Oppenheimer J H, Kistner C, Mariash C N. Proc Natl Acad Sci USA. 1984;81:974–978. doi: 10.1073/pnas.81.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]