Abstract
Bioinformatics searches of eubacterial genomes have yielded many riboswitch candidates where the identity of the ligand is not immediately obvious on examination of associated genes. One of these motifs is found exclusively in the family Streptococcaceae within the 5′ untranslated regions (UTRs) of genes encoding the hypothetical membrane protein classified as COG4708 or DUF988. While the function of this protein class is unproven, a riboswitch binding the queuosine biosynthetic intermediate pre-queuosine1 (preQ1) has been identified in the 5′ UTR of homologous genes in many Firmicute species of bacteria outside of Streptococcaceae. Here we show that a representative of the COG4708 RNA motif from Streptococcus pneumoniae R6 also binds preQ1. Furthermore, representatives of this RNA have structural and molecular recognition characteristics that are distinct from those of the previously described preQ1 riboswitch class. PreQ1 is the second metabolite for which two or more distinct classes of natural aptamers exist, indicating that natural aptamers utilizing different structures to bind the same metabolite may be more common than is currently known. Additionally, the association of preQ1 binding RNAs with most genes encoding proteins classified as COG4708 strongly suggests that these proteins function as transporters for preQ1 or another queuosine biosynthetic intermediate.
Keywords: COG4708, DUF988, noncoding RNA, pre-queuosine1, queuosine, riboswitch
INTRODUCTION
Riboswitches are structured RNAs that bind small molecule metabolites to regulate gene expression (Mandal and Breaker 2004b; Soukup and Soukup 2004; Tucker and Breaker 2005; Winkler and Breaker 2005). They typically consist of a ligand-binding aptamer and an expression platform that translates ligand binding into genetic regulation. The aptamer of each riboswitch class usually is well conserved due to constraints imposed by the ligand binding site. This conservation of aptamer sequences and structures makes bioinformatics analyses of genomic sequences an effective tool for identifying new riboswitch candidates (e.g., Rodionov et al. 2002, 2003; Barrick et al. 2004; Abreu-Goodger and Merino 2005; Corbino et al. 2005; Weinberg et al. 2007). In contrast, expression platforms may vary considerably and utilize diverse genetic regulatory mechanisms such as transcription termination and translation inhibition (Mandal and Breaker 2004b). Although expression platforms usually can be discerned by examining the sequences of individual riboswitches, their relative lack of sequence and structural conservation precludes their usefulness as targets for the bioinformatics discovery of new riboswitch classes.
We have used bioinformatics searches to discover numerous riboswitches and riboswitch candidates (e.g., Barrick et al. 2004; Corbino et al. 2005). Most recently, we reported the discovery of 22 RNA motifs using the CMfinder pipeline (Yao et al. 2007) that was applied to search several hundred bacterial genomes (Weinberg et al. 2007). Six of the 22 motifs exhibit characteristics that are common of cis-regulatory RNAs such as riboswitches. For example, these motifs are consistently positioned immediately upstream of a gene or distinct set of genes. In addition, representatives of these motifs have one of two features: a rho-independent transcription terminator or a stem that overlaps the ribosomal binding site or Shine–Dalgarno (SD) sequence for the gene located immediately downstream. For several of the motifs described, ligand identity could be inferred from the genomic context of the RNA (Weinberg et al. 2007; Wang et al. 2008; E.E. Regulski, R.H. Moy, Z. Weinberg, J.E. Barrick, and R.R. Breaker, in prep.). For other motifs, the genomic context previously provided little or no information about ligand identity. One such RNA motif we found associated only with genes for a conserved hypothetical membrane protein found in Firmicutes that is classified as COG4708 or DUF988.
We recently described a riboswitch responding to the queuosine (Q) biosynthetic precursor preQ1 (Roth et al. 2007). Q is a hypermodified nucleoside found at the wobble position of GUN anticodons in tRNAs for Tyr, Asn, Asp, and His in most bacteria (Harada and Nishimura 1972). The Q modification is known to be important for translational fidelity (Bienz and Kubli 1981; Meier et al. 1985; Urbonavičius et al. 2001). Q biosynthesis starts with GTP and proceeds through the intermediate preQ0 (7-cyano-7-deazaguanine) and preQ1 (7-aminomethyl-7-deazaguanine) in a series of enzymatic steps (Kuchino et al. 1976; Okada et al. 1978; Iwata-Reuyl 2003; Gaur and Varshney 2005; Van Lanen et al. 2005). PreQ1 is preferentially exchanged for a specific guanine in tRNA (Noguchi et al. 1982), and the remaining biosynthetic steps take place in situ (Okada et al. 1979; Slany et al. 1993). Therefore, preQ1 is the last Q precursor that exists in free form prior to insertion into the tRNA.
Representatives of the known preQ1 riboswitch class (Roth et al. 2007) are often located upstream of a Q biosynthetic operon (Reader et al. 2004), but some also have been identified upstream of other genes encoding proteins otherwise not associated with Q, and one of these gene types is classified as COG4708. Thus, we speculated that COG4708 proteins might be involved in transporting Q biosynthetic precursors. However, genes encoding COG4708 proteins in some species are associated with the citrulline biosynthesis operon (argCJBDF), and this operon is repressed by arginine in Lactobacillus plantarum (Arsène-Ploetze et al. 2005). Therefore we could not have high confidence that the new-found RNA motif associated with COG4708 in species of Streptococcaceae was a distinct class of preQ1 riboswitch without additional experimental evidence (Weinberg et al. 2007).
In the present study, we determined that the RNA associated with genes encoding COG4708 proteins binds preQ1 tightly and selectively, and displays characteristics consistent with a genetic “off” switch. Thus we propose that representatives of this RNA motif indeed serve as a second class of preQ1 riboswitches, hereafter termed preQ1-II. The preQ1-II riboswitch aptamer has a substantially different structure and molecular recognition profile than that of the previously described preQ1 riboswitch (Roth et al. 2007), now renamed preQ1-I. Thus, preQ1 joins S-adenosylmethionine (SAM) (Corbino et al. 2005) as the second example of a metabolite recognized by at least two classes of natural RNA aptamers. Moreover, the discovery of new riboswitch classes might be aided by conducting searches in the noncoding regions of mRNAs whose homologs are controlled by known riboswitches.
