Abstract
The KARP-1 (Ku86 Autoantigen Related Protein-1) gene, which is expressed from the human Ku86 autoantigen locus, appears to play a role in mammalian DNA double-strand break repair as a regulator of the DNA-dependent protein kinase complex. Here we demonstrate that KARP-1 gene expression is significantly up-regulated following exposure of cells to DNA damage. KARP-1 mRNA induction was completely dependent on the ataxia telangiectasia and p53 gene products, consistent with the presence of a p53 binding site within the second intron of the KARP-1 locus. These observations link ataxia telangiectasia, p53, and KARP-1 in a common pathway.
Ataxia telangiectasia (AT) is an autosomal recessive disorder defined primarily by a 100-fold increase in the incidence of cancer (reviewed in ref. 1). Cells derived from AT patients (AT cells) are hypersensitive to DNA damaging agents that cause DNA strand breaks, such as ionizing radiation (1). A deficiency in radiation-induced cell-cycle checkpoint controls in AT cells indicated that the AT protein likely plays a role in arresting the cell cycle in response to x-irradiation, thus allowing time for DNA repair to be carried out before cell division (2). The gene (AT mutated; ATM) responsible for the AT phenotype (3) encodes a predominately nuclear 350 kDa polypeptide homologous to a large number of phosphatidylinositol 3-kinase family members (reviewed in ref. 4). Strikingly, all of these genes appear to play important roles in maintaining genome stability, cell cycle control, and cellular responses to DNA damage. Thus, the phenotypes of AT cells and the homology of ATM to other genes known to be involved in cellular responses to ionizing radiation suggest that ATM functions in a vital DNA damage signal transduction pathway.
The observation that AT cells showed aberrant (5) or delayed (6) induction of the tumor suppressor gene, p53, in response to x-irradiation, suggested that p53 was a key downstream target of ATM (reviewed in ref. 7). This hypothesis was recently confirmed with the demonstration that ATM-deficient fibroblasts failed to up-regulate p53 following ionizing radiation exposure (8). p53 is a transcription factor and a mutation of this gene is found in more than 50% of human cancers (9), implying that p53 loss represents a fundamental step in oncogenic progression. In addition, because most of the naturally occurring mutations affect the p53 DNA binding domain, it is likely that development of tumor formation in p53−/− cells is due to the lack of appropriate p53-dependent gene expression (10). Wild-type p53 is thought to contribute to the inhibition of cell growth and tumorigenesis by its ability to strongly up-regulate the cyclin kinase inhibitor, p21, which results in cell cycle arrest (11). The cessation of cell cycling is hypothesized to allow cells time to enact DNA repair, so that mutagenic lesions are not fixed into the genome by DNA synthesis. Intriguingly, however, a p53-responsive gene involved in DNA strand break repair has not yet been identified.
Many of the genes involved in radiation-induced strand break repair have been characterized. In particular, the DNA-dependent protein kinase (DNA-PK) complex, which is composed of the Ku heterodimer and a 465 kDa catalytic subunit (DNA-PKcs) has been shown to be involved in DNA double-strand break (DSB) repair (reviewed in ref. 12). DNA-PKcs is the product of the severe combined immune deficiency (scid) gene, and mice homozygously mutated at this locus are x-ray sensitive, impaired in DNA DSB repair, and defective for lymphoid V(D)J recombination (reviewed in ref. 13). Ku is a heterodimeric protein of 70 and 86 kDa subunits that binds tightly to double-stranded DNA ends. Rodent cell lines that are mutated in the Ku86 gene are x-ray sensitive, impaired in DNA DSB repair, and defective for V(D)J recombination (reviewed in ref. 14). In addition, inactivation of the Ku86 (15, 16) or Ku70 (17) genes in either mice or murine embryonic stem cells produced the predicted deficits in ionizing radiation sensitivity, DNA DSB repair, and V(D)J recombination. Thus, DNA-PK has been unequivocally identified as an important mammalian DNA repair complex and mutations in any of the three DNA-PK subunits result in severe ionizing radiation sensitivity and V(D)J recombination defects because of impaired DNA DSB repair. Surprisingly, however, none of the subunits of the DNA-PK complex appear to be induced following x-irradiation (18).
