Abstract
Riboswitches are highly structured cis-acting elements located in the 5′-untranslated region of messenger RNAs that directly bind small molecule metabolites to regulate gene expression. Structural and biochemical studies have revealed riboswitches experience significant ligand-dependent conformational changes that are coupled to regulation. To monitor the coupling of ligand binding and RNA folding within the aptamer domain of the purine riboswitch, we have chemically probed the RNA with N-methylisatoic anhydride (NMIA) over a broad temperature range. Analysis of the temperature-dependent reactivity of the RNA in the presence and absence of hypoxanthine reveals that a limited set of nucleotides within the binding pocket change their conformation in response to ligand binding. Our data demonstrate that a distal loop–loop interaction serves to restrict the conformational freedom of a significant portion of the three-way junction, thereby promoting ligand binding under physiological conditions.
Keywords: RNA structure, gene regulation, riboswitch, chemical probing
INTRODUCTION
Recent discoveries have revealed that a substantial amount of normal cellular metabolism is controlled by trans- and cis-acting noncoding RNA sequences (Mattick and Makunin 2006). Riboswitches are one form of RNA-mediated regulation that acts by virtue of the RNA's ability to directly bind a variety of cellular metabolites (for review, see Nudler and Mironov 2004; Winkler and Breaker 2005). In bacteria, riboswitches are found in the 5′-untranslated region (5′-UTR) of mRNAs and are composed of two distinct domains (Fig. 1A). The upstream aptamer domain is responsible for specifically binding an effector ligand. Currently, there are at least 12 identified aptamer domains, each responding to a different metabolite, including amino acids (Rodionov et al. 2003; Sudarsan et al. 2003; Mandal et al. 2004), nucleobases (Mandal et al. 2003; Mandal and Breaker 2004), cofactors (Grundy and Henkin 1998; Mironov et al. 2002; Winkler et al. 2002a,b; Epshtein et al. 2003; Winkler et al. 2003), and metal ions (Dann et al. 2007). Binding of ligand to the aptamer domain is communicated to a downstream domain called the expression platform containing a genetic switch involving two mutually exclusive secondary structures. For regulation at the transcriptional level, one of these structures corresponds to a classic rho-independent transcriptional terminator, while the other is an antiterminator that allows for complete transcription of the mRNA (for review, see Winkler and Breaker 2005; Irnov et al. 2006).
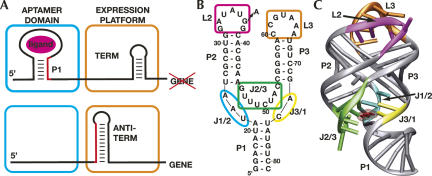
FIGURE 1.
(A) Cartoon of regulation by riboswitches. When ligand binds to the aptamer domain (cyan), this incorporates a “switching sequence” at the 3′ side of the P1 helix (red) into this domain, forcing the downstream expression platform (orange) to form a terminator element. In the absence of ligand, the “switching sequence” is available to be used to form an antiterminator that in turn allows for synthesis of the full-length mRNA. Secondary (B) and tertiary (C) structure of the aptamer domain of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain bound to hypoxanthine (red) (PDB ID 1U8D).
To understand how riboswitches specifically recognize small molecule metabolites, the structures of several aptamer domains have been solved by X-ray crystallography: the purine (Batey et al. 2004; Serganov et al. 2004), thiamine pyrophosphate (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006; Thore et al. 2006), and S-adenosylmethionine riboswitches (Montange and Batey 2006). Similarities between these structures include almost complete encapsulation of ligand by the RNA, use of peripheral tertiary structure to organize the binding pocket, and ligand-dependent conformational changes in the RNA (Gilbert et al. 2006a). Conformational changes and tertiary folding within the aptamer domain are particularly important, as they are proposed to be the link between ligand binding and the secondary structural switch that controls gene expression.
The purine riboswitch is currently the most extensively studied model system to understand the relationship between RNA folding and riboswitch function. Most of this work has focused upon a conserved loop–loop interaction that brings the P2 and P3 helices into a parallel orientation (Fig. 1B,C) and is essential for hypoxanthine binding (Batey et al. 2004). A combination of single molecule FRET (Lemay et al. 2006), NMR spectroscopy (Noeske et al. 2007), and in-line probing (Mandal et al. 2003) have demonstrated that this interaction forms in the absence of ligand binding. However, ligand-induced folding of the three-way junction must also occur, as inferred from the fact that the purine nucleobase is ∼98% solvent inaccessible when bound to the RNA (Batey et al. 2004; Serganov et al. 2004). A combination of thermodynamic and kinetic analyses has allowed us to propose a model in which the purine nucleobase initially docks with a pyrimidine residue in J3/1 (Fig. 1B,C, yellow) followed by the J1/2 and J2/3 strands (Fig. 1B,C, cyan, green) folding around the ligand to fully encapsulate the ligand (Gilbert et al. 2006b).
Chemical probing analysis combined with isothermal titration calorimetry (ITC) has revealed several new aspects of ligand-dependent folding in the purine riboswitch. First, nucleotides within the three-way junction are sufficient for binding the high-affinity ligands guanine and 2,6-diaminopurine. This demonstrates that the loop–loop interaction serves an important but non-essential supporting role during ligand recognition. Second, nucleotides in the J1/2 and J2/3 strands of the three-way junction directly involved in purine recognition and base triple formation in the P1 stem experience the largest ligand-dependent change in dynamics, thus providing a direct link between binding and gene regulation. Finally, the temperature dependence of local structure formation in the presence and absence of purine ligand demonstrates that a majority of the ligand binding pocket experiences a significant reduction in conformational freedom upon formation of the loop–loop interaction. As we will discuss, the restriction of conformational freedom in binding and catalytic sites by tertiary architecture appears to be a significant feature of RNA's ability to function in a biological context.