RESULTS AND DISCUSSION
Bioinformatics of the COG4708 RNA motif
The original description of the COG4708 RNA motif included 22 sequences from the family Streptococcaceae, of which 6 were unique (Weinberg et al. 2007). To identify additional examples of the COG4708 RNA motif, we conducted homology searches using an updated sequence database release (RefSeq25). An additional 18 sequences matching the motif were identified, bringing the total number of sequences to 40 and the number of unique sequences to 13 (Fig. 1).
FIGURE 1.
Alignment of COG4708 RNA motif sequences. Nucleotides that are present in greater than 97% and 75% of the representatives are in uppercase and lowercase, respectively (R = A, G; Y = C, U). Colored regions marked by brackets indicate secondary structure elements P1 (blue), P2 (green), P3 (orange), and P4 (violet). All the sequences depicted are unique. However, some sequences only differ in the loop regions represented by NX and thus appear identical here. Organism abbreviations: (Smu) Streptococcus mutant, (Spn-1) S. pneumoniae R6, (Spn-2) S. pneumoniae SP18-BS74, (Spn-3) S. pneumoniae SP19-BS75, (Spn-4) S. pneumoniae SP6-BS73, (Spy) S. pyogenes, (Sag-1) S. agalactiae 2630V/R, (Sag-2) S. agalactiae COH1, (Ssa) S. sanguinis, (Ssu) S. suis, (Lla-1) Lactococcus lactis subsp. lactis, (Lla-2) L. lactis subsp. cremoris SK11, (Lca) Lactobacillus casei. There are 12 additional examples of the Spy sequence, all in differing S. pyogenes strains. There are seven additional examples of Spn-1, all in differing S. pneumoniae strains. There are six additional examples of Sag-1, all in differing S. agalactiae strains. There is one additional example of Ssu, found in a different strain of S. suis, and there is an additional example of Lla-1 found in a different strain of L. lactis.
Most of the new representatives were identified in organisms such as S. pneumoniae, S. pyogenes, or Lactococcus lactis, where representative strains had previously been sequenced. Thus, some of the additional COG4708 RNAs are quite similar or identical to those already described. We also identified sequences matching the motif in S. suis, S. sanguinis, and Lactobacillus casei. All representatives of the motif were found in the 5′ untranslated region (UTR) of genes encoding the protein annotated as COG4708 or DUF988. The conserved features of the motif described previously (Weinberg et al. 2007), including the pseudoknot, loop sequences, and SD sequence within the aptamer, are present in all new representatives (Fig. 2A). This class of structured RNAs is conserved over a wider range of bacterial species than had previously been determined. In addition, both the consensus sequence pattern and the secondary structure model for the RNA are substantially more complex that those observed for the preQ1-I aptamer class (Roth et al. 2007).
FIGURE 2.
Structural modulation of COG4708 RNA by preQ1. (A) Consensus sequence and secondary structure model for the COG4708 RNA motif. Nonconserved nucleotides are represented by a solid black line or open circles, and conserved pairing elements P1–P4 are indicated. Arrows identify nucleotides present in a truncated construct that retain aptamer function (see Fig. 3). The shaded nucleotides comprise a conserved Shine–Dalgarno (SD) sequence. (B) Spontaneous cleavage products of the S. pneumoniae R6 preQ1-II aptamer representative (1–103 RNA) separated by denaturing PAGE. NR, T1, −OH, and (−) represent no reaction, partial RNase T1 digest, partial alkaline digest, and no ligand, respectively. Selected RNase T1 cleavage products (cleaves 3′ of G residues) are identified on the left. Numbered asterisks on the right indicate locations of structural modulation in response to preQ1. (C) Patterns of spontaneous cleavage of the 1–103 RNA mapped onto the predicted secondary structure. Lowercase g notations identify guanosine residues added to improve transcription, and numbered asterisks identify the numbered regions depicted in B.
COG4708 RNA motif representatives likely function as preQ1-sensing riboswitches
Based on the association of genes encoding COG4708 proteins with the previously described preQ1-I riboswitch (Roth et al. 2007), we hypothesized that a second conserved RNA motif associated with genes for COG4708 also senses preQ1. To determine whether COG4708 RNAs undergo structural changes in response to preQ1 binding, we performed in-line probing assays (Soukup and Breaker 1999; Winkler et al. 2002) on a representative of this motif in the presence and absence of preQ1. In-line probing assays exploit differing rates of spontaneous RNA cleavage due to internal phosphoester transfer. Unconstrained regions of an RNA chain typically have rates of cleavage that are faster than constrained regions so that RNA structure changes on metabolite binding generally result in changes to the RNA fragmentation pattern (Li and Breaker 1999; Soukup and Breaker 1999).
A 5′ 32P-labeled RNA corresponding to a 103-nucleotide (nt) construct encompassing the COG4708 RNA motif from S. pneumoniae R6 (hereafter, termed 1–103 RNA) was incubated with varying concentrations of preQ1, and the spontaneous cleavage products were separated by denaturing polyacrylamide gel electrophoresis (PAGE). The fragmentation pattern of the 1–103 RNA exhibits a concentration-dependent change, thus revealing that preQ1-dependent RNA structure changes occur (Fig. 2B). These findings also confirm that COG4708 RNAs are preQ1 aptamers of a distinct class that we have named preQ1-II. The cleavage pattern of the full-length 1–103 RNA construct is consistent with the secondary structure predicted based on phylogenetic analysis (Weinberg et al. 2007). The loops and joining regions show evidence of cleavage while the predicted base-paired regions remain largely protected from cleavage (Fig. 2C). Most nucleotides that exhibit reduced rates of cleavage reside in regions that are most highly conserved (Fig. 2C). The SD sequence and anti-SD that comprise P3 show some cleavage in the absence of preQ1. This cleavage is reduced upon ligand binding, suggesting that the interaction of the SD and anti-SD is stabilized in the presence of preQ1, which is expected to sequester the SD and prevent the initiation of translation. In contrast, the high level of spontaneous cleavage that occurs at nucleotides numbered 16 and lower indicates that the 5′ region of the 1–103 RNA construct remains largely unstructured. This observation, coupled with the relatively sparse distribution of conserved nucleotides among preQ1-II aptamer representatives, suggests that this region is not involved in forming critical portions of the aptamer structure.
