Abstract
Ribosomal RNAs contain numerous modifications at specific nucleotides. Despite their evolutionary conservation, the functional role of individual 2′-O-ribose methylations in rRNA is not known. A distinct family of small nucleolar RNAs, box C/D snoRNAs, guides the methylating complex to specific rRNA sites. Using a high-resolution phenotyping approach, we characterized 20 box C/D snoRNA gene deletions for altered growth dynamics under a wide array of environmental perturbations, encompassing intraribosomal antibiotics, inhibitors of specific cellular features, as well as general stressors. Ribosome-specific antibiotics generated phenotypes indicating different and long-ranging structural effects of rRNA methylations on the ribosome. For all studied box C/D snoRNA mutants we uncovered phenotypes to extraribosomal growth inhibitors, most frequently reflected in alteration in growth lag (adaptation time). A number of strains were highly pleiotropic and displayed a great number of sensitive phenotypes, e.g., deletion mutants of snR70 and snR71, which both have clear human homologues, and deletion mutants of snR65 and snR68. Our data indicate that individual rRNA ribose methylations can play either distinct or general roles in the workings of the ribosome.
Keywords: box C/D snoRNA, rRNA modification, 2′-O-ribose methylation, phenotypic profiling
INTRODUCTION
Modified nucleotides are prevalent in ribosomal RNA. The locations of these modifications are mostly evolutionarily conserved and are concentrated in functionally important domains of the ribosome, e.g., near the peptidyl transferase center (PTC) and the mRNA decoding center, manifesting their functional importance in translation (Decatur and Fournier 2002). Their importance is further supported by the fact that the machinery that creates these conserved rRNA modifications is remarkably different between bacteria and the other two domains of life. The two major types of rRNA modifications are 2′-O-ribose methylation (Nm) and isomerization of uridine to pseudouridine (Ψ) (Bachellerie et al. 2002). The number of modifications in rRNA increases with increasing evolutionary complexity; Escherichia coli has 10 Ψs and four Nms, while human rRNA contains more than 90 Ψs and 100 Nms (Ofengand and Bakin 1997). In eukaryotes, the site-specificity of these modifications is facilitated by small nucleolar RNAs (snoRNA) which guide the modifying enzymes to their nucleotide targets via a base-pairing mechanism. Box C/D snoRNAs bind fibrillarin and direct it to the sites of methylation in rRNA, while box H/ACA snoRNAs direct the binding of dyskerin/Cbf5p to the site of pseudouridylation (Kiss 2002). There are indications that rRNA modifications might play a role in human disease, since dyskerin is mutated in X-linked dyskeratosis congenita, marked by skin and bone marrow failure in humans (Heiss et al. 1998; Ruggero et al. 2003; Yoon et al. 2006). However, despite the fact that the involved snoRNA genes and their corresponding sites of modification have been well characterized, the cellular role of individual nucleotide modifications in rRNA is mostly not understood.
There are in total 76 box C/D and box H/ACA snoRNA genes annotated in the yeast snoRNA database (http://people.biochem.umass.edu/sfournier/fournierlab/snornadb/main.php) (Piekna-Przybylska et al. 2007). Genetic analyses have yielded only four lethal phenotypes, i.e., deletion mutants of NME1 (MRP), snR17a/b (U3a/b), snR30, and snR128 (U14) are inviable; however, these are all required in pre-rRNA cleavage (Hughes et al. 1987; Bally et al. 1988; Zagorski et al. 1988; Schmitt and Clayton 1993). One snoRNA mutant exhibited a clear fitness defect: the slow-growing and cold-sensitive pseudouridylation guide snR10 deletion strain (Tollervey and Guthrie 1985) that later on was also shown to be involved in pre-rRNA cleavage (Tollervey 1987). For the remaining snoRNAs, no strong link to cellular fitness has been reported despite several attempts (Parker et al. 1988; Lowe and Eddy 1999; Qu et al. 1999). However, competition assays and the use of inhibitors directly targeting the ribosomal function have revealed subtle phenotypes for some knockouts for pseudouridylation yeast snoRNA. Five yeast mutants lacking a set of box H/ACA snoRNAs guiding pseudouridylations in the peptidyl transferase center were found to show marginal but significant fitness defects (King et al. 2003). In addition, competition assays revealed subtle defects for strains lacking two highly conserved pseudouridine modifications guided by snR191 (Badis et al. 2003). However, as for the loss of 2′-O-ribose methylation with no documented impact on pre-RNA processing, no fitness defect has yet been reported.