RESULTS
Conformational flexibility in the purine riboswitch
To initially characterize the ligand-dependent folding of the purine riboswitch, we probed the Bacillus subtilis xpt-pbuX guanine riboswitch aptamer domain in 100 mM NaCl at three different temperatures, in the presence and absence of 6 mM Mg2+ and/or 10 μM hypoxanthine (Fig. 2). This was accomplished using a form of chemical probing called selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Merino et al. 2005). This technique employs N-methylisatoic anhydride (NMIA) that reacts with 2′-hydroxyl groups in dynamic regions of an RNA (Merino et al. 2005; Wilkinson et al. 2006). A particularly powerful aspect of this methodology is that the entire RNA structure can be probed simultaneously over a wide temperature range, allowing one to monitor RNA folding with nucleotide resolution as demonstrated for tRNAAsp (Wilkinson et al. 2005).
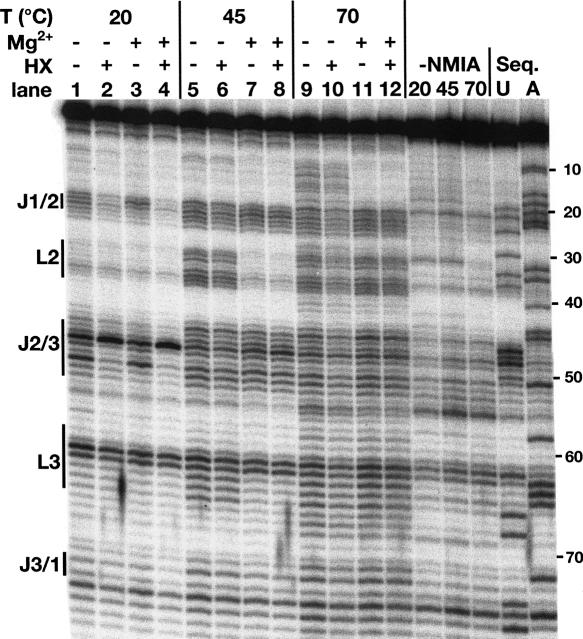
FIGURE 2.
NMIA probing of the guanine riboswitch aptamer domain at three temperatures (20°C, 45°C, and 70°C) in the presence and absence of 6 mM MgCl2 and/or 10 μM hypoxanthine. Bars to the side of the gel highlight regions of the RNA corresponding to loop and junction regions.
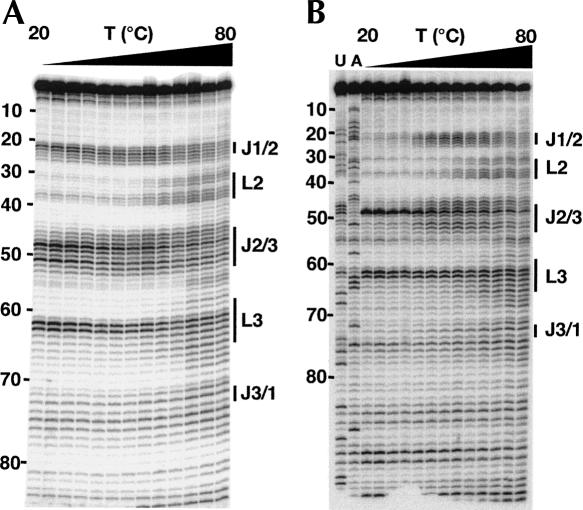
Patterns of NMIA-dependent reactivity within the RNA under varying conditions reveal changes in the secondary and tertiary structure (Fig. 2). At 20°C in the absence of ligand the most extensively modified regions of the RNA lie in J1/2, J2/3, and the turning nucleotides of L3 (G62 and U63) when compared to a control reaction without NMIA, indicating that these are the conformationally dynamic regions of the RNA. The pattern of reactivity is, however, subtly reduced upon the addition of magnesium within the 3′ side of J1/2 (Fig. 2, lanes 1,3). Addition of hypoxanthine results in protection of both J1/2 and J2/3 with the exception of U48 (Fig. 2, lanes 2,4), whose increased reactivity is consistent with the crystal structure in which it is flipped out into the bulk solvent (Batey et al. 2004; Serganov et al. 2004). A dinucleotide platform at the top of L3 that is formed by nucleotides G62 and U63 remains reactive under all conditions, a consequence of the ribose sugars adopting a reactive C2′-endo configuration in the turn. Increasing the temperature to 45°C reveals that L2 and L3 become protected from NMIA modification only in the presence of magnesium (Fig. 2, lanes 7,8). Even in the presence of magnesium, the reactivity of J1/2 and J2/3 encompasses a larger number of nucleotides reflecting a substantially increased degree of conformational freedom in the three-way junction (Fig. 2, lanes 3,7). Increasing the temperature to 70°C causes L2 and L3 to be reactive even in the presence of Mg2+ (Fig. 2, lanes 11,12), and in the absence of Mg2+ we also begin to observe reactivity within the helical regions of the RNA such as P1 (Fig. 2, lanes 9,10).
Together, these data reveal a clear interdependence of magnesium, ligand, and temperature on the RNA's ability to adopt a specific folding state and productively bind hypoxanthine. At 20°C, formation of the L2–L3 interaction is clearly independent of Mg2+ and hypoxanthine, as identical results were obtained in the presence of 2 mM EDTA (data not shown). This supports previous findings that Mg2+ is not required for ligand binding or formation of the loop–loop interaction at this temperature (Lemay et al. 2006; Noeske et al. 2007).
