Abstract
The cleavage rates of 78 hammerhead ribozymes containing structurally conservative chemical modifications were collected from the literature and compared to the recently determined crystal structure of the Schistosoma mansoni hammerhead. With only a few exceptions, the biochemical data were consistent with the structure, indicating that the new structure closely resembles the transition state of the reaction. Since all the biochemical data were collected on minimal hammerheads that have a very different structure, the minimal hammerhead must be dynamic and occasionally adopt the quite different extended structure in order to cleave.
Keywords: RNA dynamics, ribozyme, RNA crystal structure, RNA structure–function, catalytic core
INTRODUCTION
The structure of the hammerhead RNA self-cleaving motif was initially defined in 1987 to consist of three helices intersecting in a conserved catalytic core (Forster and Symons 1987). Such “minimal” hammerheads were the subject of extensive structural, biochemical, and biophysical study for more than 15 years until it was appreciated that an additional, nonconserved tertiary interaction was essential for cleavage in vivo (De la Pena et al. 2003; Khvorova et al. 2003). Such “extended” hammerheads were subsequently shown to cleave or ligate in vitro from 50- to 500-fold faster than minimal hammerheads (Canny et al. 2004, 2007; Nelson et al. 2005; Osborne et al. 2005; Roychowdhury-Saha and Burke 2006) A recent crystal structure of an extended hammerhead shows that the scissile phosphate lies in the interior of the molecule, and the attacking and leaving oxygens are nearly in an in-line configuration, suggesting that the molecule is close to the catalytically active conformation (Martick and Scott 2006). This new structure is dramatically different from several crystal structures of minimal hammerheads where the catalytic core folds into two separate domains and the scissile phosphate is on the outside of the molecule and the attacking and leaving oxygens are not in-line (Pley et al. 1994; Scott et al. 1995).
Considering that they are missing the tertiary interaction and adopt a quite different structure, minimal hammerhead ribozymes are quite efficient in catalysis. Based on the uncatalyzed rate of RNA cleavage of 10−6 min−1 (Li and Breaker 1999), the rate enhancement of minimal hammerheads is about 106-fold while the rate enhancement of extended hammerheads is between 108- and 109-fold. Thus, the minimal hammerhead shows approximately two-thirds of the catalytic efficiency of the extended hammerhead. Based on numerous biochemical experiments, Perracchi and Herschlag concluded that the crystal structure of the minimal hammerhead did not reflect the conformation that actually cleaved, but instead was in rapid equilibrium with some quite different conformation that was active (Peracchi et al. 1997, 1998). It now seems clear that their prediction was correct, and that the active conformation presumably more closely resembles the new extended hammerhead structure. The slower cleavage rate of the minimal hammerhead reflects the fact that it spends most of the time in an inactive state, but occasionally adopts the active structure. Since this conformational equilibriation occurs on a very rapid time scale compared to the rate of cleavage, it cannot be detected by experiments that measure the cleavage rate. The role of the tertiary interaction in the extended hammerhead may be to increase the fraction of time that the molecule spends in the active conformation. This would explain why the tertiary interaction varies among natural hammerheads and why it can be replaced by other tertiary interactions found by in vitro selection experiments (Saksmerprome et al. 2004).
Recent experimental data supports this dynamic model for the mechanism of minimal hammerhead catalysis. The extended hammerhead contains a tertiary Watson-Crick base pair between residues C3 and G8 that is not present in the minimal hammerhead structure. Mutational experiments performed on several different extended hammerheads show that while point mutations are inactive, other Watson–Crick base pairs at positions 3 and 8 have significant activity (Martick and Scott 2006; Przybilski and Hammann 2007; Nelson and Uhlenbeck 2008). This indicates that this pair is needed to maintain the folded structure of the catalytically active extended hammerhead. When the same mutations were made in a minimal hammerhead, a similar requirement for the formation of the 3–8 base pair was observed (Nelson and Uhlenbeck 2008). This strongly supports the idea that the minimal hammerhead must adopt a structure that resembles the extended hammerhead crystal structure before it can cleave or ligate.
In order to confirm that the new extended hammerhead structure resembles the active conformation, it will be necessary to perform “structure–function” experiments where defined chemical modifications are introduced into discrete sites in the RNA and the activity of the modified hammerhead compared with the wild type. While such experiments remain to be done, the fact that the minimal hammerhead adopts an active structure that is similar to the extended hammerhead means that it is appropriate to compare the new structure with the large body of available data on the catalytic activities of chemically modified minimal hammerheads. This comparison is the focus of this review. If the new structure does indeed reflect the conformation needed for catalysis, one would expect it would be largely consistent with the old biochemical data. As has been previously discussed in several reviews (McKay 1996; Verma et al. 1997; Blount and Uhlenbeck 2005), a substantial fraction of the biochemical data cannot be reconciled with the very different minimal hammerhead structure.