Based on bioinformatics and structural probing data, we speculate that the preQ1-II RNA motif from S. pneumoniae functions as a genetic off switch, where ligand binding stabilizes the P3 secondary structure to sequester the SD sequence and prevent gene expression. This mechanism of gene control has previously been observed for several other riboswitches (Rodionov et al. 2002; Mandal and Breaker 2004b; Fuchs et al. 2006; Wang et al. 2008). Similarly, representatives of the preQ1-I riboswitch are predicted to down-regulate expression of homologous COG4708 genes in response to preQ1 (Roth et al. 2007).
The minimal preQ1-II secondary structure is accurately predicted by bioinformatics
We constructed a series of 5′ and 3′ truncated RNAs and used in-line probing to determine whether truncated constructs retain preQ1 binding activity. An RNA containing pairing elements P1 through P4 (nucleotides 17–90) has an apparent K D for preQ1 of ∼100 nM (Fig. 3A,B). This affinity is within the range observed for other riboswitch aptamers and therefore demonstrates that the 5′-most nucleotides that remain relatively unstructured during in-line probing (Fig. 2C) are not necessary for high-affinity metabolite binding. An even shorter RNA construct (nucleotides 33–90) missing P1 has approximately the same K D as the 17–90 RNA construct, revealing that the conserved P1 element also is unlikely to be involved in molecular recognition of the preQ1 ligand, despite the covariation observed at several positions (Weinberg et al. 2007). Perhaps P1 serves a role during RNA folding or function in vivo that is not captured by our in vitro in-line probing assays.
FIGURE 3.
Ligand binding affinity of wild-type and mutant 17–90 RNAs. (A) Plot of the modulation of spontaneous RNA cleavage during in-line probing at several internucleotide linkages of 17–90 RNA versus the logarithm of the molar concentration of preQ1. (B) In-line probing gel of 17–90 RNA incubated with concentrations of preQ1 from left (0.5 nM) to right (10 μM). Arrows on the right point to bands used to generate binding curve in A. Other notations are as described in the legend to Figure 2B. (C) The sequence and structure of 17–90 RNA with disruptive and compensatory mutations M1 through M6 identified. (D) K D values for preQ1 exhibited by 17–90 RNA, 33–90 RNA, and mutant 17–90 RNAs M1 through M6 as designated in C. Open circles indicate that the actual K D value is higher than 100 μM.
To further examine the proposed secondary structure model, we created constructs that carry disruptive and compensatory mutations in each pairing element of the 17–90 RNA (Fig. 3C), and the ligand-binding activities of these constructs were assessed by using in-line probing (Fig. 3D). As expected, the disruptive mutation in P2 (M1) decreases ligand binding affinity by ∼100-fold (K D ∼10 μM), and the compensatory mutation (M2) partially rescues ligand binding affinity (K D ∼250 nM). The pattern of spontaneous cleavage products (data not shown) is consistent with structures wherein P2 is unpaired in M1 and paired in M2.
The disruptive mutations M3 and M5 located in P3 and P4, respectively, both abolish preQ1 binding (Fig. 3D). Whereas the compensatory mutation M6 (in P4) fully restores ligand binding as expected, the M4 mutant carrying compensatory changes to restore P3 base pairing does not yield a restoration of preQ1 binding. The failure of this compensatory mutation could be due to several reasons. M3 and M4 both exhibit patterns of spontaneous cleavage during in-line probing that differ from that of the wild-type RNA in the absence of ligand (data not shown), suggesting that these mutations cause considerable misfolding. Additionally, the P3 nucleotides mutated are strictly conserved due to the presence of SD and anti-SD sequences, and this region modulates upon ligand binding (Fig. 2B,C). These characteristics suggest that altering the base identities in P3 causes extensive misfolding, and that molecular contacts that are (either directly or indirectly) important for preQ1 binding might be lacking in the M4 construct.
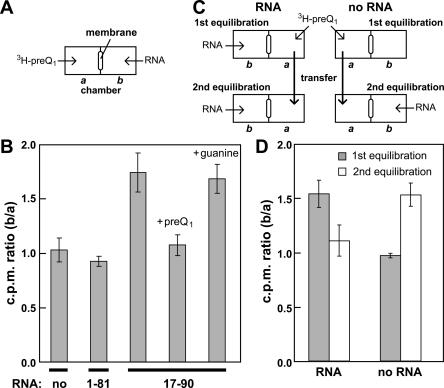
Equilibrium dialysis confirms preQ1 binding
We performed equilibrium dialysis experiments with 3H-preQ1 and the 17–90 RNA construct to confirm ligand binding and the specificity of molecular recognition using a different methodology. For equilibrium dialysis, 3H-preQ1 is added to chamber a of an equilibrium dialysis apparatus, and excess RNA is added to chamber b. The two chambers are separated by a 5000-Da molecular weight cutoff dialysis membrane (Fig. 4A). The solutions were allowed to equilibrate and the fraction of tritium in each chamber was subsequently measured by liquid scintillation counting. We observed a shift of tritium toward the RNA when 3H-preQ1 is equilibrated with the 17–90 RNA to give a ratio of radioactive signal for the two chambers (b/a) of ∼1.75 (Fig. 4B), indicating that RNA is binding 3H-preQ1 and sequestering it to chamber b. When 3H-preQ1 (1 μM) is equilibrated without RNA or with an RNA that lacks the ability to form the P3 stem (1–81 RNA), the 3H label distributes evenly (b/a ∼1). When the 17–90 RNA is allowed to equilibrate with 3H-preQ1 and establish an asymmetric distribution of radioactivity, the addition of excess unlabeled preQ1 to chamber a again causes the 3H label to redistribute. However, the addition of excess unlabeled guanine has no effect on the distribution of 3H, indicating that guanine at the concentration tested cannot compete with 3H-preQ1 for the ligand binding site in the 17–90 RNA.
