Abstract
In the yeast Saccharomyces cerevisiae, mutation of some effectors of mRNA nuclear export leads to the rapid accumulation of HSP104 RNA in transcription site-associated foci. We have screened the S. cerevisiae complement of viable gene deletion mutants for their inability to export HSP104 RNA. The 15 strains identified comprise deletions of components of the THO, Thp1p/Sac3p, and nuclear pore complexes. In all three mutant classes, retained RNA overlaps the HSP104 transcription site. Thus, an early block to HSP104 RNA export is general. Incubation of the identified deletion strains, as well as seven additional mutants, under conditions where mRNA export is blocked results in rapid dissipation of nucleolar protein and RNA constituents. Time course experiments show that dissipation of nucleolar antigens succeeds mRNA retention and is reversed when the load of nuclear mRNA ceases. Consistent with a causal role of excess nuclear mRNA, nucleolar morphology in an mRNA export mutant environment remains intact when transcription by RNA polymerase II is inhibited.
Keywords: HSP104 RNA export screen, RNA-FISH, nucleolar dissipation
INTRODUCTION
The activities of RNA polymerases I and II (RNAPI/II) in the eukaryotic nucleus are spatially separated. The major site of ribosomal RNA (rRNA) synthesis by RNAPI is the nucleolus. This nonmembranous nuclear structure forms around the highly localized rRNA genes, which, presumably due to their active rRNA synthesis, attract factors involved in rRNA processing and its assembly into ribosomal subunits (Warner 1989; Trumtel et al. 2000). In the exponentially growing Saccharomyces cerevisiae cell, the nucleolus is composed of an estimated 600–700 proteins, of which many are in dynamic exchange with the nuclear surroundings (Phair and Misteli 2000; Andersen et al. 2005). At steady state, the nucleolus makes up more than one-third of the nuclear volume (Warner 1989).
The remainder of the S. cerevisiae nucleus predominantly contains chromatin harboring RNAPII transcription units. RNAPII-derived RNAs include small nuclear (sn) and nucleolar (sno) RNAs, other noncoding RNAs of mostly unknown function, and conventional mRNAs. Molecular flux from the chromatin-rich area to the nucleolus is known to occur; e.g., snoRNAs are imported into the nucleolus where they function in rRNA biogenesis (Terns and Dahlberg 1994). Although an involvement of the nucleolus in the maturation and nuclear export of mRNA has also been suggested (Kadowaki et al. 1995; Ideue et al. 2004; Boyne and Whitehouse 2006), such a functional connection between the two compartments has not yet been firmly established. In contrast, real-time analysis in mammalian cells of GFP-decorated mRNAs suggests that mRNPs diffuse from their sites of synthesis and dock at the nuclear membrane without a substantial nucleolar phase (Shav-Tal et al. 2004).
Nuclear export of mRNA is facilitated by multiple conserved factors exercising their roles at different steps in the pathway (for recent reviews, see Vinciguerra and Stutz 2004; Cole and Scarcelli 2006; Stewart 2007). Central to this process, in S. cerevisiae and metazoans, is the essential heterodimeric mRNA export receptor Mex67p/Mtr2p (TAP/p15 in mammals), which is capable of making contacts both to constituents of the early mRNP and to nucleoporins (NUPs) of the nuclear pore complexes (NPCs). Mex67p/Mtr2p can be recruited to mRNPs by different means. In S. cerevisiae, TREX complex factors—the tetrameric THO complex, the RNA helicase Sub2p, and the RNA-binding protein Yra1p—are recruited to active genes (Strasser et al. 2002; Zenklusen et al. 2002; Abruzzi et al. 2004). Yra1p in turn interacts directly with Mex67p (Strasser and Hurt 2000; Stutz et al. 2000). An alternative route for Mex67p/Mtr2p-mRNP association is through the Thp1p/Sac3p complex. Sac3p binds directly to Mex67p and presumably connects to nascent mRNP via Thp1p, which has been implicated in transcription elongation (Gallardo and Aguilera 2001; Fischer et al. 2002). Finally, dephosphorylation of the ubiquitous mRNP factor Npl3p has also been reported to facilitate Mex67p/mRNP association (Gilbert and Guthrie 2004).
The final step in nuclear export of mRNP concerns its translocation through the NPC. This is thought to occur via interactions of Mex67p/Mtr2p with phenylalanine/glycine (FG)-repeat containing NUPs and is completed at the cytoplasmic face of the NPC by a Dbp5p/Gle1p-induced mRNP remodeling process, which involves removal of Mex67p/Mtr2p and release of the mRNP into the cytoplasm (Lund and Guthrie 2005; Alcazar-Roman et al. 2006; Weirich et al. 2006).
Consistent with the proposed role of all the abovementioned complexes and factors in mRNA nuclear export, their deletion or mutation generally results in nuclear accumulation of poly(A)+ [p(A)+] RNA within a few minutes of a shift to restrictive conditions. More surprisingly, perhaps, those mutant strains that have been tested accumulate newly synthesized heat shock (hs) RNAs, HSP104 or SSA4, in nuclear foci close to their sites of transcription (Hilleren et al. 2001; Jensen et al. 2001a,b; Libri et al. 2002; Zenklusen et al. 2002; Dunn et al. 2005). Indeed, the somewhat granular appearance of the nuclear p(A)+ RNA fluorescent in situ hybidization (FISH) signal in certain mRNA export mutants might reflect persistent mRNP retention in multiple transcription site foci (Hilleren et al. 2001). The molecular cause of this phenotype remains unknown.