Although a large number of papers have appeared reporting the cleavage properties of chemically modified minimal hammerheads, not all of these data are appropriate for comparing to a structure. Several criteria have been used to identify those experiments that clearly can be interpreted in terms of the catalytically active structure (Blount and Uhlenbeck 2005). First, it is critical that the modifications are made in a hammerhead that is substantially free of alternative conformations in order to avoid the possibility that a modification could affect a spurious conformational change rather than the hammerhead cleavage reaction. Second, the measured cleavage rate must reflect the chemical cleavage step, and not substrate binding or product dissociation steps. This is achieved by performing single turnover cleavage reactions at saturating concentrations of ribozyme (Stage-Zimmermann et al. 1998). Finally, modified nucleotides should be chosen that only make modest atomic or functional group changes and that do not change the protonation state or the stacking properties of a base. Ideal modifications either replace an atom such as N-deaza nucleotides or phosphorothioates or remove a functional group such as inosine, purine, 2-pyrimidinone, or deoxyribose substitutions, since they are least likely to disrupt the structure. However, even such modest changes have sometimes been found to cause distortions in a folded RNA structure (Salter et al. 2006). Modifications that introduce steric bulk should especially be avoided since steric clash can alter the structure in an unpredictable manner and thereby complicate interpretation. While these restrictions mean that a lot of data in the literature cannot be used, it ensures that the selected data reliably reports on modified hammerheads that are likely to be similar in structure to their unmodified counterpart.
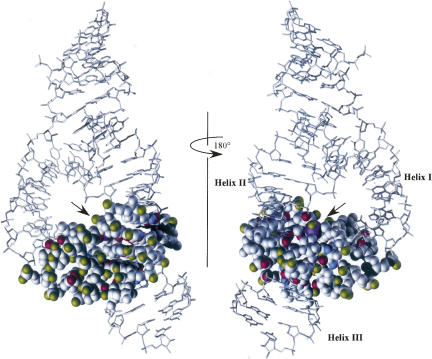
Using the above criteria, a set of 54 modified hammerheads was originally selected in 2004 for comparison with the minimal structure (Blount and Uhlenbeck 2005). Data for 24 additional modified hammerheads are included here for a total of 78 experiments that can be compared with the extended structure. The cleavage rate of each modified hammerhead was divided by the rate of the corresponding unmodified control to give a k rel value, thereby removing differences due to the slightly different buffers and temperatures used by the different laboratories. The data for the 78 modified hammerheads are summarized in Table 1. k rel values are categorized as having little or no effect (k rel > 0.1), moderate effect (0.1 > k rel > 0.01), and strong effect (k rel < 0.01). In the figures shown in this review, the functional groups associated with these categories were marked in green, blue, and red, respectively. This permits convenient visual comparison of the biochemical data with the structure (Figs. 1, 2).
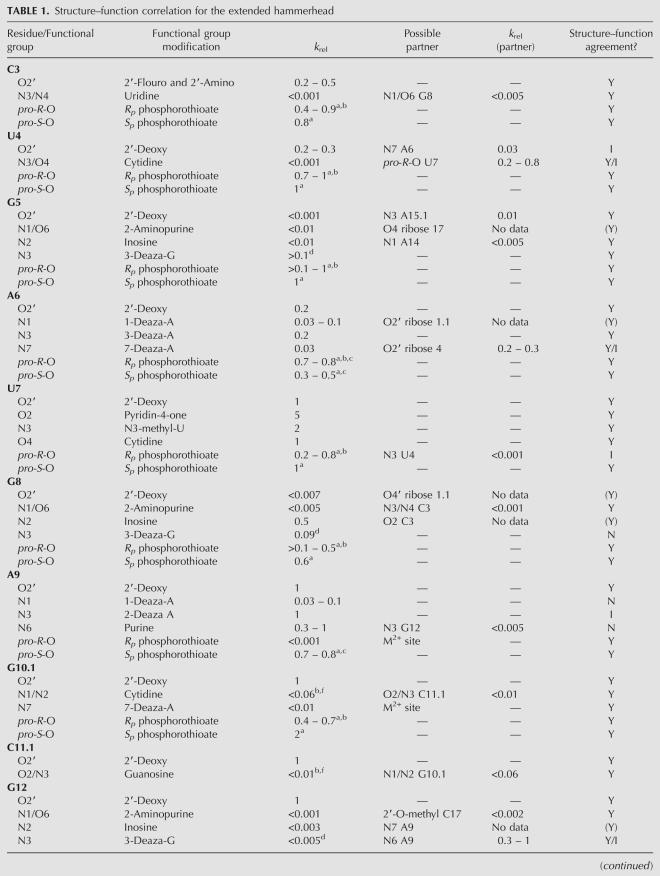
TABLE 1.