The loop–loop is important but not essential for ligand binding
There is a striking similarity between the three-dimensional architectures of the purine riboswitch (Batey et al. 2004; Serganov et al. 2004) and the natural hammerhead ribozyme (Martick and Scott 2006). Specifically, they both contain an active site embedded within a three-way junction and a peripheral loop–loop interaction that facilitates the RNA's activity. While the ribozyme functions at physiological ionic conditions with the peripheral loop–loop interaction, it requires high concentrations of Mg2+ in its absence (Khvorova et al. 2003; Canny et al. 2004). Nonetheless, only nucleotides within the three-way junction of the hammerhead are both necessary and sufficient for function; could the same be true of the purine riboswitch aptamer domain?
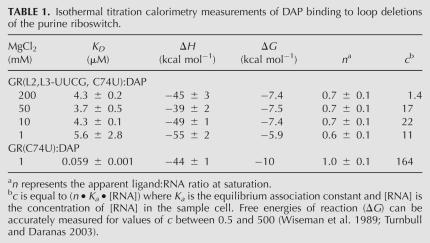
Previously, we tested the importance of the L2–L3 interaction with a minimal aptamer domain in which the terminal loops were replaced with stable UUCG tetraloops, GR(L2,L3-UUCG), to completely ablate the tertiary interaction (Batey et al. 2004). When tested for hypoxanthine binding at 10 mM Mg2+, no detectable binding was observed (Batey et al. 2004). Similar results were obtained in a mutagenic survey of the adenine riboswitch (Lemay et al. 2006). As these analyses were performed with the lower affinity ligands hypoxanthine (Kd = 760 nM) and adenine (Kd = 300 nM), we hypothesized that a minimal aptamer domain may only productively bind high affinity ligands (guanine [Kd = 3 nM] or 2,6-diaminopurine [Kd = 17 nM]) in the presence of elevated magnesium concentrations (Gilbert et al. 2006b). If the role of the distal loop–loop interaction is to influence the conformational freedom of the three-way junction, we would predict that the high affinity ligands (guanine and diaminopurine) can productively bind the minimal three-way junction. To test this by isothermal titration calorimetry (ITC), we must use a highly soluble ligand to measure potentially weak binding, limiting us to 2,6-diaminopurine (DAP). Therefore, a C74U mutation was introduced into the minimal purine riboswitch aptamer domain [GR(L2,L3-UUCG, C74U)], which previous studies have shown to be the only change required to confer a specificity switch to adenine (Mandal and Breaker 2004; Gilbert et al. 2006b; Lemay and Lafontaine 2007).
Measurement of the affinity of GR(L2,L3-UUCG, C74U) RNA for DAP reveals specific ligand binding with an apparent equilibrium dissociation constant of 4 μM in 10–200 mM MgCl2 (Table 1). Comparison of this value to both the Vibrio vulnificus add adenine riboswitch and the B. subtilis xpt-pbuX guanine riboswitch bearing the C74U mutation, each of which bind DAP with a Kd of 20 nM in 10 mM MgCl2, reveals that the L2/L3 tertiary interaction provides ∼3.3 kcal mol−1 energy to facilitate ligand binding. This value also corresponds to the ΔΔG for the most deleterious loop mutants tested in several other studies (Lemay et al. 2006; Gilbert et al. 2007). Under near physiological magnesium concentrations (1 mM), this affinity decreases ∼10-fold and the ΔΔG increases to ∼4.1 kcal mol−1. Therefore, nucleotides within the three-way junction are necessary and sufficient to specifically bind ligand while the peripheral loop–loop interaction plays an important but non-essential supporting role.
TABLE 1.
Isothermal titration calorimetry measurements of DAP binding to loop deletions of the purine riboswitch.
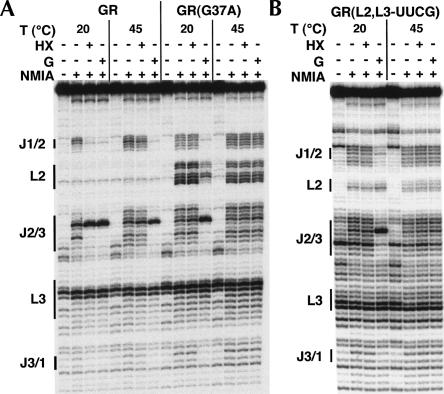
To further define the relationship between the loop–loop interaction and binding of high and low affinity ligands, chemical probing was performed on the wild-type xpt-pbuX guanine riboswitch aptamer domain (GR), an RNA containing a loop–loop mutation that ablates the tertiary interaction, GR(G37A), and an RNA in which both loops are replaced by a UUCG tetraloop, GR(L2,L3-UUCG) (Fig. 3). These three RNAs were modified at either 20°C or 45°C with 100 mM NaCl and 6 mM MgCl2 in the presence and absence of hypoxanthine or guanine. We observed that GR binds both hypoxanthine and guanine, resulting in a similar pattern of modification at 20°C (Fig. 3A); elevation of the temperature to 45°C abolishes hypoxanthine binding while guanine binding remains productive. The G37A mutant, the most deleterious point mutation in the loops (Lemay et al. 2006; Gilbert et al. 2007), does not bind hypoxanthine at 20°C but is competent to bind guanine (Fig. 3A). Notably, guanine binding results in a modest protection of the loops, indicating that ligand binding can promote the formation of this suboptimal tertiary element, as has been observed by NMR spectroscopy (Noeske et al. 2007). At 45°C, the ability to bind either ligand is abolished. A similar result is observed for the UUCG construct, which completely lacks a loop–loop interaction; however there is never a restoration of the loop–loop interaction (Fig. 3B).
FIGURE 3.