In general, many phenotypes are marginal and can therefore easily escape detection (Thatcher et al. 1998). Hence when evaluating phenotypic consequences of mutation, high precision is of paramount value. Moreover, the phenotypic response might not always be apparent at the fitness variable growth rate but could be instead reflected in other aspects of cellular physiology, such as growth lag (adaptation time) and growth efficiency. We have earlier introduced a phenotyping approach for the quantitative extraction of these three physiological parameters from Saccharomyces cerevisiae deletion strains in environmental microculture arrays (Warringer and Blomberg 2003). The precision of the method allows not only the resolution of phenotypes into effects of different aspects of cellular physiology but also the quantification of marginal phenotypes not detectable by standard, more qualitative approaches (Warringer et al. 2003; Osterberg et al. 2006).
In this report our phenotyping methodology was applied to investigate consequences of deleting individually 20 box C/D snoRNA genes. A large set of intra- and extraribosomal inhibitors was employed to screen for phenotypic effects, generating several thousand high-resolution growth curves. We found statistically significant phenotypes for all examined box C/D snoRNA mutations providing experimental evidence for their functional importance. Furthermore, we could link many of the defects in ribose methylation to specific extraribosomal agents. The information on the functional importance of individual rRNA modifications will have important implications in the analysis of the finer details of the workings of the translational machinery, in the understanding of evolutionary mechanisms that have shaped the ribosome, as well as in the understanding of some human diseases.
RESULTS AND DISCUSSION
Phenotypes in basal medium for C/D snoRNA mutants
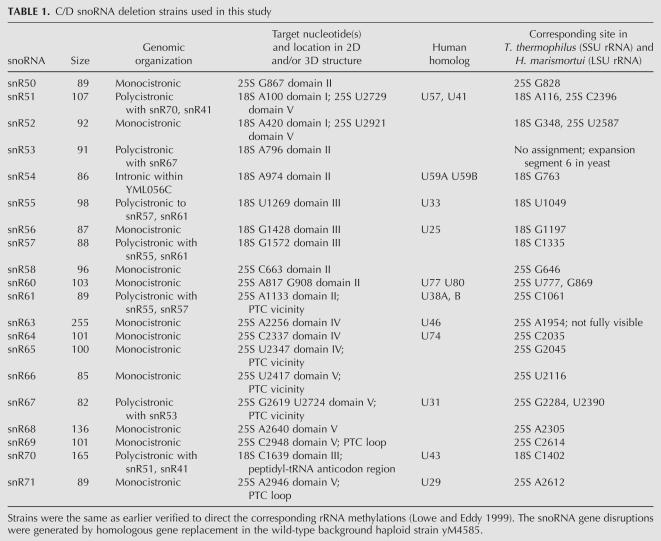
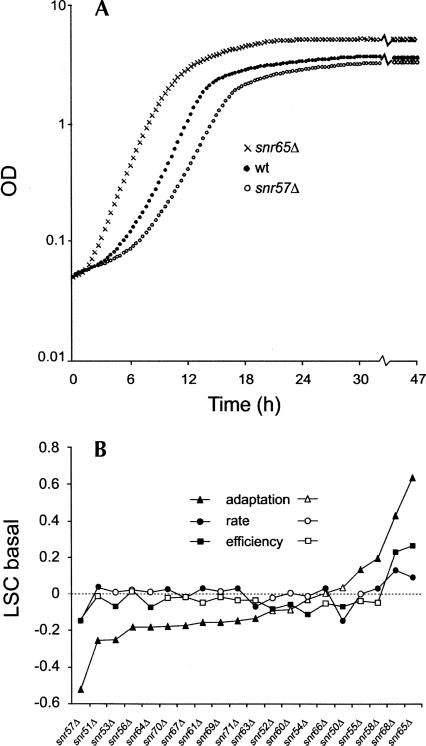
The 20 selected box C/D snoRNA deletion strains have earlier been verified with respect to their target 2′-O-ribose rRNA methylation sites (Lowe and Eddy 1999). They represent various aspects of snoRNA biology with genes distributed over 10 different chromosomes, having both intronic and extragenic gene locations, being monogenic or part of gene clusters (polycistronic), representing a wide range of sizes (78–255 nucleotides), being involved in 2′-O-ribose methylations on both single and multiple sites along the rRNA molecules, and their respective sites of modification being distributed throughout various domains of the rRNA three-dimensional (3D) structure (Table 1). High-resolution growth curves were generated by recording the optical density of cell cultures every 20 min over a 47-h period. We observed both positive and negative fitness effects in basal medium among the snoRNA knockouts. For instance, whereas snr57Δ displayed a distinct growth rate defect compared with the wild-type strain, snr65Δ showed significantly improved growth, most clearly indicated by a shorter adaptation time (Fig. 1A).