Structure–function correlation for the extended hammerhead
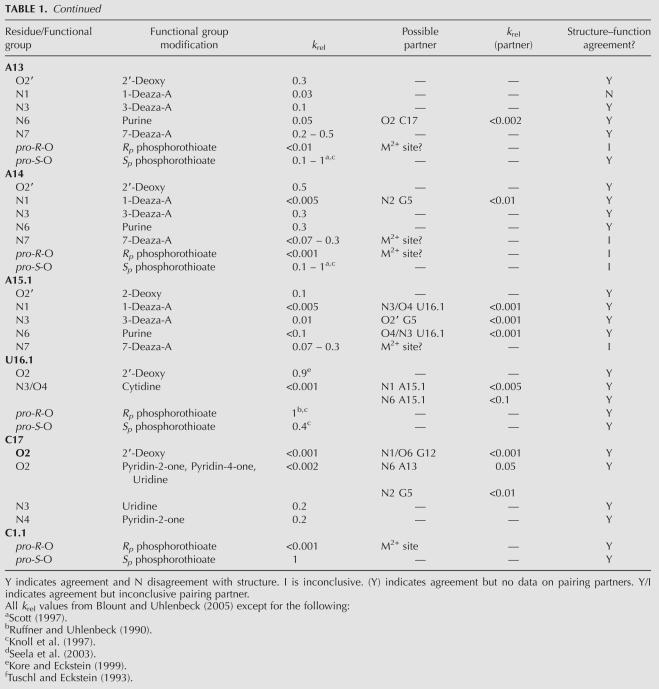
FIGURE 1.
The structure of Schistosoma mansoni hammerhead (PDB:2G07) with individual interacting nucleotides in the core color-coded based on their effect on k rel when individual functional groups were modified; red (>100-fold effect); blue (10–100-fold effect); green (<10-fold effect). Solid lines indicate hydrogen bonds where modification of one or both interacting atoms affects k rel. Dotted lines indicate potential hydrogen bonds where the biochemical data is ambiguous (see text). Solid arrow marks the cleavage site.
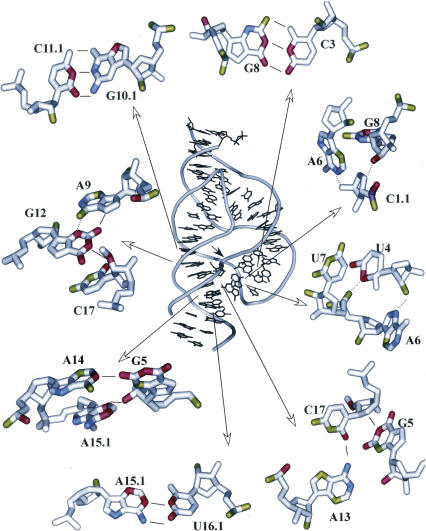
FIGURE 2.
k rel data depicted in a secondary structure with color coding identical to Figure 1.
The Schistosoma mansoni extended hammerhead used for crystallography was prepared by combining a 43-nucleotide (nt) ribozyme strand and a 20-nt substrate strand that contained a 2′-O-methyl group at the cleavage site to prevent cleavage. Hanging drop crystals appeared slowly using a buffer of 0.5 M (NH4)2SO4, 100 mM Mes pH 5.5, 1 mM MgCl2, 1.5 mM EDTA. The resulting structure was refined to a resolution of 2.2 Å (Martick and Scott 2006). For defining the hydrogen bonds that stabilize the core of this structure, we used the conservative criteria that the donor and acceptor atoms must be within 3.0 Å and be positioned in a way to permit a bond angle of >150° (Blount and Uhlenbeck 2005). However, these criteria were relaxed to 3.5 Å and >135° for the group of hydrogen bonds in the neighborhood of the cleavage site to account for the likely local perturbation of the structure induced by the 2′O-methyl group. Before comparing the structure with the biochemical data in detail, it is important to note that crystallization was done in a buffer very different than what was used in the activity assays, which could potentially alter the hydrogen-bonding pattern. Finally, we reiterate that the biochemical experiments are all on the minimal hammerhead, so requiring congruence with the extended structure assumes that they use an identical transition state arrangement.
Twenty-six modifications were made to functional groups that appear to participate in 19 hydrogen bonds present in the folded catalytic core of the extended hammerhead. Although it is hard to predict the magnitude of the effect, one would expect that these modifications would destabilize the core structure and thereby reduce the cleavage rate. Of the 26 modifications tested, 22 had either a moderate or a large effect on k rel, while four had no effect. In 10 cases both the donor and acceptor groups of a given hydrogen bond have been modified, and both showed a reduced k rel. This includes modifications which disrupt the hydrogen bonds of the G10.1–C11.1, A15.1–U16.1, and C3–G8 base pairs that are presumably critical for the stability of the catalytic core. The G10.1–C11.1 and A15.1–U16.1 pairs are further established by the fact that they can be replaced by the isosteric A10.1–U11.1 and I15.1–C16.1 pairs without effecting cleavage (Ruffner et al. 1990; Tuschl and Eckstein 1993; Ludwig et al. 1998). The C3–G8 base pair can be replaced by the other three Watson–Crick pairs, confirming its structural role (Martick and Scott 2006; Przybilski and Hammann 2007; Nelson and Uhlenbeck 2008). Since the G8I modification can still form a base pair, it is not surprising that it remains active (Ludwig et al. 1998). Thus, all the available data associated with these three base pairs agree with the structure.