NMIA probing in the presence and absence of low and high affinity ligands (hypoxanthine and guanine, respectively). (A) Aptamer domain RNA from wild-type GR and a G37A mutant and (B) a guanine riboswitch aptamer domain with loops 2 and 3 replaced by a UUCG tetraloop [GR(L2,L3-UUCG)].
The other notable feature of these experiments is that the pattern of reactivity in the three-way junction is markedly different between wild-type and loop mutant RNAs. For example, in the wild-type RNA, we observe that nucleotides U22 and A23 display a greater level of reactivity than A24–U26 in J1/2, while in J2/3 nucleotides U48–U51 are the most reactive. Abolishing the distal loop–loop interaction in both mutants results in an increased level of reactivity in both joining regions (Fig. 3A,B) that reflects the same pattern of reactivity observed for the wild-type RNA at 45°C (Fig. 2, lane 7). A lesser effect is also seen in J3/1. This demonstrates that stable formation of a distal tertiary interaction directly and significantly influences the structure of the ligand binding pocket.
Ligand-induced folding of the riboswitch aptamer domain
To investigate ligand-dependent folding of the RNA, we monitored the NMIA reactivity of the aptamer domain as a function of temperature (20°C–80°C) in the presence and absence of 10 μM hypoxanthine, 100 mM NaCl, and 6 mM MgCl2 (Fig. 4). From these data, we are able to quantify the reactivity of each nucleotide using the SAFA program (Das et al. 2005). To control for variability in loading between lanes, we used the reactivity of U63 as an internal control. This nucleotide resides at the top of L3 and displays a high degree of modification by NMIA at all temperatures in the presence and absence of hypoxanthine. Upon applying this correction factor, the scaled reactivity of each base can be plotted as a function of temperature (Fig. 5). The chemical reactivity profiles were then fit to a two-state model that allows extraction of the melting temperature for each base as previously described (Wilkinson et al. 2005).
FIGURE 4.
NMIA probing of the guanine riboswitch aptamer domain in the absence (A) and presence (B) of 10 μM hypoxanthine for quantifying the melting behavior of the RNA with nucleotide resolution. The RNA was probed in the presence of 6 mM MgCl2. Numbers to the side of the gels denote the nucleotide position, and the bars correspond to the nonhelical regions of the RNA.
FIGURE 5.
Melting profiles for several nucleotides in the aptamer domain. (A) Reactivity profile for nucleotide G37, which resides in loop 2 in the absence (triangles) and presence (circles) of hypoxanthine. The dashed line represents the best fit to a model describing a two-state transition. (B) Reactivity profile for U51, a nucleotide in J2/3. The data for reactivity in the absence of ligand (triangles) showed no transition, so they are not fit to a curve. (C) Expanded view of data in panel B. Note that at high temperatures there is a pronounced drop in reactivity; the impact of this effect was reduced with a rolling weighting function to accurately quantify the transition.
One complicating issue with this technique is that we consistently observed a decrease in reactivity for the majority of nucleotides at higher temperatures. This is easily observed for nucleotides that become fully reactive at temperatures less than 50°C, resulting in a bell-shaped melting curve that artificially elevates the predicted Tm values (Fig. 5C). This behavior was also observed in the analysis of tRNAAsp (Wilkinson et al. 2005). To ensure that this is not due to increased degradation of the RNA at higher temperatures, we ran control experiments in which the RNA was treated exactly as before, except that it was exposed to the DMSO solution without NMIA (data not shown). The background degradation pattern of the RNA did not change between 20°C and 80°C when reacted for five half-lives (data not shown). We also performed several experiments with lower NMIA concentrations to ensure that we were not overmodifying the RNA at the higher temperatures; this experiment yielded identical results to the original data (data not shown). In addition, a complication of this experiment is that the pKa of the buffer system (HEPES) changes as a function of temperature (ΔpKa/ΔT°C = 0.0014) that may also give rise to this effect. To control for this, the buffer pH was altered in each individual reaction such that, upon heating, all samples will have an identical pH (8.0). Buffer compensation, however, did not dramatically change the melting profiles in a way that would account for the unidentified effect. Therefore, to deal with this issue, for nucleotides that had well-defined upper baselines before the reactivity begins to diminish, we applied a rolling weighting function to the upper temperature points (Fig. 5C; Wilkinson et al. 2005). The Tm estimates are accurate within two degrees of the actual Tm, as demonstrated by the analysis of tRNAAsp, and thus this experiment accurately reflects bulk melting behavior observed using other techniques (Wilkinson et al. 2005).
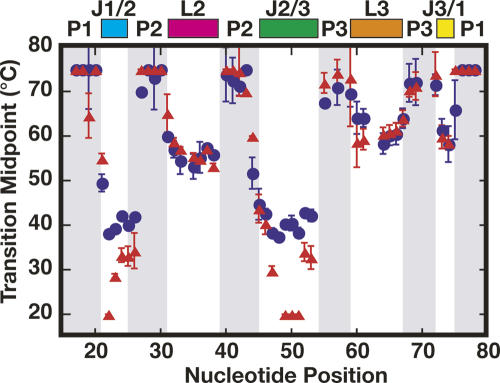
In the presence and absence of hypoxanthine, a group of nucleotides corresponding to helices P1, P2, and P3, displayed Tm values >70°C C (Fig. 6). As expected, this indicates that the A-form helices are more thermally stable than the nucleotides involved in nonhelical structures in the loop–loop interaction and three-way junction. An exception to this are bases A21 of P1 and A44 within P2 that exhibit Tm values between 50°C and 60°C. Bases within L2, L3, and J3/1 display ligand-independent melting transitions that occur between 50°C and 65°C (Fig. 6). The correlation in melting transitions for nucleotides in L2 and L3 is expected, as they form a single tertiary interaction. This behavior was also observed for nucleotides in the D- and T-loop of tRNAAsp, which also form a loop–loop interaction (Wilkinson et al. 2005). Notably, the nucleotides in J3/1 show a similar correlation to the loop nucleotides, suggesting that their conformation is likely coupled to the distant loop–loop tertiary interaction.