Recently, we demonstrated that the primate Ku86 locus was bifunctional and through the use of an upstream promoter encoded a Ku86 isoform, KARP-1 (Ku86 Autoantigen Related Protein-1; ref. 19). Here we show that KARP-1 expression is strongly induced following DNA damage and that the up-regulation of KARP-1 mRNA and protein is dependent on functional p53 and ATM gene products. These observations have important repercussions for mammalian DNA DSB repair.
MATERIALS AND METHODS
Cells.
The human HL-60 subclone, HCW-2 (20), and HeLa cells and their growth conditions have been described (19). HCT116 cells were purchased from the American Type Culture Collection and propagated as described (21). ATM−/− cell lines, GM08436A and GM01526E, and the ATM+/+ cell line, GM00130C, were purchased from the NIGMS Human Genetic Mutant Cell Repository at the Coriell Institute for Medical Research.
Electrophoretic Mobility Shift Assays.
Whole cell extracts (50 μg) were prepared (19), combined with a probe spanning the putative KARP-1 p53 response element in the presence or absence of the p53-specific antibody, pAb421 (22), and subjected to a gel mobility shift assay as described (23). The probe consisted of a 183 bp PCR fragment synthesized with [32P]-end-labeled PCR primers, 5′-AAGGGCTCGTGATCAAGTAA-3′ and 5′-CAGAGAAAATGGGATGCACA-3′. A double-stranded oligonucleotide in which nucleotide changes were introduced into the putative KARP-1 p53 binding site was made by annealing the oligonucleotide 5′-CCTAGAACCTTCTTCTAAATTAAACACCTACTCCTCCC-3′ and its complement. A double-stranded oligonucleotide encoding a dimer of the consensus p53 binding site in the p21 promoter was made by annealing the oligonucleotide 5′-TTGAACATGTCCCAACATGTTGGAACATGTCCCAACATGTTG-3′ and its complement. An identical double-stranded oligonucleotide in which nucleotide changes were introduced into the consensus p53 binding site was made by annealing the oligonucleotide 5′-GAAACCTTCCCAAACCTTTGGAAACCTTCCCAAACCTTTG-3′ and its complement.
Reverse Transcription (RT)–PCR Assays.
Cells were x-irradiated, total RNA isolated, and 20 μg was used for first strand synthesis, which was then used as a template for the PCR reaction as described (19). The KARP-1 PCR primers were 5′-ACGGCGGAATGGAGAGAATGTGCGCATGC-3′ and 5′-AACAGCTGCCTTATTCCCCGACCGCACCAT-3′. The β-actin PCR primers were 5′-ATCTGGCACCAGACCTTCTACAATGAGCTGCG-3′ and 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′. The Ku86 PCR primers were 5′-CTGTTGTGCTGTGTATGGACGTGGG-3′ and 5′-CCAGGAAGTCAGCCTGTTGAGAACC-3′. KARP-1 RT-PCR products were hybridized with a 328 bp KARP-1-specific probe generated by PCR (19). Ku86 RT-PCR products were hybridized with a 299 bp Ku86-specific probe generated by PCR with radiolabeled Ku86 PCR primers. β-actin RT-PCR products were either analyzed on ethidium bromide-stained gels or were hybridized with a β-actin gene fragment (CLONTECH).
Immunoblot Analyses.
Cytoplasmic extracts were prepared as described (23). Protein was quantitated using the Bradford assay (Bio-Rad) and 50 μg was used for each lane in 7.5% SDS/PAGE. Immunoblotting was performed with the enhanced chemiluminescence system (Amersham) by using the following antibodies: α-KARP-1 rabbit polyclonal antibody (19) and α-Ich-1L mouse mAb (Transduction Laboratories, Lexington, KY).
KARP-1 Induction with DNA Damaging Agents.