In addition to the three base pairs, there are four single hydrogen bonds where modification of both partners reduces k rel (Fig. 1). These include the bond between the N1 of A14 and the NH2 of G5, the N6 of A13 and the O2 of C17, the 2′OH of G5 with the N3 of A15.1, and the N1/O6 of G12 with the 2′OH of C17 (the site of the O'methyl substitution). While it appears that the biochemical data supports these four single hydrogen bonds in the crystal structure, the absence of compensatory experiments similar to those for the base pairs makes these results less convincing. It remains possible that both bonding partners are essential for stabilizing a different structure. Indeed, biochemical experiments originally seemed to confirm the hydrogen bond between the NH2 of G12 and the 2′OH of G8 present in the minimal hammerhead structure since both functional groups were essential (Blount and Uhlenbeck 2005). It is now clear that these two functional groups participate in hydrogen bonds with other partners in the extended hammerhead structure (Table 1). Thus, in order to definitively confirm a hydrogen bond biochemically, it is necessary to show that the hydrogen bond donor and acceptor can be exchanged. While there is a limited precedence for this in folded RNA structures (Strobel and Shetty 1997), this is technically demanding and not always possible due to limitations in the available modified nucleotides.
There are four hydrogen bonds present in the structure where disruption of one of the bonding partners shows a reduced k rel while the other has not been tested (Fig. 1). Since the substitution of the 2′OH of ribose 8 with a deoxynucleotide dramatically reduces k rel, its hydrogen bond with the 5′oxygen of C1.1 appears essential. Since the 5′oxygen of C1.1 is the catalytic leaving group, it must be essential, but has not been substituted. The 1-deaza A6 hammerhead shows a decreased k rel; however, its partner, the 2′OH of ribose 1.1, has not been modified. However, since mismatches of the 1.1–1.2 base pair dramatically reduce catalysis (Werner and Uhlenbeck 1995) it seems likely that this hydrogen bond is also essential. Substitution of G5 by 2-aminopurine dramatically reduces k rel, suggesting that either its N1 or its O6 are critical to catalysis. While the O6 is not near any atom, the proton on N1 is positioned to donate a hydrogen bond to the furanose O4 of C17. While the furanose O4 has not been modified, this type of hydrogen bond has been seen in other nucleic acid structures (Berger et al. 1996), including, interestingly, the ribose at the cleavage site of the hairpin ribozyme (Alam et al. 2005). Finally, for the single hydrogen bond between the amino group of G12 and the N7 of A9, removing the amino group by the G12I mutation reduces k rel, but no reliable k rel is available for the 7-deaza A9 hammerhead. In summary, while these four hydrogen bonds are likely to be essential for catalysis, additional experiments are required to confirm them.
Finally, six of the 26 modifications of functional groups participating in three hydrogen bonds involve cases where the modification of one pairing partner substantially reduces k rel while the other has little effect. These are the hydrogen bonds between the NH2 of A9 and the N3 of G12, between the 2′OH of ribose 4 and the N7 of A6, and between the O4/N3 of U4 and the O2P of U7. The purine 9, deoxy ribose 4, and the Rp phosphorothioate 7 modifications all reduce k rel by a modest three- to fivefold, potentially consistent with the disruption of a less critical hydrogen bond. However, modification of the three corresponding pairing partners to 3-deaza-G12, 7-deaza-A6, and C4 all show much larger, 30- to >1000-fold effects on k rel. This asymmetry in k rel values may simply be a consequence of the type of modifications made, but an alternative explanation is that the hydrogen bonds shown in the structure are not correct and a different structure forms where the three essential groups make other interactions while the three less essential groups are not involved in the folded structure. However, examination of the structure does not reveal any obvious alternative pairing schemes with better agreement of structure with function. It is also possible that the modification of the three more essential groups changes the structure of the hammerhead in some unanticipated manner and thereby artificially reduces k rel. Finally, some of the biochemical data could simply be wrong. Since these three cases are potential disagreements with the new structure, a reevaluation of the biochemical data is needed, both in the minimal and extended hammerheads.
In summary, while there may be a few potential discrepancies, the biochemical experiments that test the hydrogen bonds examined in the extended hammerhead structure generally give the expected result. When the hydrogen bond is disrupted, k rel is usually reduced. Of the 19 potential hydrogen bonds in the core of the extended hammerhead, point mutations and compensatory mutations fully confirmed eight of them. For four hydrogen bonds, separate modifications of both donor and acceptor reduce k rel, while for four others modification of one of the partners reduces k rel and the other has not been tested. Finally, there are three hydrogen bonds where one partner is essential and the other partner is much less so, an ambiguous result that must be retested. No additional hydrogen bonds with the proper angular orientation or distance are present in the core of the extended hammerhead structure. Thus, not only have all the possible hydrogen bonds been tested biochemically, but the vast majority of the data is in agreement with the structure.
The remaining 52 modifications in Table 1 were made to functional groups that were >4 Å from other residues in the extended hammerhead structure. Since they are unlikely to be participating in direct hydrogen bonding to stabilize the structure they should have no effect on catalysis. Of these 52, 42 of them indeed have no effect on k rel. As illustrated in Figure 3, the functional groups that were modified (marked in green) primarily reside on the outside surface of the core where interactions with other groups are unlikely. A few are located in a cleft within the core where helix I and helix II hinge together and form a large open space. In addition to the conservative modifications in Table 1, we note that the experiments that purposely introduced steric bulk into the 2′OH positions also agree with the extended hammerhead structure (Blount et al. 2002). Thus, this large set of modification experiments is consistent with the new structure.