In this paper, we screen the S. cerevisiae collection of viable gene deletions and identify 15 strains that accumulate HSP104 RNAs in nuclear foci. In all mRNA export mutants tested, several nucleolar antigens disassemble at the restrictive temperature. This phenotype is suppressed by second site mutation of RNAPII transcription factor genes as well as addition of the transcription drug thiolutin. We suggest that excess nuclear mRNA disturbs nucleolar morphology.
RESULTS
Screening of a yeast gene deletion bank reveals 15 strains incapable of releasing HSP104 RNA from transcription site foci
The EUROSCARF collection of about 5000 viable gene deletion strains was screened by FISH using a set of Cy3-labeled DNA oligonucleotide probes targeting the 3′ end of the HSP104 RNA (Jensen et al. 2001b). Cells grown at 25°C were temperature shifted for 20 min to 42°C and subsequently processed for FISH. Visual inspection of samples revealed two different phenotypes: (1) RNA stain all over the cell, reminiscent of a wild-type (WT) strain, or (2) the appearance of one focused HSP104 RNA “dot” per cell nucleus. Individual deletion strains with the latter phenotype were categorized according to the strength of the HSP104 RNA dot signal, ranging from (+) for strains harboring weak but reproducible dots, to (+++++) for strains harboring intensely stained dots. Fifteen strains were dot positive with variable intensities (Table 1). Protein products of the deleted genes fall into three major groups of complexes, all of which have a previous functional history in mRNA export: the THO, Sac3p/Thp1p, and nuclear pore complexes.
TABLE 1.
Summary of HSP104 RNA export screen
To examine whether the HSP104 dot phenotype reflected transcript retention at or near the HSP104 gene, as previously observed (Jensen et al. 2001b; Thomsen et al. 2003), we performed HSP104 RNA/DNA double-staining experiments. For this purpose, a representative deletion mutant (Δmft1, Δrip1, or Δsac3) from each dot-positive group was crossed to a strain harboring LacO repeats 600–700 nucleotides (nt) downstream from the HSP104 locus (Dieppois et al. 2006). Coexpression of the LacI–GFP fusion protein decorated the LacO sites, enabling dual HSP104 RNA/GFP staining (Fig. 1A). For all three deletion mutants, >90% of all cells that were successfully stained with both the HSP104 FISH probes and the GFP antibody showed partially overlapping signals (Fig. 1B). At longer exposure times, overlap of presumably nascent HSP104 RNA and GFP signals could also be observed in a WT strain. We conclude that HSP104 RNA is retained at or near its site of transcription in THO, Sac3p/Thp1p, and NPC deletion mutants.
FIGURE 1.
Retained HSP104 RNAs localize in transcription site-associated foci in Δmft1, Δrip1, and Δsac3 cells. (A) Schematic representation of the HSP104/LacO module used to localize nuclear-retained HSP104 RNA molecules. In tested strains, LacO repeats were positioned 600–700 nt downstream of the HSP104 gene, and a LacI–GFP fusion protein was coexpressed. The relative position of the four HSP104 RNA-FISH probes is indicated. (B) Triple-color RNA-FISH, LacI–GFP immunolocalization, and DAPI staining analysis of Δmft1, Δrip1, Δsac3, and WT cells fixed after a 10-min incubation at 42°C. HSP104 RNA was visualized with Cy3-labeled THJ361–364 probes, and LacI–GFP molecules bound to LacO repeats were visualized with an anti-GFP antibody. DNA was stained with DAPI. Images were merged as indicated. A longer exposure time was used to visualize nascent HSP104 RNA in WT cells.
Rapid fragmentation of nucleolar antigens in mRNA export mutants
During analyses of HSP104 RNA nuclear localization relative to other nuclear structures, we noted that antibody staining of the nucleolar antigens Nop1p and Nsr1p often appeared fragmented in mRNA transport-deficient strains as compared with the crescent-shaped nucleolus of a WT cell. Such a phenotype has previously been reported for other individual mRNA export-deficient strains (Kadowaki et al. 1994b; Dockendorff et al. 1997; Rout et al. 2000), and we therefore decided to examine this in general by conducting Nop1p staining of all 15 deletion mutants subjected to a 42°C heat shock for 20 or 40 min. In the more severe cases (e.g., the Δmft1, Δrip1, and Δsac3 strains), Nop1p fragmentation was evident in a fraction of the cells already at the 20-min time point and in all cells after 40 min of incubation at 42°C (Fig. 2, right panel; data not shown). Quantification of the Nop1p fragmentation phenotype by counting the number of positive cells after a 40-min incubation at 42°C revealed that its level of penetrance largely correlated with the severity of the HSP104 RNA retention phenotype (Table 1).
FIGURE 2.
The HSP104 RNA retention phenotype precedes Nop1p fragmentation. Triple-color HSP104 RNA-FISH, Nop1p immunolocalization, and DAPI staining analysis of Δmft1, Δrip1, Δsac3, and WT cells fixed at 25°C (left panel), or after a 10-min (middle panel) or 40-min (right panel) incubation at 42°C. HSP104 RNA and DNA were visualized as described in the legend to Fig. 1. Nop1p was detected using a Nop1p-specific antibody. Images were merged as indicated.
FISH analysis of the mRNA transport capability of S. cerevisiae temperature-sensitive mutants is often performed after incubation at the restrictive temperature for >20–30 min. Thus, data might be obtained under conditions where a part of the nucleolus is disrupted. To examine whether the HSP104 RNA localization phenotype occurred prior to or after Nop1p fragmentation, we subjected Δmft1, Δrip1, Δsac3, and WT strains to a 42°C heat shock for either 10 or 40 min. Fixed cells were processed for triple color labeling studies visualizing HSP104 RNA, Nop1p, and the DAPI-stained chromatin-rich region of the nucleus. In all three cases, Nop1p staining at the 10-min time point appeared as a coherent signal next to the DAPI stain as in WT cells (Fig. 2). In these conditions, the HSP104 RNA dot was readily detectable and overlapped with the DAPI stain but not the Nop1p-defined nucleolus (Fig. 2, middle panel). This strongly suggests that retention of HSP104 RNA in transcription site-associated foci is not a consequence of nucleolar disintegration.