HCT116 cells were treated (24) with the indicated mutagen and then subjected to an RT-PCR/Southern blot analysis as described above for KARP-1 mRNA induction at 90 min posttreatment.
RESULTS
A Functional p53 Binding Site Resides Within the Second Intron of KARP-1.
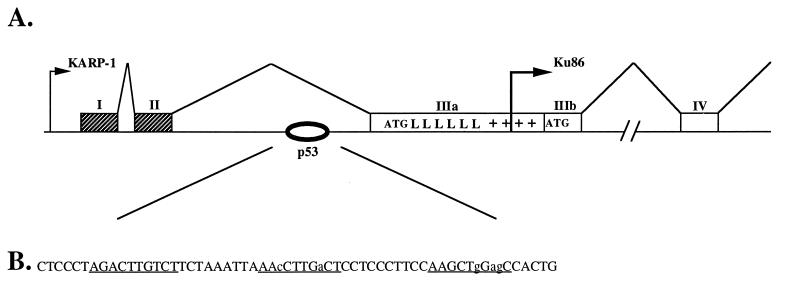
The primate Ku86 locus is bifunctional and through the use of an upstream promoter can encode an isoform termed KARP-1 (Fig. 1A). Sequence analysis of the KARP-1/Ku86 genomic locus revealed a possible p53 binding site located within the second intron of the KARP-1 gene (Fig. 1B). p53 binding sites consist of multimers of a 10 nt consensus sequence 5′-PuPuPuC(A/T)(A/T)GPyPyPy-3′ with the multimers separated by 0–13 nt (22, 25). The KARP-1 p53 binding site consisted of a consensus 10-mer separated by 9 and 10 nt, respectively, from two imperfect (8 and 7 matches, respectively) sequences (Fig. 1B). The location of the putative site, within the second intron of KARP-1 (Fig. 1B), was consistent with the locations of known functional p53 binding sites for the cyclin G (26), IGF-BP3 (27), and MDM2 (28) genes.
Figure 1.
KARP-1 is expressed from the bifunctional Ku86 locus by the use of an upstream promoter. (A) A cartoon of the Ku86 locus. Large arrows represent the starts of transcription for the KARP-1 and Ku86 genes, respectively. Hatched boxes represent noncoding exons and open boxes coding exons. The initiator methionine (ATG) for KARP-1 is shown at the beginning of exon IIIa and the ATG for Ku86 is shown at the beginning of exon IIIb. The position of the leucine zippers (L) and the basic region (+) in KARP-1 are also shown (19). The oval shows the location of the p53 binding site in the second intron of the KARP-1 transcription unit. This site is ≈1,100 nt 3′ of the KARP-1 promoter and ≈800 nt 5′ of the Ku86 promoter. (B) The putative p53 response element. The perfect (10 of 10) and the imperfect (8 of 10 and 7 of 10, respectively) p53 binding sites are underlined and the nonconsensus nucleotides are shown in lowercase letters.