TABLE 1.
C/D snoRNA deletion strains used in this study
FIGURE 1.
Box C/D snoRNA deletion mutants display growth alterations in basal medium. (A) Growth curves in basal medium for wild type (filled circles), snr65Δ (crosses), and snr57Δ (open circles). (B) The three growth variables—growth lag (adaptation time), growth rate, and growth efficiency—were extracted from growth curves. The values displayed for the mutants were normalized to the wild type and expressed on a logarithmic scale, forming a logarithmic strain coefficient (LSC) (LSC = 0 means no change compared to the wild type; LSC < 0 indicates sensitivity; and LSC > 0 indicates resistance compared to the wild type). Growth lag (triangles), growth rate (circles), and growth efficiency (squares) are shown; filled symbols indicate values that are significantly different (P < 0.001) from the wild type.
To precisely quantify and classify the growth behavior of the snoRNA knockouts, growth curves were resolved into three growth variables: growth lag (adaptation time before exponential growth), growth rate (the slope during the exponential phase of growth), and growth efficiency (the optical density increment from start to stationary phase). For each of these physiological indicators, we normalized the growth of a specific knockout strain to that of an internal wild-type control included in each experimental run, forming ratios referred to as logarithmic strain coefficients (LSCs) (Warringer and Blomberg 2003). Overall, the observed phenotypes of the box C/D snoRNA deletions during growth in basal medium tended to be subtle. Most of the statistically significant phenotypes were found for growth lag, where 75% of the mutants displayed significantly (P < 0.001) altered behavior, while growth rate and growth efficiency were unaffected for many strains (Fig. 1B). Remarkably, taking all the three different growth variables into account, all snoRNA mutants exhibited a statistically significant (P < 0.001) but mostly subtle phenotype in basal medium. Even the most sensitive deletion mutant, snr57Δ, had only a 20% increased generation time. The subtle phenotypes in combination with the main effect on adaptation time rather than on growth rate may explain why the functional importance of these snoRNA genes has eluded detection in more qualitative colony assays on solid basal medium.
Ribosome-specific antibiotics report on structural alterations in rRNA
Antibiotics that bind to the ribosome at specific sites affect protein synthesis either by directly competing with incoming translation substrates or by causing topological alterations in rRNA domains. Macrolides such as anisomycin and puromycin competitively inhibit peptide bond formation by blocking the hydrophobic crevice of the peptidyl transferase center from accepting incoming amino acid residues bound to aminoacyl tRNA (Hansen et al. 2003). Direct molecular interactions of antibiotics with key rRNA residues are important requisites for effective antibiotic action. For example, rRNA mutations in nucleotide positions G2482, A2486, C2487, and A2488 have been reported to confer anisomycin resistance because their proximity to the antibiotic binding site results in diminished ligand affinity (Hummel and Bock 1987). Since perturbing the topology of the binding pocket is enough to induce resistance-conferring phenotypes, nucleotide modifications, or lack thereof, it should also be expected to have the same effect. Indeed, it is known that many aminoglycoside-producing organisms express rRNA methylases which methylate specific rRNA residues, resulting in protection from their own antibiotic products, as in the case of paromomycin and anisomycin resistance of Serratia marcescens (Doi et al. 2004).