Interestingly, the Nop1p fragments, present in all cells of the three strains after 40 min of incubation, overlapped to a large extent with the DAPI signal (Fig. 2, right panel). This phenotype was reproduced with the nucleolar antigen Nsr1p and was also evident after a 40-min incubation of the Δmft1 and Δsac3 strains at 37°C (data not shown).
Fragmentation of nucleolar antigens correlates with nuclear accumulation of mRNA
To assay the generality of the phenotype, we also examined mutants of essential mRNA export factors for which temperature-sensitive alleles are available. After prolonged (40-min) incubation at 42°C of the mex67–5, rna14–3, and rat7–1 mutant strains, both Nop1p and Nsr1p staining appeared fragmented (Fig. 3; data not shown). Similar results were obtained using other mRNA export mutants, e.g., sub2–201, mtr2–21, and yra1–1 (data not shown). Interestingly, Nop1p staining also appeared fragmented in the pap1–1 mutant of poly(A) polymerase (Fig. 3A). Since the pap1–1 mutation leads to a significant decrease in the level of polyadenylation, yet restricts at least a subset of mRNAs to the nucleus (Hilleren et al. 2001), this suggests that Nop1p fragmentation correlates with nuclear retention of mRNAs independent of their polyadenylation status.
FIGURE 3.
Dissipation of nucleolar protein and RNA in mRNA export mutants is a general phenomenon. (A) Dual-color Nop1p immunolocalization and DAPI staining analysis of WT, mex67–5, rna14–3, rat7–1, pap1–1, Δrrp6, Δrrp47, and mtr4–1 cells fixed at 25°C (left panel), or after a 10-min (middle panel) or 40-min (right panel) incubation at 42°C. (B) Triple-color U3 RNA-FISH, Nsr1p immunolocalization, and DAPI staining analysis of Δrip1 cells fixed at 25°C (left panel), or after a 10-min (middle panel) or 40-min (right panel) incubation at 42°C. U3 RNA was visualized with a Cy3-labeled probe, Nsr1p was detected using an Nsr1p-specific antibody, and DNA was stained with DAPI.
The state of Nop1p was also assayed in strains deleted or mutated for the nuclear exosome accessory components Rrp6p, Rrp47p, and Mtr4p. In these mutants, a robust p(A)+ signal can be detected in the nucleus; however, as opposed to the mRNA export mutants examined above, this signal localizes to the non-DAPI region of the nucleus and stems from polyadenylation of stable nucleolar RNA species by the Trf4p/Trf5p poly(A) polymerases (Thomsen et al. 2005; Rougemaille et al. 2007). Prolonged incubation of the Δrrp6, Δrrp47, and mtr4–1 strains at 42°C did not alter the localization of Nop1p as compared with 25°C (Fig. 3A, three bottom rows).
We next addressed whether conditions eliciting Nop1p and Nsr1p fragmentation would also impact the localization of an RNA resident of the nucleolus. To this end, we performed RNA-FISH analysis with a probe targeting U3, a member of the box C/D family of snoRNAs that function in rRNA processing. After a 10-min incubation of Δrip1 cells at 42°C, U3 RNA largely colocalized with Nsr1p in an intact nucleolus (Fig. 3B, left panel). However, after a 40-min heat shock, the U3 RNA signal appeared fragmented, and like Nsr1p, it mainly coincided with the nuclear chromatin region stained by DAPI (Fig. 3B, right panel). Thus, both protein and RNA components of the nucleolus dissipate upon a robust block to mRNA nuclear export.
Finally, we examined whether yet another nucleolar constituent, the Nop5p protein, would also relocalize upon an mRNA export block. Indeed, staining of Nop5p exhibited a pattern similar to Nop1p and Nsr1p: differential fragmentation after prolonged incubation at 42°C in the export mutants Δmft1 and Δrip1 as compared with WT cells (Fig. 4).
FIGURE 4.
Nop5p fragmentation in the Δmft1 and Δrip1 strains. Triple-color HSP104 RNA-FISH, Nop5p immunolocalization, and DAPI staining analysis of Δmft1, Δrip1, and WT cells fixed at 25°C (left panel), or after a 10-min (middle panel) or 40-min (right panel) incubation at 42°C. HSP104 RNA, DNA, and protein were visualized as described in legend to Fig. 2.
Nucleolar antigens reassemble when the nuclear mRNA load ceases
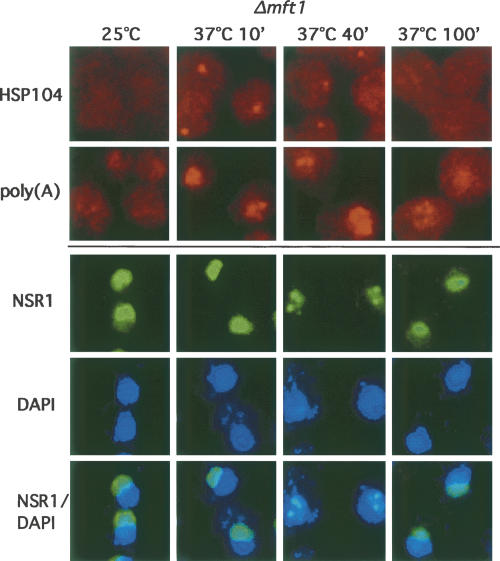
In the W303 strain background a temperature shift to 37°C leads to the transient induction of several heat-responsive genes. Possibly due to the selective role of Mft1p (i.e., the THO complex) in nuclear export of mRNAs from highly transcribed genes, the export phenotype of the Δmft1 strain wears off when heat shock transcription ceases after longer incubation at 37°C. This phenomenon is manifest by a strong nuclear accumulation of HSP104 and general p(A)+ RNA at the 10- and 40-min time points, followed by less intense nuclear staining after, e.g., 100 min (Fig. 5). We took advantage of this situation and characterized nucleolar morphology during a time course at 37°C in the Δmft1 mutant.