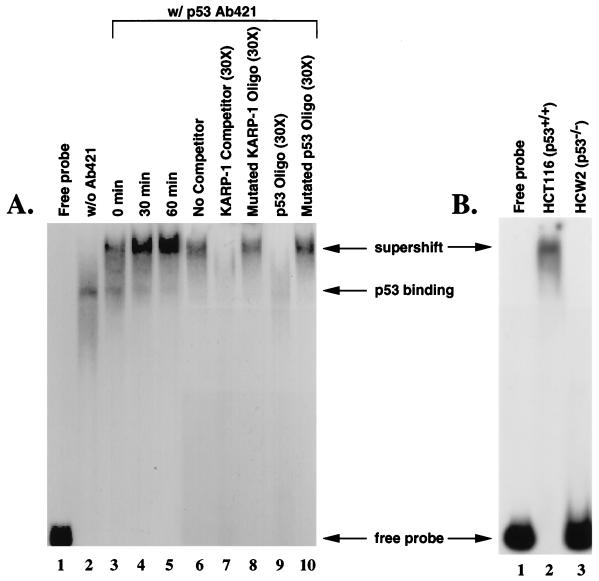
To test whether the KARP-1 site could bind p53, a 183 bp probe containing this site was used for a mobility shift assay. Nuclear extract prepared from unirradiated HCT116 cells (ref. 21; p53+/+) contained an activity that retarded the mobility of the fragment in a polyacrylamide gel (Fig. 2A, lane 2). This complex was supershifted upon inclusion of the p53-specific antibody, pAb421 (Fig. 2A, lane 3). The intensity of this supershifted complex increased when extracts were prepared from HCT116 cells 30 and 60 min post x-irradiation (15 Gy; Fig. 2A, lanes 4 and 5, respectively). This was consistent with the known induction of p53 in this cell line following x-irradiation (21). To confirm that this complex was due to p53 binding, competition experiments were carried out. A 30-fold excess of the 183 bp fragment completely competed the binding to the probe (Fig. 2A, lane 7), as did a double-stranded oligonucleotide (p53 Oligo), which contained a dimer of the consensus p53 binding site from the p21 promoter (ref. 11; Fig. 2A, lane 9). In contrast, oligonucleotides carrying mutations in either the KARP-1 p53 binding site (Mutated KARP-1 Oligo) or the p21 promoter consensus p53 binding site (Mutated p53 Oligo) were not effective competitors (Fig. 2A, lanes 8 and 10, respectively). Moreover, no complex was detected when whole cell extracts were prepared from the p53−/− cell line (20), HCW-2 (Fig. 2B, compare lanes 2 and 3), and from several other p53−/− cell lines (data not shown). Thus, a functional p53 binding site exists within the KARP-1 second intron.
Figure 2.
p53 can bind to the KARP-1 p53 response element. (A) Whole cell extracts were prepared from HCT116 (p53+/+) cells combined with a 183 bp probe containing the putative KARP-1 p53 response element in the absence (lanes 1 and 2) or presence (lanes 3–10) of the p53-specific antibody, pAb421, and subjected to a gel retardation assay: the probe incubated without cellular extract (lane 1); extracts from unirradiated cells (lanes 2 and 6); extracts from x-irradiated (15 Gy) cells prepared 0 min (lane 3), 30 min (lane 4), or 60 min (lane 5) postirradiation; addition of 30-fold excess cold 183 bp probe (lane 7); addition of 30-fold excess of a double-stranded oligonucleotide containing mutations in the KARP-1 p53 binding site (lane 8); addition of 30-fold excess of a double-stranded oligonucleotide containing a dimer of the p21 promoter consensus p53 binding site (lane 9); addition of 30-fold excess of a double-stranded oligonucleotide containing mutations in the p21 promoter consensus p53 binding site (lane 10). (B) Binding to the KARP-1 probe is not observed in p53-defective cell lines. Whole cell extracts were prepared from HCT116 (p53+/+; lane 2) and HCW-2 (p53−/−; lane 3) cells combined with a 183 bp probe containing the putative KARP-1 p53 response element in the presence of the p53-specific antibody, pAb421, and subjected to a gel retardation assay.
KARP-1 mRNA Is Induced Following DNA Damage.