FIGURE 6.
Apparent transition midpoints (Tms) for each nucleotide in the RNA in the absence (red triangles) and presence (blue circles) of hypoxanthine. Error bars represent the measured error from three independent experiments. Colored bars on the top highlight the different architectural elements of the RNA.
Melting behavior for the majority of bases located within the J1/2 and J2/3 regions display a varying dependence on the presence of ligand (Fig. 6). In the presence of hypoxanthine, bases U22–C26 of J1/2 and A45–C53 of J2/3 are characterized by a narrow range of Tm values between 38°C and 45°C. In the absence of hypoxanthine, nucleotides A23–C26 of J1/2 and U47/A52–C53 of J2/3 display Tm values ∼10°C lower than observed in the presence of ligand. Since these values reside between 28°C and 33°C, these nucleotides are restricted in their dynamics as compared to adjacent nucleotides (U22 and U48–U51; see below). The conformational restriction of these nucleotides is due to formation of the L2–L3 interaction, as supported by the markedly enhanced reactivity of J1/2 and J2/3 when it is ablated (Fig. 3A,B). Bases U22 and U48–U51 display Tm values more than 20°C lower than observed in the presence of ligand. Actual Tm values are not known for these bases in the absence of ligand, as they are fully reactive at the lowest temperature tested; thus they are assigned as 20°C. The identities of these bases are significant, as U22 and U51 are directly involved in hydrogen bonding with hypoxanthine. Also, bases U49 and C50 participate in the formation of two base triples with the top two base pairs of the P1 helix. This region of P1 contains the switching sequence that determines secondary structure formation in the expression platform and subsequently the on/off state of the riboswitch. The large change in Tm upon ligand binding as well as a low Tm in the absence of ligand are entirely consistent with our previously proposed model in which J1/2 and J2/3 are the conformationally dynamic regions of the junction that fold around the ligand following its initial recognition by the specificity pyrimidine C74 in J3/1 (Gilbert et al. 2006b).
DISCUSSION
A key feature of the purine riboswitch is a distal loop–loop interaction that is important for ligand binding under physiological conditions (Batey et al. 2004; Lemay et al. 2006). The use of RNA tertiary structure to facilitate its biological function is a common feature of small ribozymes (Murchie et al. 1998; Khvorova et al. 2003; Zamel et al. 2004), RNP assembly (Recht and Williamson 2004; Talkington et al. 2005), RNA–RNA interactions (Hodak et al. 2005), as well as for riboswitches (Batey et al. 2004; Montange and Batey 2006; Serganov et al. 2006; Thore et al. 2006). For example, the natural hammerhead and hairpin ribozymes require peripheral tertiary elements to achieve maximal catalytic rates under physiological Mg2+ conditions (0.1–1 mM) (Murchie et al. 1998; Fedor 1999; Khvorova et al. 2003). A comparison of the structures of the natural and minimal hammerhead ribozymes indicates that the role of tertiary architecture is to promote an active site conformation in the ground state that is significantly closer to that of the transition state (Martick and Scott 2006). Kinetic analysis of wild-type and minimal hammerhead ribozymes directly demonstrates that distal structural elements enhance catalysis by shifting the population of RNAs toward a catalytically active state (Canny et al. 2004).
The purine riboswitch displays a similar architecture to the hammerhead ribozyme, suggesting that they may respond comparably to the loss of distal tertiary structural features. Accordingly, our binding studies reveal that complete removal of the loop–loop interaction of the purine riboswitch aptamer domain, akin to the minimal hammerhead construct, does not completely destroy activity under in vitro conditions. Loss of this feature decreases the binding affinity for 2,6-diaminopurine by ∼3.3 kcal mol−1 and requires elevated Mg2+ concentrations to achieve the highest affinity of binding. Furthermore, increased concentrations of magnesium cannot fully compensate for the loss of the loop–loop interaction; at high magnesium concentrations (10–200 mM) the binding affinity for 2,6-diaminopurine plateaus at ∼100-fold lower than wild type at 1 mM Mg2+. It is important to note, however, that under biological conditions the loop–loop interaction is essential for proper riboregulation. This is underscored by the fact that nucleotides in both L2 and L3 that form two base quadruples are >97% conserved across all known purine riboswitches (Griffiths-Jones et al. 2005; Barrick and Breaker 2007). Structural organization of the aptamer domain by the loop–loop interaction may serve to increase the rate at which the ligand productively binds, which is likely a critical feature of kinetically driven riboswitches (Wickiser et al. 2005a).
Clearly, the tertiary interaction facilitates ligand binding in a fashion that magnesium cannot, despite the fact that each RNA achieves the same bound conformation in the junction. One interpretation is that, like the hammerhead ribozyme, distal structural elements increase the populations of active conformers. This view is supported by a comparison of the reactivity pattern of nucleotides in the three-way junction in wild-type and loop mutant RNAs. Two very different types of mutants in the loop show increased patterns of reactivity in J1/2 and J2/3, indicating that loss of the L2–L3 interaction increases the conformational dynamics of the junction, despite the presence of 6 mM Mg2+. This is consistent with the finding that certain nucleotides in the junction are highly phylogenetically conserved because they play important roles in maintaining a binding-competent free state (Gilbert et al. 2007; Lemay and Lafontaine 2007). Together, this argues that the unliganded binding pocket is biased toward conformations via tertiary interactions and conserved junction sequence elements that favor purine binding; but, what exactly is the “structure” of the free state?