FIGURE 5.
Fragmentation of nucleolar antigens is reversible. HSP104, poly(A)+ RNA-FISH, Nsr1p immunolocalization, and DAPI staining analysis of Δmft1 cells fixed after incubation at 25°C or after 10, 40, or 100 min at 37°C. Poly(A)+ RNA was visualized with the Cy3-labeled and LNA-modified THJ790 dT20 probe (Thomsen et al. 2005). HSP104 RNA, Nsr1p protein, and DNA were detected as described in legend to Figs. 1 and 3. Nsr1p and DAPI images were merged as indicated.
Remarkably, at the 100-min time point, the nucleolus, exemplified by Nsr1p, returned to its normal shape and position (Fig. 5, third row, last column). Interestingly, the degree of Nsr1p disassembly seems to correlate with the extent of nuclear p(A)+ RNA load (Fig. 5, cf. NSR1 and poly(A) rows; data not shown) rather than with the extent of HSP104 transcription/retention (Fig. 5, cf. NSR1 and HSP104 rows; data not shown). Identical results were obtained when staining for the Nop1p protein (data not shown).
Dissipation of nucleolar antigens depends on active RNAPII transcription
If disassembly of part of the nucleolus is triggered by a heavy nuclear load of retained mRNA during an export block, a lowering of the transcription activity induced by heat shock should ease the phenotype. To test this prediction, we analyzed Nop1 localization in the Δrip1 and Δmft1 mutant backgrounds when combined with mutation of transcription factors Rad3p (TFIIH component) or Srb4p (Mediator component). In the background of both employed alleles, rad3–6.4 and srb4–138, RNAPII-dependent transcription is markedly decreased at the restrictive temperature (Thompson and Young 1995; Holstege et al. 1998; Jensen et al. 2004).
The Nop1p fragmentation phenotypes of the Δrip1 and Δmft1 single mutants after prolonged incubation at 42°C were completely suppressed by introduction of the rad3–6.4 and srb4–138 mutations (Fig. 6A,B). This result was evident at the levels of both Nop1p (Fig. 6A) and U3 (Fig. 6B) RNA localization. Moreover, similar results were obtained when introducing the rad3–6.4 mutation into the rna14–3 strain (data not shown).
FIGURE 6.
Dissipation of nucleolar antigens in mRNA export mutants depends on on-going RNAPII transcription. (A) Dual-color Nop1p immunolocalization and DAPI staining analysis of Δrip1, Δrip1/rad3–6.4, Δmft1, Δmft1/rad3–6.4, and Δmft1/srb4–138 cells fixed after a 15-min or 60-min incubation at 42°C as indicated. (B) Dual-color U3 RNA-FISH and DAPI staining analysis of Δrip1 and Δrip1/rad3–6.4 cells fixed after a 15-min or 60-min incubation at 42°C as indicated. (C) Poly(A)+ RNA, Nsr1p, and DAPI staining of Δmft1 or Δrip1 cells incubated for 40 min at 42°C in the absence (first and third rows) or the presence (second and fourth rows) of 50 μg/mL thiolutin. U3 RNA, poly(A)+ RNA, and Nsr1p were visualized as described in legends to Figs. 1–5. Images were merged as indicated.
Finally, we tested the effect of the transcription inhibitor thiolutin to assess whether it would mimic the effects of the Rad3p and Srb4p mutations. The transcription down-regulatory effect of the drug was verified by a dramatic decrease in the nuclear p(A)+ RNA-FISH signals in thiolutin-treated Δmft1 and Δrip1 cells compared with their untreated counterparts (Fig. 6C, first column). When evaluating nucleolar morphology by Nsr1p immunostaining and DAPI overlay, the presence of thiolutin in the Δmft1 and Δrip1 contexts clearly decreased the extent to which the Nsr1p signal spread into the chromatin region (Fig. 6C). The less potent effect of thiolutin, as compared with the rad3–6.4 and srb4–138 mutations, might be because it also has an inhibitory effect on RNAPI transcription (Jimenez et al. 1973). Nevertheless, taken together, our data strongly indicate that a general decrease in RNAPII-dependent transcription prevents disassembly of nucleolar antigens.