FIGURE 4.
Analysis of metabolite binding by the 17–90 RNA aptamer using equilibrium dialysis. (A) Loading of the equilibrium dialysis apparatus with 3H-preQ1 and RNA. Chambers a and b are separated by a 5000 MWCO membrane. (B) Results of equilibrium dialysis with chamber b lacking RNA (no), a truncated and inactive derivative of 17–90 RNA (1–81) and 17–90 RNA. For the competition experiments, excess unlabeled preQ1 and guanine were added to previously equilibrated apparatuses containing 17–90 RNA and 3H-preQ1. Error bars represent the standard deviation of three independent experiments. (C) Method of equilibrium dialysis to determine whether unshifted 3H-labeled material can be bound by RNA. For the first equilibration, H3-preQ1 is equilibrated with 17–90 RNA (left) or in the absence of RNA (right). Aliquots are removed from each side of the apparatus to assess tritium distribution. For the second equilibration, buffer from the chamber lacking RNA (chamber a that contains unshifted H3-label) was transferred to a new apparatus and reequilibrated with chamber b containing 17–90 RNA. Aliquots again are removed to reassess tritium distribution. (D) Results of the equilibrium dialysis experiment described in C. Error bars represent the standard deviation of three independent experiments.
Although the trends observed during equilibrium dialysis are consistent with the binding functions of the preQ1-II aptamer observed by in-line probing, the extent of 3H-label shifted is not as large as expected. The concentration of RNA used should have provided excess binding sites for 3H-preQ1 and, given the measured K D of the aptamer for its ligand, should have been sufficient to saturate all preQ1 molecules with RNA. Therefore, the b/a ratio should have been greater when equilibration occurred in the absence of unlabeled preQ1 competitor.
To examine this discrepancy further, we conducted experiments where the contents of chamber a containing the unshifted 3H-labeled material remaining after equilibration with the 17–90 RNA was transferred to chamber a of a new apparatus opposite a newly prepared sample of 17–90 RNA, and the two chambers were allowed to reequilibrated (Fig. 4C). As expected the 3H-labeled material distributed evenly between both chambers (Fig. 4D), indicating that the molecular source of the unshifted portion of the 3H is not bound by the RNA and is unlikely to be preQ1. From this analysis, we estimated that only ∼25% of the 3H-labeled material is preQ1. Consistent with this conclusion are the results of a purity analysis of the 3H-labeled preQ1 sample used for equilibrium dialysis. Approximately 82% of the 3H-label is derived from solvent (see Materials and Methods), and no other UV-absorbing compounds are present in the mixture. In summary, the equilibrium dialysis experiments also indicate that an RNA construct carrying the core of the COG4708 RNA motif functions as a selective aptamer for preQ1.
Selective recognition of preQ1 by 17–90 RNA
The molecular recognition determinants of the preQ1-II aptamer from S. pneumoniae were established by measuring the binding affinities of the 17–90 RNA for a series of preQ1 analogs. To assess recognition of the aminomethyl group, we tested preQ0 (7-cyano-7-deazaguanine), 7-(N,N′-dimethylaminomethyl)-7-deazaguanine, and 7-carboxamide-7-deazaguanine. Both the dimethylaminomethyl analog and preQ0 exhibit K D values of ∼500 nM, indicating that there is no substantial steric occlusion occurring at the terminus of the alkylamine group, and that this group is unlikely to function as a hydrogen bond donor (Fig. 5A). Rather, the productive interaction to the aminomethyl group might involve the amino group as a hydrogen bond acceptor. Although such interactions are rare, the nitrogen atom of an amino group is predicted (Luisi et al. 1998) to serve as a hydrogen bond acceptor in some other nucleic acid structures.
FIGURE 5.
Molecular recognition analysis of the 17–90 preQ1-II aptamer. (A) Comparison of the molecular recognition characteristics of 17–90 RNA (circles) and the preQ1-I riboswitch (squares) for preQ1 and ligand analogs. K D values for the preQ1-I riboswitch were published previously (Roth et al. 2007). Shading on chemical structures identifies differences between the analog and preQ1. Open symbols designate the highest concentrations of ligand tested in in-line probing assays, indicating that the K D values are higher than this concentration. (B) Summary of the molecular recognition contacts inferred from the data in A.
The carboxamide analog of preQ1 is not bound by the RNA even when the analog is present at 10 μM. The simplest explanation for this observation is that the carbonyl group is too large to be accommodated by the binding pocket, although we cannot rule out the possibility that the presence of the keto oxygen alters the chemical characteristics of the alkylamine sufficiently to disrupt binding.
As was observed by using equilibrium dialysis (Fig. 4B), the 17–90 RNA strongly discriminates against guanine (K D ∼10 μM). The guanine analog 7-deazaguanine is bound with only a slightly better affinity, suggesting that little selectivity for preQ1 is derived from the absence of a nitrogen atom at position 7. The lack of 7-methylguanine binding at concentrations as high as 1 mM is likely due to the absence of a hydrogen bond donor at position 9 rather than to any disruption of molecular recognition contacts at position 7.