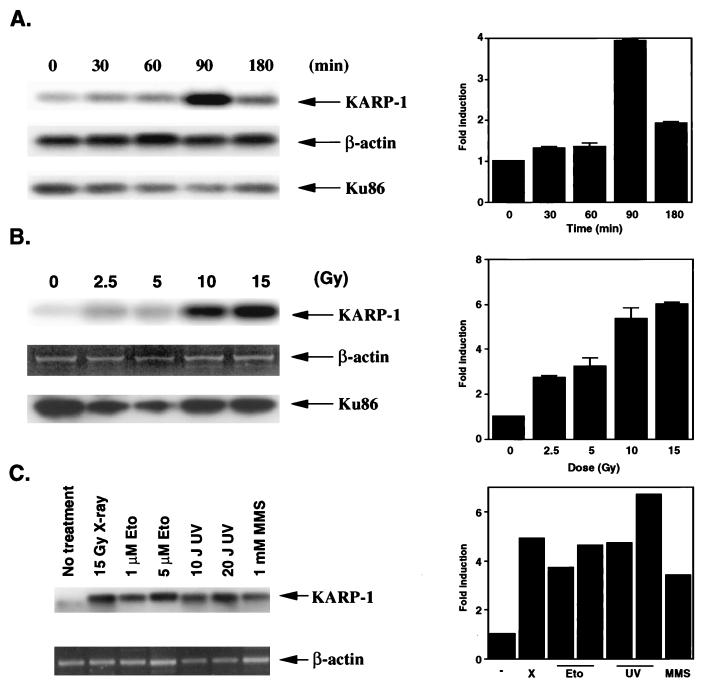
Because p53 is an important transcription factor for the cellular response to ionizing radiation (7), we investigated whether KARP-1 gene expression could be stimulated by x-irradiation. The immortalized human diploid fibroblast cell line, HCT116, was used for these experiments, because these cells have been shown to be completely normal in their response to ionizing radiation (21). KARP-1 message level was quantitated by an RT-PCR/Southern blot analysis at subsequent times after exposure of HCT116 cells to 10 Gy of x-irradiation. As controls, β-actin and Ku86 RT-PCR/Southern reactions were performed. Strong induction (4-fold) of KARP-1 mRNA was observed at 90 min after x-irradiation (Fig. 3A). Therefore, HCT116 cells were next x-irradiated using different doses, and at 90 min after x-irradiation the KARP-1 message level was determined (Fig. 3B). KARP-1 mRNA induction appeared to be dose dependent with a maximum of 6-fold stimulation at the highest dose (15 Gy) tested and a reproducible 2-fold induction even at the lowest dose (2.5 Gy) tested. In contrast, Ku86 message levels did not increase following x-irradiation at any dose or time tested (Fig. 2). Thus, KARP-1 expression was inducible by x-irradiation, whereas expression of Ku86 was not.
Figure 3.
(A) KARP-1 gene expression is induced following x-irradiation. (Left) HCT116 cells were x-irradiated (10 Gy) and then at the indicated times postirradiation RT-PCR reactions followed by Southern hybridizations were performed. (Right) The fold-induction of KARP-1 mRNA was quantitated by using β-actin levels as a reference. (B) KARP-1 induction is dose dependent. (Left) HCT116 cells were x-irradiated at the indicated doses and at 90 min postirradiation RT-PCR reactions followed by Southern hybridizations were performed as described. For the β-actin sample, only the ethidium bromide profile is shown. (Right) The fold-induction of KARP-1 mRNA was quantitated by using β-actin levels as a reference. The average of two independent experiments is shown for A and B. (C) KARP-1 gene expression is induced by other types of DNA damage. (Left) HCT116 cells were x-irradiated (X, 15 Gy) or treated with etoposide (Eto, 1 or 5 μM), UV irradiation (UV, 10 or 20 J/m2), or methyl methanesulfonate (MMS, 1 mM) and then at the indicated times posttreatment RT-PCR reactions followed by Southern hybridizations were performed. (Right) The fold-induction of KARP-1 mRNA was quantitated by using β-actin levels as a reference.
Because p53 is induced by many DNA damaging agents and not exclusively x-irradiation (7), we exposed HCT116 cells to etoposide (Eto), UV-irradiation (UV), and methyl methanesulfonate (MMS) and determined the KARP-1 message levels 90 min later (Fig. 3C). KARP-1 mRNA levels were significantly (4- to 7-fold) increased under all these conditions, consistent with p53 mediating the induction.
KARP-1 mRNA Induction Is p53- and ATM-Dependent.