FIGURE 3.
Space-filling model of the core, color coded and oriented in the same manner as Figure 1.
However, there are 10 cases where a modification shows a significantly reduced k rel despite the fact that the modified functional group is not sufficiently close to any other atom to form a direct hydrogen bond. Since many factors could contribute to a reduced k rel, these cases do not necessarily disagree with the structure but each one must be carefully considered. Three of the 10 cases are almost certainly associated with the binding of a divalent metal ion that is needed to maintain the folded structure and may play a role in catalysis. As shown in Figure 4A, when manganese is soaked into the crystals of the extended hammerhead, it binds to a site involving inner sphere coordination with the essential N7 of G10.1 and the essential pro-R-oxygen of phosphate 9. The presence of this metal ion has been confirmed by biochemical experiments using both the minimal and extended hammerheads which show that when an Rp phosphorothioate is introduced at P9 k rel is strongly reduced, but cleavage can be restored by the addition of the thiophilic Cd2+ ion (Ruffner and Uhlenbeck 1990; Wang et al. 1999; Osborne et al. 2005). The Rp phosphorothioate at P1.1 can also be rescued with the addition of cadmium ion in both the minimal and extended hammerheads, suggesting that it also coordinates directly with a metal ion (Ruffner and Uhlenbeck 1990; Scott and Uhlenbeck 1999; Wang et al. 1999; Derrick et al. 2000; Osborne et al. 2005). However, no such direct coordination is seen in the extended structures, and the metal ion at P9 is a bit too far (4.9 Å) from the pro-R-oxygen of P1.1 to even coordinate through a water molecule. Thus, either a second unobserved metal ion binds to P1.1, or the structure rearranges slightly in the transition state (Wang et al. 1999). Indeed, if P1.1 were repositioned about 0.5 Å closer to the pro-R-oxygen of P9 and 1.1 Å closer to N7 of G10.1, a single metal ion could be modeled into this site. In either case, these three essential functional groups are unlikely to represent a disagreement with the structure.
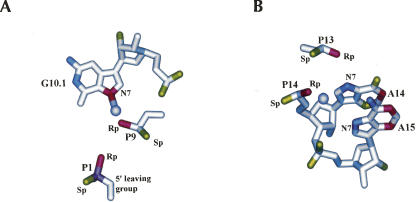
FIGURE 4.
Manganese ions (lavender) near P9 (A) and P14 (B) in the catalytic core (PDB:20EU).
It is possible that four of the remaining seven essential functional groups that are distant from other atoms may be involved in binding additional essential metal ions. As shown in Figure 4B, the essential N7 of A14 and the pro-R-oxygen of P13 as well as the essential N7 of A15 and the essential pro-R-oxygen at P14 could each potentially coordinate a metal ion. Indeed, when manganese ions are soaked into the extended hammerhead crystals, a single hydrated metal ion is observed within 2 Å of the pro-R-oxygen of P14, consistent with an inner sphere contact. However, no coordination with the N7 of A15 is observed. While 31P NMR data indicates that a metal ion is bound to the P13 of the minimal hammerhead (Hansen et al. 1999), no data are available for the extended hammerhead. Biochemical evidence for the metal ions coordination to either of these sites is weak. Since the inactive pro-R-phosphorothioate substitutions at P13 and P14 are not reactivated by the addition of cadmium ions (Ruffner and Uhlenbeck 1990), their reduced k rel may be a consequence of the known tendency of phosphorothioate substitutions to disrupt RNA folding (Smith and Nikonowicz 2000; Suzumura et al. 2000, 2004; Lupták and Doudna 2004) rather than the result of blocking metal ion binding. Furthermore, although two labs prepared hammerheads with 7-deaza-A at A14 and A15, one reported no effect on k rel (Seela et al. 1993) while the other reported a modest decrease in k rel (Fu and McLaughlin 1992) that was much less than the effect on k rel by the 7-deaza-G 10.1 hammerhead which disrupts the well-established P9 metal binding site (Pley et al. 1994; Scott et al. 1995, 1996). However, both labs used hammerheads with very stable helices and determined cleavage rates with multiple turnover conditions where k obs is likely to partially reflect product release instead of only cleavage chemistry (Stage-Zimmermann et al. 1998), so it is possible that the k rel values are much greater. Thus, it is unclear whether or not these four essential functional groups are involved in metal ion binding or represent a disagreement with the structure.