NMR spectroscopy and in-line probing suggest the entire three-way junction region of the RNA is highly disorganized in the absence of ligand (Mandal et al. 2003; Noeske et al. 2005). However, some aspect of the binding pocket must be organized to serve as an initial docking site for productive ligand binding. Using thermodynamic and kinetic approaches, we proposed that the ligand-binding pocket is preorganized with respect to J3/1 yet dynamic in J1/2 and J2/3 (Gilbert et al. 2006b) and the results of the current investigation strongly support this model. Consistent with this model, the high degree of correlation in observed transition midpoints between J1/2 and J2/3 in the bound state suggests that they simultaneously undergo a single folding event when the purine nucleobase productively binds the RNA.
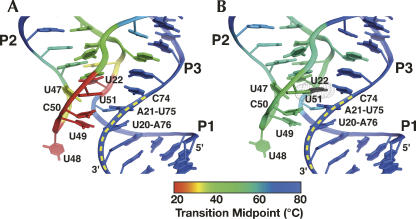
Within J1/2 and J2/3 we also observe a pronounced gradation of transition midpoints in the absence of ligand (Fig. 6). Plotting these midpoints onto the structure of the bound RNA (Fig. 7) reveals that nucleotides directly involved in ligand binding and formation of base triples with P1 are the most dynamic in the absence of ligand and become significantly more ordered in the presence of ligand. In contrast, nucleotides above the purine-binding site (A23–C26 of J1/2 and U47/A52–C53 of J2/3) display much smaller changes in observed Tm in response to ligand binding, suggesting that this region of the junction is conformationally restricted prior to ligand binding. This is consistent with the observation that the fluorescence of 2-aminopurine at the 24 position undergoes a small change between the bound and unbound forms of the RNA (Rieder et al. 2007).
FIGURE 7.
Superposition of apparent transition midpoints (Tms) on the structure of the three-way junction in the (A) absence and (B) presence of ligand. Blue represents regions with high melting transitions (> 65°C), green for intermediate Tms (45°C–65°C) and red for low Tms (< 30°C).
Conformational restriction of the upper region of the junction (G46–C53•A23 and U22–A52•A73 base triples) explains how the distal loop–loop interaction facilitates ligand binding. The L2–L3 interaction brings P2 and P3 parallel to one another, inducing formation of an important hydrogen bonding contact between the U25–A45 and C54–G72 pairs at the base of these helices adjacent to the junction. Parallel positioning of these two helices further allows J2/3 to act more as a terminal loop through formation of a closing G46–C53 Watson–Crick pair along with interactions with A23 and A24 of J1/2 to stabilize the top of the junction. Concurrently, J3/1 is organized via coaxial stacking of P1 and P3, presenting the critically important pyrimidine 74 for initial docking by the ligand. However, it is important to emphasize that the above describes one of many conformations accessible by the free state; the distal tertiary interaction only serves to increase the populations of productive states in solution. Thus, the loop–loop interaction serves to conformationally restrict the free state, thereby increasing the number of productive collision events. This is in contrast to a “predetermined” induced fit binding mechanism in which specific base pairs in the junction are obligatorily and stably formed prior to ligand binding (Ottink et al. 2007).
The lack of a strict requirement for a “predetermined” three-way junction is clearly evident by the fact that there is not an absolute requirement for the distal tertiary interaction during productive binding. Comparison of the wild-type RNA at various temperatures reveals that at 45°C the reactivity pattern reflects a formed L2–L3 interaction, but the pattern in J1/2 and J2/3 is identical to that at 70°C. It is also clear that 10 μM hypoxanthine does not appear to bind the RNA at 45°C, revealing that the increased disorder of the three-way junction is extremely deleterious for this low affinity ligand. Guanine, on the other hand, is able to bind the junction at elevated temperatures because it has ∼3 kcal mol−1 more binding energy available to overcome the enhanced flexibility of the binding pocket, allowing it to form the interactions observed in the crystal structure. For this ligand, the loss of conformational restriction of the junction by either mutations or temperature does not prohibit productive binding, indicating that preorganization is not an absolute requirement for the RNA's activity.
Another critical feature of riboswitch mediated genetic regulation is a strong linkage between ligand-dependent conformational changes and the secondary structural switch of the downstream expression platform. The observations made in this study reveal two aspects of the mechanism by which this occurs. First, an increased population of productive binding states within the three-way junction is conferred by the loop–loop interaction and has a dramatic effect on the rate at which ligand binds the RNA. Previous studies have noted that the bimolecular association rate constant between the nucleobase ligand and RNA is slow (∼105 M−1 s−1) (Wickiser et al. 2005a; Gilbert et al. 2006b). Without populating productive states, ligand binding would be far too slow for effective riboregulation to occur, especially in light of findings that suggest some riboswitches are kinetically controlled (Wickiser et al. 2005a,b). This is supported by the observation that increasing magnesium concentrations has a greater effect on the ligand association rate constant rather than the dissociation rate constant (Gilbert et al. 2006b). Second, this model clearly shows how ligand binding and the secondary structural switch are intimately coupled. In the limited folding event of the three-way junction, ligand binding and formation of base triples between J2/3 and P1 likely occur in a concerted fashion. This is evident by the fact that the melting transitions for all nucleotides in J1/2 and J2/3 are ∼40°C in the presence of ligand. Since the base triples are proposed to stabilize the 3′ side of the P1 helix to prevent its incorporation into the competing antiterminator element, this mechanism yields a clear picture as to how the RNA couples ligand binding to the secondary structural switch. Like allosterically regulated proteins, riboswitches directly couple ligand binding to conformational changes to confer biological function.