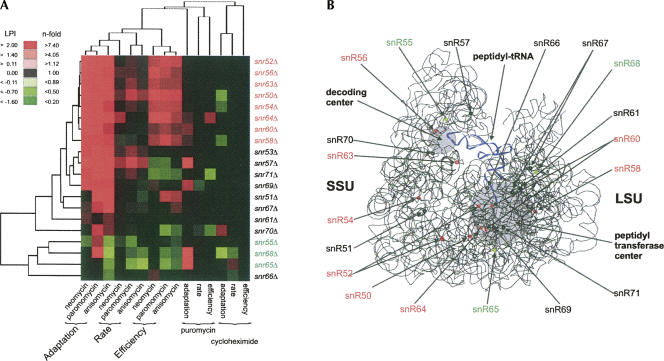
We applied antibiotics that are known to specifically bind to and inhibit the ribosome and assayed for growth inhibition to assess the influence on rRNA conformation from the lack of 2′-O-ribose methylations. The concentrations of the various antibiotics were set to confer about the same growth rate reduction in the wild type. The antibiotics concentrations should furthermore be low to minimize pathological side effects that might occur at high concentrations. We quantified the specific gene-by-environment interaction by comparing the LSC values with and without antibiotics, forming for each phase of growth a relative measure that is normalized for general growth defects in the mutant. This term we refer to as logarithmic phenotypic index (LPI). Except for puromycin and cycloheximide, which resulted in few and only minor gene–antibiotic interactions, we uncovered a great number of antibiotic-dependent phenotypic responses for almost all the snoRNA knockout strains. A large fraction of the snoRNA deletions is resistant to neomycin, paromomycin, and anisomycin (Fig. 2A). The highly similar phenotypic effects among the snoRNA knockouts elicited by neomycin and paromomycin confirm the identical modes of action of these antibiotics. Surprisingly, a similar phenotypic response to neomycin and paromomycin was also displayed by anisomycin, despite the fact that it belongs to a different mechanistic class of antibiotics. This might be explained by the fact that these two classes of antibiotics actually affect the binding of the aminoacyl tRNA, albeit on opposite ends (Fig. 2B). Whereas paromomycin and neomycin both exert their influence during the decoding of the mRNA at the anticodon end of the aminoacyl tRNA in the small ribosomal subunit, anisomycin blocks the delivery of amino acids carried by the same A-site bound aminoacyl tRNA in the large ribosomal subunit. This result implies that perturbing key sites in the ribosome, albeit in different spatial positions, could manifest the same phenotypic consequence and reflect similar outcome of parallel mechanistic events during translation.
FIGURE 2.
Ribosomal inhibitors differently affect growth of the snoRNA deletion mutants. The ribosomal inhibitors neomycin, paromomycin, anisomycin, cycloheximide, and puromycin were administered at concentrations that elicited a <20% growth rate reduction in the wild type. The growth of the snoRNA mutants in the presence of the ribosomal inhibitor was normalized for any general growth defects for the mutants in basal medium, forming the logarithmic phenotypic index (LPI): LPI = LSCinhibitor − LSCbasal. (A) Two-way hierarchical clustering of LPI for the three growth variables and inhibitors. Only statistically significant (P < 0.001) values are displayed in color; LPI < 0 (green) and LPI > 0 (red). Two of the clusters have been indicated; the snoRNA deletions that confer mainly improved growth compared to the wild type are indicated as resistant (red), and the ones that confer mainly growth worse than the wild type are indicated as sensitive (green). (B) 3D structural model of the rRNA portion of the ribosome with the small subunit to the left (SSU) and the large subunit to the right (LSU). Positions of the 2′-O-ribose methylations are indicated; red and green indicate clustering in A. The two functionally important domains, peptidyl transfer center and decoding center, are indicated with gray circles, and a tRNA molecule that stretches between these two centers is indicated in blue.
A seemingly baffling result was the discrepancy between the phenotypic profiles of mutants within the same class of antibiotics known to bind in the same crevice in the ribosome. For instance, snr65Δ and snr68Δ both displayed sensitive phenotypes toward anisomycin while both mutants exhibited resistance and/or no effect toward puromycin, the latter being a structural analog of anisomycin. Both antibiotics have also been shown to share extensive overlap in their binding to the same hydrophobic crevice in the peptidyl transferase center (Hansen et al. 2003). Nevertheless, the same structural study pointed out that anisomycin and puromycin approach the hydrophobic crevice from opposite directions. Hence, perturbing the entry site of one antibiotic should not necessarily affect the entrance of the other and could account for the observed difference in the phenotypic profiles we observed for anisomycin and puromycin.