Fragmentation of nucleolar antigens is not due to impaired export of ribosomal subunits
It was recently demonstrated that in addition to its role in mRNA export, the Mex67p/Mtr2p heterodimer also functions in the nuclear export of the ribosomal large 60S subunit (Yao et al. 2007). It was therefore possible that ribosome export was affected in other mRNA export mutants, either directly, or via titration of ribosome export factors by excess nuclear mRNA levels. This in turn could slow recycling of ribosome assembly factors, leading to dissipation of nucleolar antigens. To test for this possibility, we assayed the cellular localization of Rps3–GFP (40S small subunit reporter), Rpl25–GFP (60S large subunit reporter), and Nmd3–GFP (60S subunit export receptor) in WT, Δrip1, mex67–5, and Δmft1 strains at 25°C, or after a 40-min heat treatment at 37°C or 42°C. At 25°C, all three GFP-reporter proteins distributed to both the nucleus and the cytoplasm of all four strains (Fig. 7A, left panels). Moreover, this distribution of Rps3–GFP was unaltered upon increasing the temperature, showing that nuclear export of 40S rRNA subunits is functional in all mutant contexts. In contrast, nuclear export of Rpl25–GFP was clearly decreased at 42°C, to some extent in WT cells, but even more so in the three mRNA export mutants (Fig. 7A, right panels). However, despite these export phenotypes, nucleolar disruption cannot solely be explained by this effect, as, e.g., at 37°C in the Δmft1 and mex67–5 strains, where Nop1p/Nsr1p/Nop5p rapidly disassemble (Fig. 5; data not shown), neither 40S rRNA nor 60S rRNA subunit transport defects were detected (Fig. 7A).
FIGURE 7.
Production and nuclear export of ribosomal subunits under conditions of nucleolar dissipation. (A) Localization of Rps3–GFP, Rpl25–GFP, and Nmd3–GFP in WT, Δrip1, mex67–5, and Δmft1 cells at 25°C (left columns), or after a 40-min incubation at 37°C (middle columns) or 42°C (right columns). (B) Northern blotting of indicated ribosomal and SCR1 RNAs from 7 μg of total RNA purified from WT, Δrip1, mex67–5, and Δmft1 cells grown at 25°C, or temperature-shifted for 40 min to 37°C or 42°C. 20S, 18S, 27S, and 25S RNAs were separated on 1.2% agarose gels containing formaldehyde. 5.8S and SCR1 RNAs were separated on an 8% polyacrylamide gel.
At 42°C, the Nmd3–GFP fusion protein appeared fragmented. However, this phenotype is presumably distinct from the Nop1p/Nsr1p/Nop5p fragmentation phenotype, as it was also evident in WT cells.
To test for more specific effects of an mRNA export block on rRNA processing, we compared, by Northern blotting analysis, the state of various rRNA species between WT and Δrip1, mex67–5, and Δmft1 cells at 25°C, or after a 40-min heat treatment at 37°C or 42°C. Except for 18S rRNA, which was largely unaffected by temperature change, levels of other rRNAs decreased in all strains by increasing temperature (Fig. 7B). This is presumably because of the substantial decrease in levels of transcripts encoding ribosomal proteins upon heat shock (Warner 1989). Importantly, although rRNA levels are higher in the WT strain compared with the three mRNA export mutants, this relative difference was apparent already at 25°C. Thus, the dissapation of nucleolar antigens at the restrictive temperatures (37°C or 42°C) does not seem to severely affect rRNA processing in these mutants.
DISCUSSION
In the present study, we have screened the 83% of S. cerevisiae gene knockouts that are viable and identified 15 genes whose deletion individually disables HSP104 RNA release from its transcription site focus. Both “early” and “late” export factors were identified: (1) all of the four THO complex components, presumed to couple transcription to mRNA export by stimulating cotranscriptional maturation of mRNP (Schneiter et al. 1999; Jimeno et al. 2002; Libri et al. 2002; Strasser et al. 2002; Zenklusen et al. 2002); (2) the Sac3p/Thp1p dimer, which has a reported role in transcription but also localizes to the NPC (Fischer et al. 2002; Gallardo et al. 2003; Lei et al. 2003); and finally, (3) nine components of the NPCs. Fourteen of the corresponding protein products of the respective deletions have previously been assigned a role in mRNA export based on the nuclear accumulation of bulk p(A)+ RNA upon their mutation/deletion (Saccharomyces Genome Database: www.yeastgenome.org). Moreover, we have previously detected retained hsRNA in five of the deletion strains (the four THO deletions as well as Δrip1) (Jensen et al. 2001b; Libri et al. 2002). The fact that only factors with a previous history in mRNA transport and/or NPC integrity are identified in the screen suggests that the HSP104 RNA dot-formation phenomenon is highly specific. Furthermore, it indicates that additional nonessential genes with a role in HSP104 RNA export are unlikely to exist.
The most robust HSP104 RNA retention phenotype was observed in deletions of genes encoding THO and Sac3p/Thp1p complex components as well as in strains deleted for RIP1/NUP42, NUP84, NUP100, and NUP133. A role of the two former complexes in HSP104 RNA export can be rationalized by their association with the Mex67p/Mtr2p dimeric mRNA transport receptor; i.e., the THO complex connects to Mex67p via its association with Sub2p and the Mex67p-interacting protein Yra1p, and Sac3p binds directly to Mex67p. Of the NUPs, Rip1p/Nup42p is important for the final step in mRNA export, presumably by providing a platform on the cytoplasmic side of the NPC from where the RNA helicase Dbp5p, and its Rip1p-binding activator Gle1p, can facilitate mRNP remodeling and subsequent cytoplasmic release (Lund and Guthrie 2005; Alcazar-Roman et al. 2006; Weirich et al. 2006). Nup84p, Nup100p, and Nup133p are all situated in the vicinity of Rip1p/Nup42p in the molecular architecture of the yeast NPC (Rout et al. 2000). Of the weaker HSP104 RNA export mutants, Nup60p has no functional history in mRNA export. However, Nup60p has been reported to anchor Nup2p, whose deletion leads to HSP104 RNA retention (Table 1), to the nuclear face of the NPC, providing a possible explanation for the appearance of the Δnup60 strain in our screen (Denning et al. 2001).