There are three remaining cases where essential functional groups do not appear to be close enough to another atom to make a productive interaction. The reduced k rel values of 3-deaza-G8, 1-deaza-A9, and 1-deaza-A13 suggest that the ring nitrogens of these residues may act as hydrogen bond acceptors. However, the N3 of G8 is not close to any possible hydrogen bond donor, and the closest hydrogen bond donor to the N1 of A9 is the 2′OH of G12 (3.7 Å), and the closest donor to the N1 of A13 is the 2′OH of A6 (4.2 Å). Since these 2′-hydroxyls can be changed to a deoxynucleotide without changing k rel, it is unlikely that they participate in hydrogen bonds. There are several explanations that may account for these three apparent disagreements between the functional data and the structure. The first is that the altered electronic structure of the 3-deaza-G or 1-deaza-A nucleotides disrupts the folding of the extended hammerhead in some other manner instead of simply removing the nitrogen. For example, since A9 and A13 participate in stacking interactions with A14 and A15.1 that are believed to be critical for catalysis, the altered stacking properties of 1-deaza-A (Seela et al. 1998) may subtly effect the positioning of A14 and A15.1. Similarly, the 3-deaza-G8 modification may weaken the critical C3–G8 base pair. Alternatively, the deaza modifications may, in some way, stabilize the A9–G12 and A13–G8 base pairs that form in the minimal hammerhead. Any mutations which favors the minimal hammerhead structure will reduce the time spent in the extended structure and thereby reduce k rel. Finally, the crystal structure shows the arrangement of atoms in an uncleaved hammerhead that is blocked from cleavage by a 2′-O-methyl group. In contrast, the biochemical data reports on the transition state of an active hammerhead. Thus, it is possible that either distortions caused by the 2′-O-methyl group or small structural changes that occur on the way to the transition state may account for the need for the N3 nitrogen at G8 and the N1 nitrogens at A9 and A13.
This review summarizes the results of experiments that measure the cleavage rates of 78 different chemically modified minimal hammerhead ribozymes. The data represents the efforts of more than 12 different laboratories over a period of nearly 20 years, with many of the experiments performed multiple times. It is noteworthy that all of the experiments were performed well before the Schistosoma crystal structure appeared. Considering that the choice of modifications was unbiased by this structural information, the experimental “coverage” of the structure is excellent. With the exception of five cases, all 38 hydrogen bond donors and acceptors that participate in the folding of the hammerhead core were tested. In addition, 52 sites in the core were modified that did not appear to contribute to stabilizing the folded structure. As summarized in this review, the overall agreement of the biochemical data with the structure is astonishingly good, considering the disparate data set. With the exception of seven ambiguities and three outright disagreements, all of the functional data is simply explained by the Schistosoma structure. This agreement contrasts dramatically with the earlier comparison of functional data with the minimal hammerhead structure where less than half of the experiments could be reconciled (Blount and Uhlenbeck 2005).
The clear congruence between the structure and the biochemical data documented in this review permits two important conclusions to be made. First, the structure of the 2′-O-methyl blocked Schistosoma hammerhead must closely reflect the transition state of the hammerhead reaction. This fact is very important for designing experiments directed at deducing the catalytic mechanism. Second, since all the biochemical data were determined with a minimal hammerhead, it is clear that despite its quite different structure, the minimal hammerhead must go through the same transition state as the Schistosoma hammerhead. This means that the minimal hammerhead must be very dynamic, only adopting the active structure infrequently. One calculation estimates that the minimal hammerhead is in the active conformation <0.2% of the time (Nelson and Uhlenbeck 2008).
It will be important to confirm the above conclusions by making selected modifications in an extended hammerhead and measuring k rel. Emphasis should be placed on making modifications at those sites discussed above that were not previously tested or were ambiguous. It would also be useful to confirm some of the critical core hydrogen bonds by performing acceptor–donor swaps using appropriate modified nucleotides. In addition, the binding sites of essential metal ions must be better established and distinguished from the more global ionic strength effects. This will probably best be done using direct spectroscopic methods instead of functional group substitutions to avoid making unintended structural changes.
The minimal hammerheads will continue to be a valuable system to more carefully study the role of dynamics in RNA catalysis. It will be useful to directly measure the dynamics of the conformational isomerization that is proposed to occur between the minimal and extended hammerhead core structures, perhaps by using single molecule FRET experiments with fluorophores carefully placed within the core. Another interesting approach is to use the minimal and extended hammerhead structures to design modifications that would destabilize the minimal structure and not effect (or even stabilize) the extended structure. Such modifications would be expected to enhance the cleavage rate of the minimal hammerhead compared to the wild-type sequence. An example of such a modification that was found accidentally in the minimal hammerhead involved introducing a pyridin-4-one at position 7 that appears to enhance cleavage by destabilizing the A14–U7 pair in the minimal hammerhead structure and not affecting the structure of the extended hammerhead (Burgin et al. 1996; Nelson and Uhlenbeck 2006). It will be interesting to see whether a pyridin-4-one at position 7 or any other activating modification would also enhance the rate of cleavage when introduced into an extended hammerhead. This would support the idea that extended hammerheads also dynamically sample an inactive state that resembles the minimal hammerhead structure a significant fraction of the time (Nelson and Uhlenbeck 2008).