From the remaining purine compounds tested, it is clear that the amine at position 2 is critical for ligand binding (Fig. 5A). 2,6-diaminopurine (DAP) is the only compound that lacks a guanine-like Watson–Crick base-pairing edge but retains the ability to be measurably bound by the RNA at the concentrations tested. Interestingly, compounds carrying either a keto oxygen (guanine) or an amine (DAP) at position 6 of the purine ring yield near identical K D values. However, no binding is detected for 2-aminopurine, which differs from guanine and DAP by lacking an exocyclic functional group at position 6. Thus, the keto group of guanine and the amino group of DAP at position 6 appear to contribute equally to molecular recognition. Although there are more complex possibilities, the simplest explanation for this observation is that both the keto group of preQ1 and the nitrogen atom of the DAP amine at position 6 serve as hydrogen bond acceptors.
A summary of the putative molecular recognition contacts made by the 17–90 RNA and its preQ1 ligand (Fig. 5B) reveals differences compared to the molecular recognition contacts identified for preQ1-I aptamers (Roth et al. 2007). Recognition of the aminomethyl moiety at the 7 position is substantially different, as the relative specificities of the 7-dimethylaminomethyl and 7-carboxamide analogs are reversed for the two aptamer classes. In addition, a preQ1-I riboswitch aptamer forms a much tighter interaction with guanine, resulting in significantly less specificity for preQ1 over guanine. Similar to the preQ1-I riboswitch, the amine at position 2 appears to be critical for ligand binding for 17–90 RNA. However, the analog binding data suggest that the preQ1-II aptamer does not form a contact with the N1 position of the purine ring. This differs from the Watson–Crick base pair proposed (Roth et al. 2007) to form between the ligand and the preQ1-I aptamer (see additional discussion below). It is clear from the existing data that the distinctive sequences and structural elements of preQ1-II aptamers fold to form a binding pocket that is distinct from that made by preQ1-I aptamers.
Watson–Crick base pairing is unlikely to account for ligand recognition
Riboswitches binding guanine and adenine have been found to use canonical Watson–Crick base pairs for selective ligand binding (Batey et al. 2004; Mandal and Breaker 2004a; Serganov et al. 2004; Noeske et al. 2005; Gilbert et al. 2006). A single point mutation of the purine riboswitch aptamer is sufficient to switch aptamer specificity between guanine and adenine (Mandal and Breaker 2004a). Similarly, the specificity of a riboswitch binding 2′-deoxyguanosine can be altered by a comparable mutation (Kim et al. 2007). An analogous mechanism appears to be used in preQ1-I riboswitch aptamers, where mutation of a conserved cytidine to uridine alters ligand specificity from preQ1 to DAP (Roth et al. 2007).
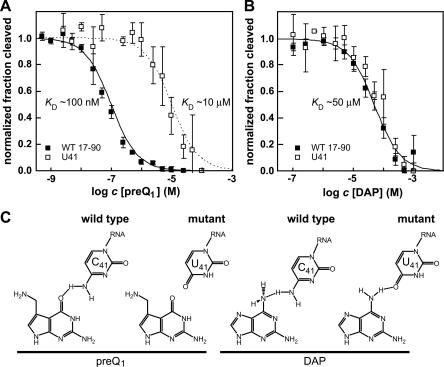
To assess whether the 17–90 RNA uses a similar mechanism to promote ligand specificity, we mutated the sole universally conserved unpaired cytidine (C33) (Figs. 1, 2A) to uridine. Although there are additional strictly conserved cytidine residues present in preQ1-II aptamers (C32 and C51), these reside within predicted base-paired elements and thus are unlikely to be forming a canonical base pair with the ligand. However, we observed that the mutant RNA carrying a U at position 33 behaves very similarly to the wild-type RNA and exhibits a K D for preQ1 of ∼250 nM (data not shown). Therefore this residue is unlikely to form a canonical base pair with the ligand. This observation is not surprising given that this area of the RNA does not modulate on ligand binding (Fig. 2B,C) and that P1 is not necessary for ligand binding.
We also examined nucleotide C41 as a candidate for base-pair formation with preQ1. This nucleotide is proximal to the conserved core of the aptamer and modulates upon ligand binding (Fig. 2). Only one representative of the 40 known COG4708 RNAs was found wherein C41 is mutated to a U residue (Fig. 1). A 17–90 RNA construct carrying this C-to-U change at position 41 exhibits an ∼2 orders of magnitude loss in affinity for preQ1 (Fig. 6A), indicating that this residue indeed is important for ligand binding. However, the mutant construct does not undergo a change in specificity to favor analogs with an adenine base-pairing face (data not shown). These results suggest that C41 also might not form a canonical base pair with the ligand.
FIGURE 6.
Altered ligand discrimination by a mutant 17–90 aptamer. Plot depicting the modulation of RNA structure by in-line probing of wild-type (C41) and mutant (U41) 17–90 RNA at various concentrations of (A) preQ1 and (B) DAP. Points represent average normalized fraction cleaved for several modulating bands, and error bars represent the standard deviation of this average. (C) Possible aptamer–ligand interaction that is consistent with the molecular recognition model and mutational analysis data for the preQ1-II aptamer from S. pneumoniae R6.
Although the U41 mutant binds preQ1 more poorly than the wild-type 17–90 RNA, this mutant binds DAP with essentially the same affinity as wild type (Fig. 6B). In contrast, guanine binding is no longer detectable with the U41 mutant (data not shown). These observations suggest that the nucleotide at position 41 forms a contact with the ligand in a manner consistent with the molecular recognition model for preQ1-II aptamers (Fig. 5B). The exocyclic amino group of C41 could donate a hydrogen to the keto group of preQ1, but the keto group in place of this amine in U41 cannot form an equivalent hydrogen bond (Fig. 6C). Therefore, as is observed, the mutant RNA should bind preQ1 more poorly than the wild-type RNA (Fig. 6A). In contrast, the exocyclic amino group of C41 can donate a hydrogen to the nitrogen atom of the exocyclic amine at the 6 position of DAP to form a hydrogen bond. Likewise, the corresponding keto group of U41 can serve as a hydrogen bond acceptor and allow formation of an equivalent hydrogen bond with the same exocyclic amine at the 6 position of DAP (Fig. 6C). This arrangement might explain why we observe near identical K D values for DAP binding by the C41 and U41 RNAs (Fig. 6B).