The 15 deletion strains reported here add to the dozen or so mRNA export mutants, which retain hsRNAs, SSA4, or HSP104, in nuclear dots (Hilleren et al. 2001; Jensen et al. 2001a,b; Libri et al. 2002; Zenklusen et al. 2002; Dunn et al. 2005; data herein and data not shown). The differential localization of the mutated or deleted factors, e.g., transcription machinery- or NPC-associated, reinforces the notion that regardless of the physical location of the defect, hsRNA retention may be caused by the similar molecular phenotype (Jensen et al. 2001a). A possibility is that bulk mRNA retained in the nucleoplasm upon a persistent export block sequesters important factor(s) required for mRNP release from transcription site foci. The identification of such a factor(s) will be an important contribution to our understanding of the early stages in mRNP nuclear export.
Based on the yeast strains analyzed in this study, the correlation between the inability to export mRNA and fragmentation of nucleolar antigens is strong. This is because model strains of those that we coin “mRNA export mutants” in the present paper also have been directly shown to accumulate substantial amounts of hyperadenylated mRNA in their nuclei (Hilleren et al. 2001; Jensen et al. 2001b). Moreover, Northern blotting of rRNAs and snoRNAs in these mutants shows no sign of polyadenylation of these species (Fig. 7B; data not shown). Thus, at least the major part of the nuclear p(A)+ signal in these strains must arise from mRNAs. As an anecdotal point, the mtr3–1 and mtr4–1 mutant alleles of the two nuclear exosomal factors Mtr3p and Mtr4p were first identified in the original p(A)+ RNA-FISH isolation of mRNA transport-defective (mtr) mutants (Kadowaki et al. 1994a). Like for the Δrrp6 and Δrrp47 mutants, the p(A)+ signal accumulating in nuclei of these strains turns out to localize outside the DAPI-stained chromatin nuclear region and instead overlaps with Nop1p and Nsr1p (Thomsen et al. 2005; Carneiro et al. 2007; Rougemaille et al. 2007; data not shown). The majority of the p(A)+ signal in these mutants is produced by the Trf4p poly(A) polymerase and most likely stems from the adenylation of rRNA and other stable nucleolar RNAs (Rougemaille et al. 2007). Thus, mtr3–1 and mtr4–1 are not bona fide mRNA export mutants. Consistently, exosome mutants do not retain HSP104 RNA in their nuclei (Rougemaille et al. 2007; data not shown), and the nucleolus remains intact in the Δrrp6, Δrrp47, and mtr4–1 strains, even after a 40-min incubation at 42°C (Fig. 3A).
The extent to which the nucleolus fragments during an mRNA export block is not clear. Nop1p, Nsr1p, Nop5p, and U3 RNA all operate quite early in the ribosome assembly pathway, and therefore it is possible that their dissipation is due to alteration of one specific early process. This idea is somewhat at odds with Northern blotting analysis of rRNA processing intermediates, which failed to identify a maturation step that was specifically inhibited. Attempts to directly address the question by analyzing the behavior of nucleolar proteins involved in later steps of rRNA biogenesis revealed that, although normal nucleolar localization of these factors (i.e., Nmd3p and Rrp6p) was indeed disturbed in mRNA export mutants, this was also the case in a WT strain at high temperature (Fig. 7A; data not shown). Investigations by, e.g., electron microscopy, will be needed to analyze the more general impact on nucleolar morphology under these different conditions.
Although we cannot exclude that reduced nuclear export of transcript(s) encoding factors required for nucleolar integrity underlies the observed correlation, the rapidity by which the localizations of Nop1p, Nsr1p, Nop5p, and U3 RNA are affected suggests that the connection between retained mRNA and dissolution of nucleolar antigens is more direct. Interestingly, the transient nature of nuclear mRNA accumulation in the Δmft1 mutant at 37°C reveals that nucleolar antigens reassemble when the nuclear load of p(A)+ RNA wears off (Fig. 4). Taking this together with the observation that mRNA retention preceedes nucleolar disruption, and that the latter phenotype requires RNAPII transcription, we suggest that the excess nuclear load of p(A)+ RNA is at least one of the triggers causing nucleolar antigens to dissipate. This view is supported by our general observation that the major portion of the Nop1p, Nsr1p, and Nop5p immunofluorescent signals seemingly “move” to DAPI-stained chromatin upon nucleolar disassembly. However, our data do not support the simple interpretation that Nop1p/Nsr1p/Nop5p are merely attracted to the extra binding sites provided by retained mRNA, as visualized by p(A)+ or HSP104-RNA-FISH. This is because neither immunocytochemical nor UV cross-linking experiments show a significant interaction between p(A)+ RNA and Nop1p, Nsr1p, and Nop5p under these conditions (data not shown). Rather, RNA and protein signals occasionally overlap, but neither a strong tendency to cluster nor to be excluded from the same nuclear volume can be observed. These observations contradict an earlier report, showing Nop1p/p(A)+ RNA colocalization in the nonchromatin region of the yeast mtr2–1 mutant nucleus, suggesting mRNA movement to the nucleolar space (Kadowaki et al. 1994b). Whether this apparent discrepancy reflects an mtr2–1-specific phenotype, or is due to the fact that the reported colocalization experiment was performed at relatively long (90-min) incubation times at the restrictive temperature, remains to be determined.
Nuclear export of ribosomal subunits takes place under conditions of Nop1p/Nsr1p/Nop5p mislocalization (Fig. 7A). Thus, inefficient ribosome transit is unlikely to be causing nucleolar disruption. A more likely trigger could be that ribosomal proteins imported from the cytoplasm never make it to the nucleolus because they are depleted by the excess nuclear mRNA in these mutants. This might affect aspects of pre-ribosome assembly and lead to dissolution of nucleolar proteins already residing in stable complexes. In such a scenario, Nop1p/Nsr1p/Nop5p nucleolar dissipation would be a secondary event, which could also explain the lack of mRNA association of these factors.