The likelihood that the Schistosoma core structure closely resembles the transition state of hammerhead catalysis opens the way to experiments directed at understanding the catalytic mechanism of the hammerhead. One important goal will be to determine whether any of the nucleobases near the scissile phosphate such as G12, A6, or G5 donate or abstract a proton during the reaction cycle (Han and Burke 2005). There is clear evidence that nucleobases participate in the mechanisms of the hairpin and hepatitis delta ribosomes, although exactly how they function remains unclear (Das and Piccirilli 2005; Kuzmin et al. 2005; Perrotta et al. 2006; Nakano and Bevilacqua 2007). Another goal will be to better understand how the metal ion bound to P9 participates in the reaction. Is it simply structural or does it play a role in the mechanism, such as helping to neutralize the scissile phosphate in the transition state? Such mechanistic studies must also explain the increase in the rate of cleavage with pH. While chemically modified hammerheads may be useful in these studies, a critical challenge will be to show that the modification directly affects the chemical mechanism rather than altering the structure or the conformational dynamics of the molecule.
A final interesting avenue of hammerhead research will be to understand the surprising diversity in the catalytic rates and the cleavage/ligation equilibria observed among natural hammerheads (De la Pena et al. 2003; Khvorova et al. 2003; Canny et al. 2004, 2007; Nelson et al. 2005; Osborne et al. 2005; Przybilski et al. 2005; Carbonell et al. 2006; Roychowdhury-Saha and Burke 2006). Since most of these hammerheads share the same catalytic core, the differences must in some way be due to the structure or stability of the different tertiary interactions that hold helices I and II in proximity. In addition, there are structural variations of the catalytic core that seem to impact the activity by an unknown mechanism (De la Pena and Flores 2001).
In conclusion, it is clear that the hammerhead ribozyme will continue to be a useful system to understand the structure and dynamics of RNA.
ACKNOWLEDGMENTS
The project described above was supported by R01GM36944-22 from the National Institutes of Health, and 1C06RR018850-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its content is solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.912608.
REFERENCES
- Alam, S., Grum-Tokars, V., Krucinska, J., Kundracik, M.L., Wedekind, J.E. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Berger, I., Egli, M., Rich, A. Inter-strand C-H…O hydrogen bonds stabilizing four-stranded intercalated molecules: Stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. 1996;93:12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, K.F., Uhlenbeck, O.C. The structure–function dilemma of the hammerhead ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Blount, K.F., Grover, N.L., Mokler, V., Beigelman, L., Uhlenbeck, O.C. Steric interference modification of the hammerhead ribozyme. Chem. Biol. 2002;9:1009–1016. doi: 10.1016/s1074-5521(02)00218-1. [DOI] [PubMed] [Google Scholar]
- Burgin A.B., Jr, Gonzalez, C., Matulic-Adamic, J., Karpeisky, A.M., Usman, N., McSwiggen, J.A., Beigelman, L. Chemically modified hammerhead ribozymes with improved catalytic rates. Biochemistry. 1996;35:14090–14097. doi: 10.1021/bi961264u. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., Pardi, A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Pardi, A. Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry. 2007;46:3826–3834. doi: 10.1021/bi062077r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A., De la Pena, M., Flores, R., Gago, S. Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: Differences in co- and post-transcriptional self-cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res. 2006;34:5613–5622. doi: 10.1093/nar/gkl717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S.R., Piccirilli, J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Flores, R. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: Implications for the design of more efficient ribozymes. J. Biol. Chem. 2001;276:34586–34593. doi: 10.1074/jbc.M103867200. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Gago, S., Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick, W.B., Greef, C.H., Caruthers, M.H., Uhlenbeck, O.C. Hammerhead cleavage of the phosphorodithioate linkage. Biochemistry. 2000;39:4947–4954. doi: 10.1021/bi000146a. [DOI] [PubMed] [Google Scholar]
- Forster, A.C., Symons, R.H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Fu, D.J., McLaughlin, L.W. Importance of specific adenosine N7-nitrogens for efficient cleavage by a hammerhead ribozyme. A model for magnesium binding. Biochemistry. 1992;31:10941–10949. doi: 10.1021/bi00160a001. [DOI] [PubMed] [Google Scholar]
- Han, J., Burke, J.M. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Hansen, M.R., Simorre, J.P., Hanson, P., Mokler, V., Bellon, L., Beigelman, L., Pardi, A. Identification and characterization of a novel high affinity metal-binding site in the hammerhead ribozyme. RNA. 1999;5:1099–1104. doi: 10.1017/s1355838299990738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., Jayasena, S.D., Amgen, I. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Knoll, R., Bald, R., Furste, J.P. Complete identification of nonbridging phosphate oxygens involved in hammerhead cleavage. RNA. 1997;3:132–140. [PMC free article] [PubMed] [Google Scholar]