CONCLUSIONS
PreQ1 is the second example of a metabolite recognized by more than one class of natural aptamers. Four different aptamer classes recognizing S-adenosylmethionine (AdoMet or SAM) have been identified to date. SAM-I or S-box (Epshtein et al. 2003; McDaniel et al. 2003; Winkler et al. 2003), SAM-II (Corbino et al. 2005), SAM-III or SMK-box (Fuchs et al. 2006), and SAM-IV (Weinberg et al. 2007) often regulate the same genes in different organisms (e.g., SAM synthase, homoserine O-succinyltransferase). The discovery of a second natural preQ1 aptamer class shows that SAM is not a special case and that additional metabolites may be bound by multiple distinct RNA architectures to control similar sets of genes in different organisms.
The association of two preQ1-binding RNAs with the membrane protein classified as COG4708 or DUF988 strongly suggests this protein is a transporter for a queuosine biosynthetic intermediate. Membrane proteins of unknown function whose genes are associated with the TPP riboswitch have subsequently been shown to be involved with TPP and thiamin transport (Rodionov et al. 2002; Winkler et al. 2002; Schyns et al. 2005), illustrating how characterization of RNA genetic control elements can lead toward description of protein function. Likewise, knowledge of the function of a protein encoded by an mRNA can aid in establishing the ligand identity of an associated riboswitch. Although the precise function of the COG4708 protein has not been experimentally confirmed, its common association with preQ1-I riboswitches implicated preQ1 as the ligand for the preQ1-II riboswitch candidate.
Interestingly, in many bacteria there are numerous genes with homology with those associated with preQ1 biosynthesis or transport, yet these genes have no established control mechanisms. Therefore, we speculated that the mRNAs for such genes could carry other preQ1 riboswitch classes. To assess the potential for new preQ1 aptamer discovery, we examined the intergenic regions upstream of several COG4708 genes in organisms that lack preQ1-I and preQ1-II aptamers and whose genomes have been sequenced.
The COG4708 gene is narrowly distributed in bacteria, with all but three examples occurring in the classes Bacilli and Clostridia (Fig. 7). There is one example of COG4708 in a γ-proteobacteria (Rubrobacter xylanophilus) and there are two examples found in Archaea (Thermophilum pendens and Staphylothermus marinus). Genes corresponding to COG4708 proteins are associated with preQ1 aptamers in 30 of 46 instances (Fig. 7). Several organisms contain two copies of the gene, and a riboswitch is typically associated with only one of these copies, although in Alkaliphilus metalliredigens both copies of the gene are preceded by a preQ1-I riboswitch. In many organisms where the gene for COG4708 is not associated with a known preQ1-sensing RNA, the intergenic region directly upstream of the ORF is greater than 50 base pairs (bp). This is sufficient space for an RNA aptamer to be located in the 5′ UTR of the mRNA. However, such regions typically display little to no detectable homology with any other sequence, suggesting that they either lack structured RNA or that they do not carry an aptamer that has homologs in many other bacteria.
FIGURE 7.
Taxonomy tree of species carrying genes for COG4708 proteins. Species containing two copies of the gene are marked by an asterisk. Species where the gene encoding COG4708 is preceded by the preQ1-I riboswitch are indicated by I; species where the gene is preceded by a preQ1-II motif are indicated by II.
We used in-line probing assays to examine the intergenic regions upstream of ORFs encoding COG4708 proteins in Moorella thermoacetica, Clostridium difficile, Oenococcus oeni, and Geobacillus kaustophilus and upstream of both COG4708 copies in Streptococcus thermophilus. None of the RNAs tested show structural modulation in the presence of preQ1, suggesting that they do not contain preQ1-binding riboswitches. However, several different factors, including RNA misfolding due to interference from extraneous 5′ or 3′ sequence as well as differences between in vivo and in vitro conditions, can complicate in-line probing assays.
In contrast to most other identified riboswitches, the preQ1-II aptamer is very narrowly distributed, predominantly occurring in bacteria from the Streptococcaceae family. This narrow distribution has precedent, as SAM-III also appears limited to the same group of bacteria (Fuchs et al. 2006). However, the bioinformatics searches that identified the preQ1-II riboswitch candidate did not uncover SAM-III (Yao et al. 2007; Weinberg et al. 2007), suggesting that there may be other RNA elements unique to this family that have yet to be identified. As genomic information continues to become available, however, bioinformatics searches could be used to discover additional rare structured RNA elements of which some could function as metabolite-sensing riboswitches. The recently described riboswitch class that recognizes 2′-deoxyguanosine is known to be present in only one species of bacteria (Kim et al. 2007), and this rarity precludes discovery of such riboswitches by most bioinformatics search strategies. Therefore, a combination of computational searching and the individual analysis of the UTRs of mRNAs that lack riboswitch control only in some organisms appears to be a promising strategy for new riboswitch discovery.
MATERIALS AND METHODS
Chemicals and DNA oligonucleotides
The chemical syntheses of preQ1, preQ0, 7-(N,N′-dimethylaminomethyl)-7-deazaguanine, 7-carboxamide-7-deazaguanine, and 7-aminomethyl-7-deazaadenine were described previously (Roth et al. 2007). The remaining purine compounds and other chemicals were purchased from Sigma-Aldrich unless noted otherwise. 3H-preQ1 was obtained as previously described (Hurt et al. 2007). Synthetic DNAs were prepared by Sigma-Genosys.