MATERIALS AND METHODS
RNA preparation
A DNA template for in vitro transcription of the B. subtilis xpt-pbuX aptamer domain was generated using PCR amplification from a series of overlapping oligonucleotides. All RNA constructs contain both the 5′ and 3′ structure cassettes, as previously described (Wilkinson et al. 2006), to allow for primer binding outside of the RNA sequence of interest. The DNA sequence of the wild-type aptamer domain RNA (GR) used for chemical probing in this study is as follows (boldface denotes the aptamer domain RNA sequence): GCGGAATTCTAATACGACTCACTATAGGCCTTCGGGCCAACGCGGACATATAATCGCGTGGATATGGCACGCAAGTTTCTACCGGGCACCGTAAATGTCCGACTATGTCCGCGTCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC. The loop mutants are essentially the same sequence.
DNA was amplified in a standard PCR reaction (Sambrook and Russel 2001) containing 1 μM outer oligonucleotides, 10 nM inner oligonucleotides, 2 mM dNTPs, 10× ThermoPol reaction buffer (NEB), and 1 unit of Taq polymerase. RNA was synthesized using an in vitro transcription reaction containing 1× transcription buffer (30 mM Tris-HCl at pH 8.0, 20 mM DTT, 0.01% [v/v] Triton X-100, 3.4 mM spermidine), 32 mM MgCl2, 4 mM each rNTP, 0.25 mg/mL T7 RNA polymerase, 50 nM DNA template (Milligan et al. 1987; Doudna 1997). RNA was precipitated at −20°C overnight in 70% (v/v) ethanol, then centrifuged at 3000g for 30 min. The pellet was resuspended in buffer containing 0.1 mM EDTA and 8 M urea and the RNA purified on a 12% polyacrylamide gel (29:1 acrylamide:bisacrylamide). RNA was visualized by UV shadowing, excised from the gel, and electroeluted into 0.5× TBE buffer, followed by concentration and buffer exchange into 0.5× TE buffer using an Amicon 10-kDa MWCO microconcentrator.
Temperature dependent RNA modification with NMIA
Two picomoles RNA as measured by UV absorbance were suspended in 12 μL 0.5× TE and denatured at 90°C for 2 min, then refolded on ice for 10 min. Six microliters of folding buffer (333 mM HEPES at pH 8.0, 20 mM MgCl2, 333 mM NaCl) and ligand were then added to the RNA and incubated at room temperature for 20 min. Reactions performed in the absence of magnesium were later supplemented with 3 mM magnesium to allow for efficient reverse transcription. During incubation, the RNA was separated into two 9-μL reactions in 100 μL PCR tubes for plus and minus N-methylisatoic anhydride (NMIA, Sigma-Aldrich) reactions. A 130-mM NMIA stock is prepared in anhydrous DMSO (Sigma-Aldrich) immediately prior to performing each modification reaction. Upon addition of 1 μL of NMIA stock, the reaction is allowed to proceed five half-lives according to the equation
where t 1/2 is the half-life of NMIA at temperature T in aqueous solution (Merino et al. 2005; Wilkinson et al. 2005, 2006). After modification is complete, 3 μL of 32P 5′-end labeled reverse transcription primer (∼0.3 μM, 5′-GAACCGGACCGAAGCCCG) is added and allowed to incubate at 65°C for 5 min. A second incubation at 35°C for 20 min is sufficient to promote primer annealing, followed by a 1-min incubation at 52°C to facilitate efficient primer extension. Addition of 6 μL enzyme mix (250 mM KCl, 167 mM Tris-HCl at pH 8.3, 1.67 mM each dNTP, 17 mM DTT, 0.33 units Superscript III) initiates the extension reaction and is stopped after 5 min by the addition of 1 μL 4 M NaOH. The reaction is incubated for 5 min at 90°C followed by an additional 5-min incubation in the presence of 29 μL acid stop mix (4:25 [v/v] mixture of 1 M unbuffered Tris-HCl and stop dye [85% formamide, 0.5× TBE, 50 mM EDTA at pH 8.0, 0.1% (w/v) bromophenol blue and xylene cyanol]). Sequencing reactions are performed exactly as previously described for an NMIA modification reaction; however, NMIA is excluded. Addition of 3 μL 10 mM ddATP, ddTTP, ddCTP, or ddGTP to 6 μL enzyme mix is sufficient to generate strong sequencing stops. All samples are separated on a 0.33-mm-thick 12% polyacrylamide gel (29:1 acrylamide:bisacrylamide, 1× TBE, 8 M urea). Two micorliters of sequencing reactions or modification reaction are loaded and electrophoresed at 55 W for 5–6 h.
Analysis of NMIA accessibility data
Upon gel drying, phosphorimaging was used to quantify individual band intensities, and Semi Automated Footprinting Analysis software (SAFA) (Das et al. 2005) was used to generate output densities. Following scaling to the reactivity of U63, data for each nucleotide were fit to a model describing a unimolecular melting transition
 |
where I is the band intensity, ΔHvH is the van't Hoff enthalpy, Tm is the transition midpoint, T is temperature, and A and b are the transition amplitude and initial intensity (Wilkinson et al. 2005). Data exhibiting a significant decrease in reactivity at elevated temperatures were weighted to allow for fitting in Kaleidagraph. van't Hoff enthalpy values (ΔHvH) were found to display a wide range of values and are not considered to be useful in our analysis due to sparse temperature sampling (data points were take every 5°C for practical considerations). To visualize melting behavior, B-factor values of the 1U8D PDB text file were replaced with scaled, inverse Tm values.