The phenotypic heat map for intraribosomal inhibitors revealed antibiotics resistances for all three physiological indicators for a good proportion of the strains (Fig. 2A), while a smaller group of mutants, like snr55Δ and snr68Δ, displayed mostly sensitive phenotypes. The differential sensitivity to ribosome-specific antibiotics among the box C/D snoRNA mutants would indicate that lack of individual ribose methylations apparently results in different structural alterations in rRNA. Spatial mapping of the corresponding modification sites onto the 3D structural models of the ribosome revealed a fairly scattered distribution of both the “antibiotic resistant” and “antibiotic sensitive” modifications (Fig. 2B). This lack of correlation between the tight phenotypic responses of the box C/D snoRNA deletions and the spatial location of the corresponding modified residues probably reflects the highly complicated nature of allosteric physical interactions in the ribosome, i.e., local structural change in rRNA can trigger significant conformational changes in distant parts of the ribosomal structure (Bashan et al. 2003). We conclude that the set of antibiotics used as intraribosomal perturbants indicated both different and long-ranging rRNA structural effects as a result of lack of site-specific 2′-O-ribose methylations.
C/D snoRNA genes are differentially important during extraribosomal stresses
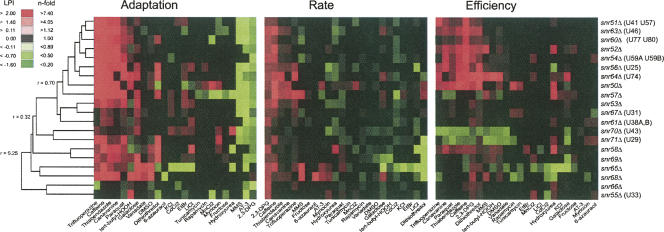
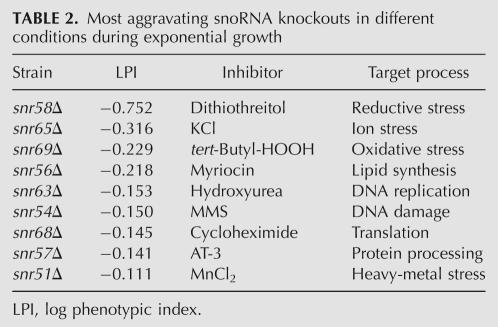
To investigate the functional importance of 2′-O-ribose rRNA methylations in a wider cellular and physiological context, we subjected the 20 box C/D snoRNA gene deletion strains to 24 environmental challenges. These extraribosomal perturbants represented inhibitors with a wide target profile, such as inhibition/influence on signaling (caffeine, rapamycin), DNA synthesis and repair (hydroxyurea, MMS), and glycosylation (tunicamycin). We also included external stresses that have a wide array of cellular targets and thus affect various aspects of cell physiology, e.g., redox imbalance (paraquat, tert-butyl-HOOH), metal toxicity (LiCl, MnCl2), and osmotic/ionic stress (KCl). This experimental design will reveal functional interactions between individual 2′-O-ribose methylations on rRNA and a wide set of cellular features. We found a surprisingly large number of gene–chemical interactions to the snoRNA genes; a total of 427 significant resistant and 236 significant sensitive phenotypes were identified (Fig. 3; phenotypic data are available upon request from the corresponding author). Many of these phenotypes were marginal in nature and would not have been identified by less precise measurements. However, even if we apply a threshold of 50% phenotypic difference in relation to the wild type (LPI < −0.35; 0.35 < LPI), we still get 33 sensitive significant phenotypes, indicating frequent strong functional links between methylation on rRNA and certain cellular features. In fact, when only analyzing growth rate, a variable which earlier has failed to reveal growth alterations for these box C/D snoRNA deletions, our wide environmental array scored several substantial phenotypes (Table 2).
FIGURE 3.
The snoRNA deletion mutants respond differently to wide array of extraribosomal growth inhibitors and stresses. A one-way hierarchical cluster on LPI is displayed; only statistically significant (P < 0.001) better (red) and worse (green) growth than the wild type is indicated. Clustering was performed on all data, but the three different growth variables are displayed separately. The r values by the tree to the left indicate the Pearson correlation at that branch point. The corresponding human rRNA methylations are indicated after the strain name.
TABLE 2.