A somewhat more provocative suggestion would be that some factors are recruited from the nucleolus to the nucleoplasm in a functionally relevant manner, and that return of these proteins to their steady-state environment is inhibited when mRNA export is blocked. The idea that nucleolar components partake in mRNP reactions has previously been put forward (Schneiter et al. 1995). While the present data do not allow us to address this issue, a previous argument against a role for at least Nop1p in mRNA export is the inability to detect accumulation of nuclear p(A)+ RNA in a number of Nop1p mutant backgrounds (Tollervey et al. 1991, 1993).
The eukaryotic nucleus is a dynamic entity harboring factors that constantly roam the nuclear space. Nucleolar morphology is most likely determined by the transient functional and physical interactions of its constituents, and seemingly small changes can have profound structural consequences; e.g., deficient capping of snoRNAs leads to loss of nucleolar organization (Colau et al. 2004). The dramatic changes in molecular environment that occur in S. cerevisiae mRNA export mutants therefore reinforce the need for examining mRNA-related phenotypes at the earliest possible time point after transfer to restrictive conditions.
MATERIALS AND METHODS
Yeast strains, manipulations, and growth conditions
Specific yeast strains utilized in this study are shown in Table 2. Strain manipulations were done by standard procedures. For growth conditions involving a temperature shift, strains were grown at 25°C to an OD600 = 0.1–0.3 in YM-1 medium followed by addition of an equal volume of preheated medium to achieve the desired growth temperature of 37°C or 42°C. In the case of inhibition of RNAPII transcription by thiolutin, cells were grown at 25°C, pelleted by centrifugation, and resuspended in 42°C warm YM-1 medium containing 50 μg/mL thiolutin.
TABLE 2.
Saccharomyces cerevisiae strains used in this study
The EUROSCARF gene deletion bank was screened by pooling five strains at a time and subjecting the sample to a 42°C heat shock for 20 min with subsequent processing for HSP104 RNA-FISH analysis. In the case of nuclear retention of HSP104 RNA, strains were subsequently assayed individually.
In situ hybridization, immunolocalization, and GFP-localization analysis
Fixation and preparation of cells for FISH analysis were done as previously described (Jensen et al. 2001b). For screening of the EUROSCARF strains, HSP104 RNA was detected using the previously described Cy3-labeled HSP104 oligonucleotide probes: THJ203, THJ204, THJ205, and THJ206 (Jensen et al. 2001b). Combined RNA-FISH/immunolocalization analyses were performed as described (Thomsen et al. 2003), using the THJ361: 5′-gctgaaxagacgaaxcattgaaxagagcaaacaaxatatggtctxgcgct, THJ362: 5′-ctxtccccaaagcaxaacttggagxtatctccgcaggxgcaggttgctgxt, THJ363: 5′-atagxcgtaacggcccxtctcaatxagattctgxaggtaagggacxgatcc, and THJ364: 5′-gtggaxgttgatgaxccgaagccaattttxgagccaacgxcaaaatcgtxag oligonucletide probes (X denotes amino-modified T residue amenable to Cy3 conjugation), and antibodies toward Nop1p, Nop5p, and Nsr1p (EnCor Biotech. Inc.) or GFP (Santa Cruz). The THJ361–364 probes target the 5′ end of the HSP104 transcript and were used because they yield a more robust signal reproducibly tolerating the subsequent immunolocalization procedure. Poly(A)+ RNA detection was carried out using the LNA-modified dT20 probe as described (Thomsen et al. 2005), and U3 RNA detection was performed as described (Verheggen et al. 2001).
For transport assays of GFP-fused proteins, WT, Δrip1, mex67–5, and Δrip1 strains were transformed with plasmids expressing Rps3–GFP, Nmd3–GFP, or Rpl25–GFP as described (Yao et al. 2007).
Imaging
Images were acquired on an Olympus BX51 microscope equipped with a cooled Olympus DP50 CCD camera and analySIS software. Image handling was done in Adobe Photoshop.
Northern blotting analysis
Total RNA was purified using the “hot phenol” method. RNAs were detected using the following 5′end-labeled oligonucleotide probes: 20S: (002: 5′-GCTCTTTGCTCTTGCC-3′), 18S: (008: 5′-CATGGCTTAATCTTTGAGAC-3′), 27S: (003: 5′-TGTTACCTCTGGGCCC-3′), 25S: (007: 5′-CTCCGCTTATTGATATGC), 5.8S: (o5.8S: 5′-TGCGTTCTTCATCGATGCGAGAACC-3′), and SCR1: (oScR1: 5′-GTCTAGCCGCGAGGAAGG-3′), as previously described (Briggs et al. 1998; Oeffinger et al. 2004; Panse et al. 2006).
ACKNOWLEDGMENTS
We thank the following individuals for providing reagents: Edouard Bertrand, Francoise Stutz, Andres Aguilera, Michael Rosbash, Chuck Cole, Domenico Libri, Daniela Roser, and Ed Hurt. We thank Manfred Schmid, Pascal Preker, Fransisco Malagon, and Domenico Libri for comments on the manuscript, and three anonymous reviewers for fruitful suggestions to the manuscript. This work was supported by the Danish National Research Foundation (Grundforskningsfonden) and the Novo Nordisk Foundation. C.S. was the recipient of a postdoctoral fellowship from the FEBS association.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.718708.