Isothermal titration calorimetry (ITC)
RNAs were prepared for ITC by dialysis for 16 h against 1 L of a buffer solution containing 50 mM K-HEPES (pH 7.5), 100 mM KCl, and the indicated MgCl2 concentration at 4°C. Hypoxanthine and 2,6-diaminopurine (HX and DAP; Sigma-Aldrich) were dissolved in the buffer that the RNA was dialyzed against to ensure an exact buffer match. Concentrations of the ligand and RNA solutions were determined using spectrophotometer readings at 260 nm for RNA, 278 nm for DAP (ε278 = 10,200 M−1 cm−1), and 249.5 nm for HX (ε249.5 = 10,700 M−1 cm−1) with the result that DAP and HX were ∼8× more concentrated than the RNA. Prior to titration, the RNA and purine samples were degassed for 10 min at 25°C. All ITC experimental parameters were set at 30°C, a reference power of 5 μcal sec−1, an initial delay of 60 sec, and a titration of either sixty 5 μL (for higher concentrations of MgCl2) or thirty 10 μL injections (for lower MgCl2 concentrations) at an injection rate of 0.5 μL sec−1 with individual injections spaced 240 sec apart and a stir speed of 300 rpm. Data collected from each experiment were fit to a single-site binding isotherm that yielded values for the change in enthalpy, ΔH, and the association constant, Ka (Pierce et al. 1999; MicroCal 2003).
ACKNOWLEDGMENTS
The authors thank the members of the Batey laboratory, Jeffrey Kieft, Art Pardi, and Marty Fedor for insightful discussions throughout this project. This work was supported by an American Cancer Society Research Scholar Grant to R.T.B. and by a Creative Training in Molecular Biology Grant to C.D.S. (T32 GM 07135).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.736908.
REFERENCES
- Barrick, J.E., Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Gilbert, S.D., Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., Pardi, A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Dann C.E., 3rd, Wakeman, C.A., Sieling, C.L., Baker, S.C., Irnov, I., Winkler, W.C. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Das, R., Laederach, A., Pearlman, S.M., Herschlag, D., Altman, R.B. SAFA: Semiautomated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. Preparation of homogeneous ribozyme RNA for crystallization. Methods Mol. Biol. 1997;74:365–370. doi: 10.1385/0-89603-389-9:365. [DOI] [PubMed] [Google Scholar]
- Edwards, T.E., Ferré-D'Amaré, A.R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Epshtein, V., Mironov, A.S., Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor, M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.D., Montange, R.K., Stoddard, C.D., Batey, R.T. Structural studies of the purine and SAM binding riboswitches. Cold Spring Harb. Symp. Quant. Biol. 2006a;71:259–268. doi: 10.1101/sqb.2006.71.015. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.D., Stoddard, C.D., Wise, S.J., Batey, R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006b;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.D., Love, C.E., Edwards, A.L., Batey, R.T. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., Bateman, A. Rfam: Annotating noncoding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Henkin, T.M. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- Hodak, J.H., Downey, C.D., Fiore, J.L., Pardi, A., Nesbitt, D.J. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc. Natl. Acad. Sci. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov, A., Kertsburg, A., Winkler, W.C. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: General principles and prospects for discovery. Cold Spring Harb. Symp. Quant. Biol. 2006;71:239–249. doi: 10.1101/sqb.2006.71.021. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Lemay, J.F., Lafontaine, D.A. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M.J., Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., Breaker, R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Lee, M., Barrick, J.E., Weinberg, Z., Emilsson, G.M., Ruzzo, W.L., Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J.S., Makunin, I.V. Noncoding RNA. Hum. Mol. Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Merino, E.J., Wilkinson, K.A., Coughlan, J.L., Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- MicroCal. MicroCal; Northhampton, MA: 2003. VP-ITC microcalorimeter user's manual. [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Montange, R.K., Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Murchie, A.I., Thomson, J.B., Walter, F., Lilley, D.M. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol. Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- Noeske, J., Richter, C., Grundl, M.A., Nasiri, H.R., Schwalbe, H., Wohnert, J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske, J., Buck, J., Furtig, B., Nasiri, H.R., Schwalbe, H., Wohnert, J. Interplay of “induced fit” and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler, E., Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ottink, O.M., Rampersad, S.M., Tessari, M., Zaman, G.J., Heus, H.A., Wijmenga, S.S. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;12:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, M.M., Raman, C.S., Nall, B.T. Isothermal titration calorimetry of protein–protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- Recht, M.I., Williamson, J.R. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Rieder, R., Lang, K., Graber, D., Micura, R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., Gelfand, M.S. Regulation of lysine biosynthesis and transport genes in bacteria: Yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russel, D.W. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- Serganov, A., Yuan, Y.R., Pikovskaya, O., Polonskaia, A., Malinina, L., Phan, A.T., Hobartner, C., Micura, R., Breaker, R.R., Patel, D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov, A., Polonskaia, A., Phan, A.T., Breaker, R.R., Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan, N., Wickiser, J.K., Nakamura, S., Ebert, M.S., Breaker, R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington, M.W., Siuzdak, G., Williamson, J.R. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore, S., Leibundgut, M., Ban, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Turnbull, W.B., Daranas, A.H. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Wickiser, J.K., Cheah, M.T., Breaker, R.R., Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005a;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wickiser, J.K., Winkler, W.C., Breaker, R.R., Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005b;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.A., Merino, E.J., Weeks, K.M. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.A., Merino, E.J., Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler, W., Nahvi, A., Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002a;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Cohen-Chalamish, S., Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 2002b;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Wiseman, T., Williston, S., Brandts, J.F., Lin, L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Zamel, R., Poon, A., Jaikaran, D., Andersen, A., Olive, J., De Abreu, D., Collins, R.A. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]