Most aggravating snoRNA knockouts in different conditions during exponential growth
Hierarchical clustering of the deletion strain phenotypes revealed functional links between certain rRNA ribose methylations (Fig. 3). Interestingly, the methylations earlier identified as influencing the rRNA topology in a similar way (indicated as antibiotic resistant in Fig. 2A), exhibited very similar phenotypic profiles also to the extraribosomal challenges. This further supports that lack of these ribose methylations will similarly alter the functioning of the ribosome. In addition, many of the ribose methylations in this resistant group have clear human homologs, indicating strong evolutionary conservation for a functionally consistent set of modifications (Fig. 3). Another interesting phenotype cluster is formed between snr70Δ and snr71Δ, which in particular exhibited frequent growth efficiency defects. Apparently, the lack of these ribose modifications results in ribosomes that under a wide array of growth challenges, e.g., redox imbalance imposed by paraquat, DTT, or tert-butyl-HOOH, negatively influences the cellular energy balance. SnR70 guides the methylation of a nucleotide in the P site of the small subunit, just below the site where the mRNA codon is positioned to interact with the anticodon of the peptidyl tRNA, while snR71 guides the methylation of a nucleotide that is in the peptidyl transferase loop itself. Since these modifications are conserved in human rRNA, the corresponding sites potentially link redox imbalance to ribosomes in higher eukaryotes as well.
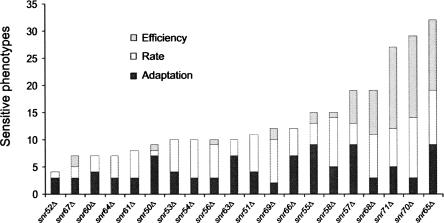
The snoRNA deletions resulted in numerous phenotypic sensitivities. This was particularly clear for the already mentioned snr70Δ and snr71Δ mutants, which were only exceeded by snr65Δ that displayed a great number of growth lag, rate, and efficiency defects (Fig. 4). The snr65Δ strain is defective in ribose methylation on nucleotide U2347 close to the peptidyl transfer center, a defect that resulted in phenotypes in several ionic challenges like Cd2+, K+, and Li+. Moreover, snr65Δ is an interesting yeast specific ribose methylation mutant that in basal medium out-competes the wild type at all three growth variables (Fig. 1). Changes in rRNA modifications were recently reported to sometimes result in increased rate of peptidyl transfer and enhanced affinity for tRNA binding (Baxter-Roshek et al. 2007), mechanistic changes that could result in alterations in translational fidelity. An altered ribosome could be envisaged to have drastic impact on other cellular activities during external stress via several mechanisms, e.g., differential translation of specific mRNAs, altered protein functionality by mistranslation, or general impact on physiology from diminished translational rate. The latter is in line with our observation that inhibition of translation by addition of cycloheximide generally suppresses the phenotypic effect from other stress conditions (J. Warringer, D. Anevski, B. Liu, A. Blomberg, in prep.).
FIGURE 4.
Certain mutants display a great number of sensitive phenotypes. The number of significantly (P < 0.001) sensitive phenotypes is indicated for each of the three growth variables separately: growth lag (black), growth rate (white), and growth efficiency (gray).
CONCLUDING REMARKS
Although the biochemical activity of box C/D snoRNAs has been known for a long time, guiding 2′-O-ribose methylation at specific nucleotides in rRNA, the cellular role played by most of these modifications in a larger physiological context is not clear. One reason for this lack of functional information is that phenotypic characterization of individual ribose methylations has largely been without success in earlier reports. It has been proposed that most individual modifications contribute a small, nonessential benefit, but in aggregate a large benefit is provided by the full ensemble of modifications. Here we identify phenotypic consequences of all tested individual box C/D snoRNA deletions. However, it cannot at this stage be excluded that some of the snoRNA genes could also be part of other processes than rRNA modification (Schoemaker and Gultyaev 2006). We noted that rRNA ribose methylation appeared to be frequently important during the adaptation phase and that many phenotypes, irrespective of growth phase, were marginal. Given these results, relying only on growth rate measurements or solid media colony assays would certainly miss many phenotypic effects.
The consensus view regarding the function of rRNA modifications is that they are “fine tuners” of RNA structure. Structural and thermodynamic changes in the rRNA are believed to extend globally in the whole ribosome and not to be confined to the vicinity of the modified site (Decatur and Fournier 2002). From the mechanistic viewpoint, the ribosome is a highly mechanical molecular machine, e.g., it performs a ratchet-like movement during translation. Modified nucleotides are probably analogous to “ball bearings” that are strategically placed in the moving parts of the machine. The fact that the rRNA modifications are highly conserved means that natural selective pressures maintain modifications in those particular positions for optimal ribosomal performance. Our phenotypic data indicate a potential for natural selection also to favor these modifications under various nonoptimal growth conditions. In further support of such specific evolutionary importance, a selective advantage in a natural biological context has been shown to be conferred by pseudouridylation of RNA (Charette and Gray 2000). Indeed archaea with higher optimum growth temperature appear to contain more guide RNAs and more modifications (Kirpekar et al. 2005).