RNA preparation
The portion of the 5′ UTR of the gene encoding COG4708 corresponding to the preQ1-II (COG4708 RNA) motif was PCR amplified from S. pneumoniae R6 using the following primers: 5′-GGAATTCAACAAGTCTAACAGAAAAGTAGAAAGGCGGGC-3′, 5′-TTTTGTCATTTTTTCTCCTTTAACGTCTGGATAACTCTCAAAAGC-3′. The resulting double-stranded DNA was subsequently cloned into pCR2.1 using a TOPO TA cloning kit (Invitrogen). The plasmid insert was sequenced (The W. M. Keck Foundation Biotechnology Resource Center, Yale University) and used as template for PCR amplification of DNA fragments encoding the desired RNA preceded by a T7 promoter sequence and including two guanosine residues to improve transcription efficiency. Disruptive and compensatory mutations were introduced into PCR products using mutant primers for PCR. PCR products were transcribed in vitro using T7 RNA polymerase and the resulting RNA purified by denaturing 6% PAGE using methods similar to those described elsewhere (Seetharaman et al. 2001).
DNA fragments corresponding to the constructs described from S. thermophilus were constructed by PCR (Dillon and Rosen 1990) from sets of synthetic oligonucleotides based on the genomic sequence. DNA fragments corresponding to the intergenic regions upstream of genes encoding COG4708 proteins from M. thermoacetica, O. oeni (PSU-1), C. difficile, and G. kaustophilus were amplified by PCR using the respective genomic DNAs and primers containing the T7 promoter sequence in the appropriate position. O. oeni and C. difficile genomic DNA samples were obtained from ATCC. RNA molecules were prepared by in vitro transcription as described above.
In-line probing assays
RNA molecules prepared by in vitro transcription were dephosphorylated with alkaline phosphatase (Roche Diagnostics), radiolabeled with [γ-32P]ATP (GE Biosciences) and T4 polynucleotide kinase (New England Biolabs) according to the manufacturers' instructions. The 5′ 32P-labeled RNAs were purified by PAGE as described above. In-line probing reactions containing ∼1 nM of 5′ 32P-labeled RNAs were assembled with and without ligand as described (Soukup and Breaker 1999). Reaction mixtures containing 50 mM Tris-HCl (pH 8.3 at 23°C), 20 mM MgCl2, and 100 mM KCl were incubated at 25°C for ∼40 h. For RNAs originating from M. thermoacetica and G. kaustophilus, incubations were also conducted at 58°C for 30 min and 60°C for 15 min, respectively. Samples were separated by denaturing 10% PAGE, and the imaging and quantification were carried out using a Molecular Dynamics PhosphorImager and ImageQuaNT software.
The K D values were determined by conducting in-line probing assays using a series of ligand concentrations. The normalized fraction of RNA cleaved at the sites indicated for each analysis was calculated at each ligand concentration, and a standard binding curve was fit to the points. Concentrations of most compounds examined varied between 1 nM and 1 mM, with the exception of preQ1, guanine, 7-carboxamide-7-deazaguanine, and 7-aminomethyl-7-deazaadenine, where the highest concentrations tested were 100 μM, 100 μM, 10 μM, and 30 μM, respectively.
Equilibrium dialysis
A 30 μL mixture containing 50 mM Tris-HCl (pH 8.3 at 23°C), 20 mM MgCl2, 100 mM KCl, and 1 μM of 3H-labeled preQ1 was placed in one chamber of a DispoEquilibrium Dialyzer (ED-1, Harvard Bioscience) and the same volume containing the same constituents minus the 3H-labeled preQ1 was added to the opposing side. When noted, the latter solution also contained either 5 μM 17–90 RNA or 5 μM 1–81 RNA. After equilibrating for 14 h, a 5 μL aliquot was removed from each side of the apparatus to determine the distribution of 3H-label in the two chambers by liquid scintillation counting. For competitive binding experiments, 3 μL of the buffer mixture described above containing 1 mM unlabeled compound was added to the chamber lacking RNA, and an equivalent volume of buffer was added to the RNA-containing chamber. The chambers were allowed to equilibrate for an additional 8 h before the distribution of 3H-label was reassessed as described above.
HPLC of the 3H-labeled preQ1 showed a single UV-active peak consistent with preQ1. To assess where the 3H-label not incorporated into preQ1 originated, a 40 μL aliquot of the 3H-preQ1 solution (∼2 mM) was mixed with 8 mg of decolorizing carbon and 200 μL water. The slurry was subsequently separated by centrifugation and the supernatant removed and filtered (0.22 μm). HPLC analysis of the supernatant revealed that no preQ1 was present in the aqueous portion, indicating that the carbon had completely adsorbed the heterocycle. Tritium present in the supernatant portion and the carbon residue was measured by liquid scintillation counting.
Bioinformatics
Homology searches were conducted on RefSeq25 (Pruitt et al. 2007) and the COG4708 RNA motif consensus sequence was determined as previously described (Weinberg et al. 2007). Instances of COG4708 (DUF988) genes were identified through the Pfam database (release 22.0) (Finn et al. 2006). A BLAST search of the COG4708 protein sequence from several organisms against the NCBI genomic database failed to yield any additional sequences. We limited examples to fully sequenced genomes and determined whether the preQ1-I or preQ1-II riboswitches were present in the intergenic regions upstream of the ORF encoding COG4708 proteins. Highly similar strains or isolates of the same species were removed from our listing to reduce redundancy.
ACKNOWLEDGMENTS
We thank Dr. Zasha Weinberg for help setting up RNA homology searches, Dr. Melinda Pettigrew at the Yale Medical School for S. pneumoniae R6 genomic DNA, and Dr. Hideto Takami at the Extremophiles Research Program, Japan Agency for Marine-Earth Science and Technology for G. kaustophilus genomic DNA. This project was funded by grants from the NIH (R33 DK07027 and GM 068819) and the Howard Hughes Medical Institute. M.M.M. is supported by an NIH post-doctoral fellowship (F32GM079974).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.937308.