- Kore, A.R., Eckstein, F. Hammerhead ribozyme mechanism: A ribonucleotide 5′ to the substrate cleavage site is not essential. Biochemistry. 1999;38:10915–10918. doi: 10.1021/bi991332n. [DOI] [PubMed] [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Cottrell, J.W., Fedor, M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Li, Y.F., Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- Ludwig, J., Blaschke, M., Sproat, B.S. Extending the cleavage rules for the hammerhead ribozyme: Mutating adenosine15.1 to inosine15.1 changes the cleavage site specificity from N16.2U16.1H17 to N16.2C16.1H17. Nucleic Acids Res. 1998;26:2279–2285. doi: 10.1093/nar/26.10.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupták, A., Doudna, J.A. Distinct sites of phosphorothioate substitution interfere with folding and splicing of the Anabaena group I intron. Nucleic Acids Res. 2004;32:2272–2280. doi: 10.1093/nar/gkh548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, D.B. Structure and function of the hammerhead ribozyme: An unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- Nakano, S., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: A mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Shepotinovskaya, I., Uhlenbeck, O.C. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry. 2005;44:14577–14585. doi: 10.1021/bi051130t. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Uhlenbeck, O.C. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Uhlenbeck, O.C. Minimal and extended hammerheads utilize a similar dynamic reaction mechanism for catalysis. RNA. 2008;14:43–54. doi: 10.1261/rna.717908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, E.M., Schaak, J.E., Derose, V.J. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi, A., Beigelman, L., Scott, E.C., Uhlenbeck, O.C., Herschlag, D. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- Peracchi, A., Karpeisky, A., Maloney, L., Beigelman, L., Herschlag, D. A core folding model for catalysis by the hammerhead ribozyme accounts for its extraordinary sensitivity to abasic mutations. Biochemistry. 1998;37:14765–14775. doi: 10.1021/bi980867y. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Wadkins, T.S., Been, M.D. Chemical rescue, multiple ionizable groups, and general acid-base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Przybilski, R., Hammann, C. The tolerance to exchanges of the Watson–Crick base pair in the hammerhead ribozyme core is determined by surrounding elements. RNA. 2007;13:1625–1630. doi: 10.1261/rna.631207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybilski, R., Graf, S., Lescoute, A., Nellen, W., Westhof, E., Steger, G., Hammann, C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana . Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury-Saha, M., Burke, D.H. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner, D.E., Uhlenbeck, O.C. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 1990;18:6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner, D.E., Stormo, G.D., Uhlenbeck, O.C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Saksmerprome, V., Roychowdhury-Saha, M., Jayasena, S., Khvorova, A., Burke, D.H. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter, J., Krucinska, J., Alam, S., Grum-Tokars, V., Wedekind, J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E.C. University of Colorado; Boulder: 1997. A phosphorothioate analysis of the nonbridging phosphate oxygens in the hammerhead ribozyme. Ph.D. thesis, [Google Scholar]
- Scott, E.C., Uhlenbeck, O.C. A reinvestigation of the thio effect at the hammerhead cleavage site. Nucleic Acids Res. 1999;27:479–484. doi: 10.1093/nar/27.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Murray, J.B., Arnold, J.R., Stoddard, B.L., Klug, A. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- Seela, F., Mersmann, K., Grasby, J.A., Gait, M.J. 7-Deazaadenosine- oligoribonucleotide building-block synthesis and autocatalytic hydrolysis of base-modified hammerhead ribozymes. Helv. Chim. Acta. 1993;76:1809–1820. [Google Scholar]
- Seela, F., Debelak, H., Usman, N., Burgin, A., Beigelman, L. 1-Deazaadenosine: Synthesis and activity of base-modified hammerhead ribozymes. Nucleic Acids Res. 1998;26:1010–1018. doi: 10.1093/nar/26.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela, F., Debelak, H., Andrews, L., Beigelman, L. Synthesis and enzymic hydrolysis of oligoribonucleotides incorporating 3-deazaguanosine: The importance of nitrogen-3 atom of single conserved guanosine residues on the catalytic activity of the hammerhead ribozyme. Helv. Chim. Acta. 2003;86:2726–2740. [Google Scholar]
- Smith, J.S., Nikonowicz, E.P. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T.K., Uhlenbeck, O.C. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, S.A., Shetty, K. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl. Acad. Sci. 1997;94:2903–2908. doi: 10.1073/pnas.94.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzumura, K., Warashina, M., Yoshinari, K., Tanaka, Y., Kuwabara, T., Orita, M., Taira, K. Significant change in the structure of a ribozyme upon introduction of a phosphorothioate linkage at P9: NMR reveals a conformational fluctuation in the core region of a hammerhead ribozyme. FEBS Lett. 2000;473:106–112. doi: 10.1016/s0014-5793(00)01499-x. [DOI] [PubMed] [Google Scholar]
- Suzumura, K., Takagi, Y., Orita, M., Taira, K. NMR-based reappraisal of the coordination of a metal ion at the pro-Rp oxygen of the A9/G10.1 site in a hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:15504–15511. doi: 10.1021/ja0472937. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Eckstein, F. Hammerhead ribozymes: Importance of stem–loop II for activity. Proc. Natl. Acad. Sci. 1993;90:6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S., Vaish, N.K., Eckstein, F. Structure–function studies of the hammerhead ribozyme. Curr. Opin. Chem. Biol. 1997;1:532–536. doi: 10.1016/s1367-5931(97)80049-x. [DOI] [PubMed] [Google Scholar]
- Wang, S., Karbstein, K., Peracchi, A., Beigelman, L., Herschlag, D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- Werner, M., Uhlenbeck, O.C. The effect of base mismatches in the substrate recognition helices of hammerhead ribozymes on binding and catalysis. Nucleic Acids Res. 1995;23:2092–2096. doi: 10.1093/nar/23.12.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]