MATERIALS AND METHODS
Yeast strains
All yeast strains were kindly provided by Dr. Sean Eddy's laboratory (Washington University School of Medicine, St. Louis, MO). These were the same strains used in characterizing the computationally identified methylation guide box C/D snoRNAs and have been verified to direct the corresponding rRNA methylations (Table 1; Lowe and Eddy 1999). The snoRNA gene disruptions were generated by homologous gene replacement in the wild-type background haploid strain yM4585 (MATa;his3Δ200;lys2–801;leu2–3,2–112;trp1–901;tyr1–501;URA3+;ADE2+;CANS). Strains were immediately revived in rich media after receipt, and cells were stored in 20% (w/v) glycerol solution at –80°C until use.
Media and growth conditions
Strains deep-frozen (−80°C) in 20% glycerol were inoculated in 350 μL of SD medium (0.14% yeast nitrogen base, YNB; Difco) without amino acids, 0.5% ammonium sulfate, and 1% succinic acid; 2% (w/v) glucose; supplemented with complete amino acids per the manufacturer's recommendations (CSM; Bio 101 Systems), pH 5.8, and incubated for ∼72 h at 30°C (referred to as the pre-preculture). This procedure was repeated once (second incubation ∼48 h, referred to as the preculture). For experimental runs, strains were inoculated to an OD of 0.03–0.1 in 350 μL of SD medium and cultivated for 47 h in a Bioscreen analyzer C (Labsystems OY, Vantaa, Finland) during environmental stress at the following concentrations: 6-azauracil (200 μg/mL), AT-3 (315 mM), CdCl2 (47.5 μM), canavanine (1 μg/mL), cycloheximide (0.035 μg/mL), EtBr (45 μg/mL), hydroxyurea (8 mg/mL), KCl (1.45 M), MnCl2 (10 mM), myriocin (2 μg/mL), tunicamycin (1 μg/mL), DMSO (1.5%), anisomycin (0.5 μg/mL), sodium orthovanadate (0.47 mM), tert-butyl-H2O2 (0.35 mM), trifluoperazine (50 μM), rapamycin (0.6 μg/mL), paraquat (300 μg/mL), LiCl (100 mM), caffeine (1.00 mg/mL), fructose (2%, no glucose present), galactose (2%, no glucose present), MMS (0.0015%), neomycin (4 mM), puromycin, paromomycin (12 mg/mL), 2,3-DPG (13 mM), thiabendazole (0.1 μg/mL), and DTT (1.6 mM). Concentrations were selected as to represent approximately equal growth inhibitory effects on the reference strain with no more than 20% growth rate reduction. Optical density was measured using a wide-band (450–580 nm) filter. Incubation was at 30.0°C (±0.1°C) with 10 min of preheating time. Plates were subjected to shaking at highest shaking intensity with 60 s of shaking every other minute. OD measurements were taken every 20 min during a 47-h period. Strains were run in duplicate on separate plates, with four wild-type strains in randomized (single) positions on each plate.
Growth analysis
Growth lag (adaptation time), growth rate, and growth efficiency as well as logarithmic strain coefficients (LSCs) and logarithmic phenotypic indexes (LPIs) were calculated as earlier described (Warringer and Blomberg 2003; Warringer et al. 2003). We performed tests of the null hypothesis that LPI equals 0 separately for each strain and separately for adaptation time, rate of growth, and stationary phase. Statistical significance (P < 0.001) was calculated as earlier described (Warringer et al. 2003).
Data clustering
Function assignment by nearest-neighbor joining (Eisen et al. 1998) was performed using LPI values and agglomerative hierarchical clustering with an uncentered similarity metric (Pearson correlation coefficient).
3D mapping of modified residues
The modified RNA residues were mapped into the combined 3D models of the high-resolution X-ray structures of Thermus thermophilus 16S rRNA (2.8 Å) (Selmer et al. 2006) for the small subunit and Haloarcula marismortui 23S rRNA (2.4 Å) (Ban et al. 2000) for the large subunit. The PDB files (PDB ID: 2j00 and 1ffk) were downloaded from the Protein Data Bank (http://www.rcsb.org) (Berman et al. 2000). All visual and structural manipulations were performed using the extensible molecular modeling system, UCSF Chimera v. 2184 (Pettersen et al. 2004) (http://www.cgl.ucsf.edu/chimera/).