REFERENCES
- Abruzzi, K.C., Lacadie, S., Rosbash, M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Roman, A.R., Tran, E.J., Guo, S., Wente, S.R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Andersen, J.S., Lam, Y.W., Leung, A.K., Ong, S.E., Lyon, C.E., Lamond, A.I., Mann, M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Boyne, J.R., Whitehouse, A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. 2006;103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, M.W., Burkard, K.T., Butler, J.S. Rrp6p, the yeast homolog of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′-end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Carneiro, T., Carvalho, C., Braga, J., Rino, J., Milligan, L., Tollervey, D., Carmo-Fonseca, M. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colau, G., Thiry, M., Leduc, V., Bordonné, R., Lafontaine, D.L.J. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol. Cell. Biol. 2004;24:7976–7986. doi: 10.1128/MCB.24.18.7976-7986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, C.N., Scarcelli, J.J. Transport of messenger RNA from the nucleus to the cytoplasm. Curr. Opin. Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Denning, D., Mykytka, B., Allen, N.P., Huang, L., Al, B., Rexach, M. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 2001;154:937–950. doi: 10.1083/jcb.200101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois, G., Iglesias, N., Stutz, F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff, T.C., Heath, C.V., Goldstein, A.L., Snay, C.A., Cole, C.N. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, E.F., Hammell, C.M., Hodge, C.A., Cole, C.N. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes & Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, T., Strasser, K., Racz, A., Rodriguez-Navarro, S., Oppizzi, M., Ihrig, P., Lechner, J., Hurt, E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, M., Aguilera, A. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 2001;157:79–89. doi: 10.1093/genetics/157.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, M., Luna, R., Erdjument-Bromage, H., Tempst, P., Aguilera, A. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- Gilbert, W., Guthrie, C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- Gorsch, L.C., Dockendorff, T.C., Cole, C.N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., Jensen, T.H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., Young, R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ideue, T., Azad, A.K., Yoshida, J., Matsusaka, T., Yanagida, M., Ohshima, Y., Tani, T. The nucleolus is involved in mRNA export from the nucleus in fission yeast. J. Cell Sci. 2004;117:2887–2895. doi: 10.1242/jcs.01155. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Boulay, J., Rosbash, M., Libri, D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001a;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., Rosbash, M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001b;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Boulay, J., Olesen, J.R., Colin, J., Weyler, M., Libri, D. Modulation of transcription affects mRNP quality. Mol. Cell. 2004;16:235–244. doi: 10.1016/j.molcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Jimenez, A., Tipper, D.J., Davies, J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae . Antimicrob. Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno, S., Rondon, A.G., Luna, R., Aguilera, A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Chen, S., Hitomi, M., Jacobs, E., Kumagai, C., Liang, S., Schneiter, R., Singleton, D., Wisniewska, J., Tartakoff, A.M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994a;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Hitomi, M., Chen, S., Tartakoff, A.M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell. 1994b;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Schneiter, R., Hitomi, M., Tartakoff, A.M. Mutations in nucleolar proteins lead to nucleolar accumulation of poly(A)+ RNA in Saccharomyces cerevisiae . Mol. Biol. Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P., Stern, C.A., Fahrenkrog, B., Krebber, H., Moy, T.I., Aebi, U., Silver, P.A. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell. 2003;14:836–847. doi: 10.1091/mbc.E02-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., Jensen, T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M.K., Guthrie, C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Oeffinger, M., Dlakic, M., Tollervey, D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes & Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse, V.G., Kressler, D., Pauli, A., Petfalski, E., Gnadig, M., Tollervey, D., Hurt, E. Formation and nuclear export of preribosomes are functionally linked to the small-Ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R.D., Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Rougemaille, M., Gudipati, R.K., Olesen, J.R., Thomsen, R., Seraphin, B., Libri, D., Jensen, T.H. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., Chait, B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Kadowaki, T., Tartakoff, A.M. mRNA transport in yeast: Time to reinvestigate the functions of the nucleolus. Mol. Biol. Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Guerra, C.E., Lampl, M., Gogg, G., Kohlwein, S.D., Klein, H.L. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., Darzacq, X., Shenoy, S.M., Fusco, D., Janicki, S.M., Spector, D.L., Singer, R.H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Strasser, K., Hurt, E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz, F., Kantor, J., Zhang, D., McCarthy, T., Neville, M., Rosbash, M. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes & Dev. 1997;11:2857–2868. doi: 10.1101/gad.11.21.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz, F., Bachi, A., Doerks, T., Braun, I.C., Seraphin, B., Wilm, M., Bork, P., Izaurralde, E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M.P., Dahlberg, J.E. Retention and 5′-cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Thompson, C.M., Young, R.A. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, R., Libri, D., Boulay, J., Rosbash, M., Jensen, T.H. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae . RNA. 2003;9:1049–1057. doi: 10.1261/rna.5170303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, R., Nielsen, P.S., Jensen, T.H. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11:1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Carmo-Fonseca, M., Hurt, E.C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Jansen, R., Kern, H., Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Trumtel, S., Leger-Silvestre, I., Gleizes, P.E., Teulieres, F., Gas, N. Assembly and functional organization of the nucleolus: Ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell. 2000;11:2175–2189. doi: 10.1091/mbc.11.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen, C., Mouaikel, J., Thiry, M., Blanchard, J.M., Tollervey, D., Bordonne, R., Lafontaine, D.L., Bertrand, E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra, P., Stutz, F. mRNA export: An assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. Synthesis of ribosomes in Saccharomyces cerevisiae . Microbiol. Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich, C.S., Erzberger, J.P., Flick, J.S., Berger, J.M., Thorner, J., Weis, K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Yao, W., Roser, D., Kohler, A., Bradatsch, B., Baßler, J., Hurt, E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Zenklusen, D., Vinciguerra, P., Wyss, J.C., Stutz, F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]