REFERENCES
- Abreu-Goodger, C., Merino, E. RibEx: A web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33:W690–W692. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsène-Ploetze, F., Nicoloff, H., Bringel, F. Lactobacillus plantarum ccl gene is non-essential, arginine-repressed and codes for a conserved protein in Firmicutes. Arch. Microbiol. 2005;183:307–316. doi: 10.1007/s00203-005-0774-9. [DOI] [PubMed] [Google Scholar]
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial gene control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Gilbert, S.D., Montagne, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bienz, M., Kubli, E. Wild-type tRNATyrG reads the TMV RNA stop codon, but Q base-modified tRNATyrQ does not. Nature. 1981;294:188–190. doi: 10.1038/294188a0. [DOI] [PubMed] [Google Scholar]
- Corbino, K.A., Barrick, J.E., Lim, J.L., Welz, R., Tucker, B., Puskarz, I., Mandal, M., Rudnick, N.D., Breaker, R.R. Evidence for a second class of S-adenosylmethiothine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon, P.J., Rosen, C.A. A rapid method for the construction of synthetic genes using the polymerase chain-reaction. Biotechniques. 1990;9:298–300. [PubMed] [Google Scholar]
- Epshtein, V., Mironov, A.S., Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D., Mistry, J., Schuster-Böckler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., Moxon, S., Marshall, M., Khanna, A., Durbin, R., et al. Pfam: Clans, webtools, and services. Nucleic Acids Res. 2006;24:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, R.T., Grundy, F.J., Henkin, T.M. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthesis. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Gaur, R., Varshney, U. Genetic analysis identifies a function for the queC (ybaX) gene product at an initial step in the queuosine biosynthetic pathway in Escherichia coli . J. Bacteriol. 2005;187:6893–6901. doi: 10.1128/JB.187.20.6893-6901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, S.D., Stoddard, C.D., Wise, S.J., Batey, R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Harada, F., Nishimura, S. Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first position of the anticodons of these transfer ribonucleic acids. Biochemistry. 1972;11:301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- Hurt, J.K., Olgen, S., Garcia, G.A. Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res. 2007;35:4905–4913. doi: 10.1093/nar/gkm473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Reuyl, D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg. Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- Kim, J.N., Roth, A., Breaker, R.R. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl. Acad. Sci. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino, Y., Kasai, H., Nihei, K., Nishimura, S. Biosynthesis of the modified nucleoside Q in transfer RNA. Nucleic Acids Res. 1976;3:393–398. doi: 10.1093/nar/3.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- Luisi, B., Orozco, M., Sponer, J., Luque, F.J., Shakked, Z. On the potential role of the amino nitrogen atom as a hydrogen bond acceptor in macromolecules. J. Mol. Biol. 1998;279:1123–1136. doi: 10.1006/jmbi.1998.1833. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004a;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004b;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- McDaniel, B.A., Grundy, F.J., Artsimovitch, I., Henkin, T.M. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, F., Suter, B., Grosjean, H., Keith, G., Kubli, E. Queuosine modification of the wobble base in tRNA HIS influences “in vivo” decoding properties. EMBO J. 1985;4:823–827. doi: 10.1002/j.1460-2075.1985.tb03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske, J., Richter, C., Grundle, M.A., Nasiri, H.R., Schwalbe, H., Wohnert, J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. 2005;104:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, S., Nishimura, Y., Hirota, Y., Nishumura, S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycoslyase. J. Biol. Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- Okada, N., Noguchi, S., Nishimura, S., Ohgi, T., Goto, T., Crain, P.F., McCloskey, J.A. Structure of a nucleoside Q precursor isolated from E. coli tRNA: 7-(aminomethyl)-7-deazaguanine. Nucleic Acids Res. 1978;5:2289–2296. doi: 10.1093/nar/5.7.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, N., Noguchi, S., Kasai, H., Shindo-Okada, N., Ohgi, T., Goto, T., Nishimura, S. Novel mechanism of post-transcriptional modification of tRNA. J. Biol. Chem. 1979;254:3067–3073. [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., Maglott, D.R. NCBI reference sequence (RefSeq): A curated nonredundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader, J.S., Metzgar, D., Schimmel, P., de Crécy-Lagard, V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., Gelfand, M.S. Comparative genomics of thiamin biosynthesis in prokaryotes. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., Gelfand, M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Roth, A., Winkler, W.C., Regulski, E.E., Lee, B.W.K., Lim, J., Jona, I., Barrick, J.E., Ritwik, A., Kim, J.N., Welz, R., et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- Seetharaman, S., Zivarts, M., Sudarsan, N., Breaker, R.R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- Schyns, G., Potot, S., Geng, Y., Barbosa, T.M., Henriques, A., Perkins, J.B. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis . J. Bacteriol. 2005;187:8127–8136. doi: 10.1128/JB.187.23.8127-8136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov, A., Yuan, Y.R., Pikovskaya, O., Polonskaia, A., Malinina, L., Phan, A.T., Hobartner, C., Micura, R., Breaker, R.R., Patel, D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slany, R., Bosl, M., Crain, R.F., Kersten, H. A new function of S-adenosylmethionine: The ribosyl moiety of AdoMet is the precursor of the cyclopentenediol moiety of the tRNA wobble base queuine. Biochemistry. 1993;32:7811–7817. doi: 10.1021/bi00081a028. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, J.K., Soukup, G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Tucker, B.J., Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Urbonavičius, J., Qian, Q., Durand, J.M.B., Hagervall, T.G., Bjork, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen, S.G., Reader, J.S., Swairjo, M.A., de Crécy-Lagard, V., Lee, B., Iwata-Reuyl, D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.X., Lee, E.R., Rivera, D., Lim, J., Breaker, R.R. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell. 2008 doi: 10.1016/j.molcel.2008.01.012. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, Z., Barrick, J.E., Yao, Z., Roth, A., Kim, J.N., Gore, J., Wang, J.X., Lee, E.R., Block, K.F., Sudarsan, N., et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Yao, Z., Barrick, J., Weinberg, Z., Neph, S., Breaker, R., Tompa, M., Ruzzo, W.L. A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]