ACKNOWLEDGMENT
We thank Sean Eddy for providing the snoRNA deletion strains. Luciano Fernandez-Ricaud helped in processing the raw data in the PROPHECY database. J.E. and J.W. are financially supported by the National Research School in Genomics and Bioinformatics.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.845808.
REFERENCES
- Bachellerie, J.P., Cavaille, J., Huttenhofer, A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Badis, G., Fromont-Racine, M., Jacquier, A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally, M., Hughes, J., Cesareni, G. SnR30: A new, essential small nuclear RNA from Saccharomyces cerevisiae . Nucleic Acids Res. 1988;16:5291–5303. doi: 10.1093/nar/16.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Zarivach, R., Schluenzen, F., Agmon, I., Harms, J., Auerbach, T., Baram, D., Berisio, R., Bartels, H., Hansen, H.A., et al. Ribosomal crystallography: Peptide bond formation and its inhibition. Biopolymers. 2003;70:19–41. doi: 10.1002/bip.10412. [DOI] [PubMed] [Google Scholar]
- Baxter-Roshek, J.L., Petrov, A.N., Dinman, J.D. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette, M., Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- Decatur, W.A., Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Doi, Y., Yokoyama, K., Yamane, K., Wachino, J., Shibata, N., Yagi, T., Shibayama, K., Kato, H., Arakawa, Y. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 2004;48:491–496. doi: 10.1128/AAC.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Moore, P.B., Steitz, T.A. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Heiss, N.S., Knight, S.W., Vulliamy, T.J., Klauck, S.M., Wiemann, S., Mason, P.J., Poustka, A., Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Hughes, J.M., Konings, D.A., Cesareni, G. The yeast homolog of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel, H., Bock, A. 23S Ribosomal RNA mutations in halobacteria conferring resistance to the anti-80S ribosome targeted antibiotic anisomycin. Nucleic Acids Res. 1987;15:2431–2443. doi: 10.1093/nar/15.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T.H., Liu, B., McCully, R.R., Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- Kirpekar, F., Hansen, L.H., Rasmussen, A., Poehlsgaard, J., Vester, B. The archaeon Haloarcula marismortui has few modifications in the central parts of its 23S ribosomal RNA. J. Mol. Biol. 2005;348:563–573. doi: 10.1016/j.jmb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M., Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Ofengand, J., Bakin, A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria, and chloroplasts. J. Mol. Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- Osterberg, M., Kim, H., Warringer, J., Melen, K., Blomberg, A., von Heijne, G. Phenotypic effects of membrane protein overexpression in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2006;103:11148–11153. doi: 10.1073/pnas.0604078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., Simmons, T., Shuster, E.O., Siliciano, P.G., Guthrie, C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: Viable sextuple mutant. Mol. Cell. Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska, D., Decatur, W.A., Fournier, M.J. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.H., Henras, A., Lu, Y.J., Zhou, H., Zhou, W.X., Zhu, Y.Q., Zhao, J., Henry, Y., Caizergues-Ferrer, M., Bachellerie, J.P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., Grisendi, S., Piazza, F., Rego, E., Mari, F., Rao, P.H., Cordon-Cardo, C., Pandolfi, P.P. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.E., Clayton, D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker, R.J., Gultyaev, A.P. Computer simulation of chaperone effects of Archaeal C/D box sRNA binding on rRNA folding. Nucleic Acids Res. 2006;34:2015–2026. doi: 10.1093/nar/gkl154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.T., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Thatcher, J.W., Shaw, J.M., Dickinson, W.J. Marginal fitness contributions of nonessential genes in yeast. Proc. Natl. Acad. Sci. 1998;95:253–257. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Guthrie, C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985;4:3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer, J., Blomberg, A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae . Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- Warringer, J., Ericson, E., Fernandez, L., Nerman, O., Blomberg, A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, A., Peng, G., Brandenburger, Y., Zollo, O., Xu, W., Rego, E., Ruggero, D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Zagorski, J., Tollervey, D., Fournier, M.J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol. Cell. Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]