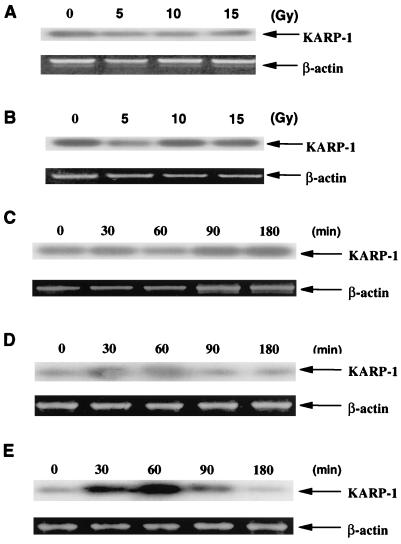
The status of KARP-1 mRNA induction in cell lines deficient or defective for p53 function was next examined. HCW-2 cells are a subclone of HL-60 cells, which are known to be deficient in p53 because of a deletion in the gene (20). HeLa cells contain a wild-type p53 gene, but its function, inclusive of almost all DNA damage responses, is seriously impaired because they have been infected with a human type-18 papillomavirus (29). Strikingly, HCW-2 (Fig. 4A) and HeLa (Fig. 4B) cells showed no induction of KARP-1 message after increasing doses of x-ray irradiation, consistent with the hypothesis that the induction of KARP-1 message requires a wild-type p53 gene product.
Figure 4.
KARP-1 induction is dependent on functional p53 and ATM gene products. (A) HCW-2 (p53−/−) and (B) HeLa (p53-defective) cell lines were x-irradiated at the indicated doses and at 90 min postirradiation RT-PCR reactions (β-actin) followed by Southern hybridizations (KARP-1) were performed as described. (C) GM08436A (ATM−/−), (D) GM01526E (ATM−/−), and (E) GM00130C (ATM+/+) cells were x-irradiated at 10 Gy and at the indicated times postirradiation RT-PCR reactions (β-actin) followed by Southern hybridizations (KARP-1) were performed as described.
Because ATM is thought to lie upstream of p53 in the DNA damage signal transduction pathway (1), we investigated the requirement for the ATM gene product in KARP-1 induction. GM08436A (Fig. 4C) and GM01526E (Fig. 4D) AT cells, which were derived from independent AT patients, exhibited no KARP-1 induction following x-irradiation with 10 Gy. Because these cell lines were of lymphoblastoid origin and the original demonstration of KARP-1 inducibility was carried out with HCT116 fibroblastic cells (Fig. 3), we also examined a lymphoblastoid cell line (GM00130C) derived from a wild-type individual. KARP-1 was strongly induced (6-fold) in GM00130C cells, although the time of induction was approximately 30 min faster than in HCT116 cells (Fig. 4E). Thus, only in a wild-type ATM background was timely KARP-1 mRNA induction following DNA damage detectable.
KARP-1 Protein Accumulates Following DNA Damage.
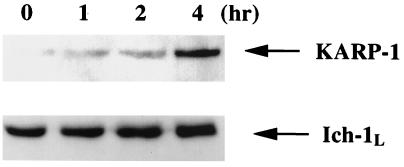
To investigate whether the KARP-1 protein is also induced by x-irradiation, cytoplasmic extracts were prepared from HCT116 cells at various times after x-ray exposure and analyzed by immunoblotting. Following x-irradiation, the amount of KARP-1 protein increased significantly over time (Fig. 5). To show that the induction was specific for KARP-1 protein, this blot was stripped and then reprobed with an antibody specific for Ich-1L (caspase-2), which showed no such induction (Fig. 5). This experiment was independently repeated two additional times by using either Fas-ligand or Ku70 as controls, and neither protein was induced by x-irradiation (data not shown). In contrast, KARP-1 protein levels were always increased (5.6 ± 0.7-fold). Thus, KARP-1 is a protein whose expression levels increase following DNA damage.
Figure 5.
KARP-1 protein is induced following x-irradiation. HCT116 cells were x-irradiated at 15 Gy and at the indicated times cytoplasmic extracts were prepared and then subjected to immunoblot analysis using a KARP-1 polyclonal antibody. Subsequently, the blot was stripped and probed with an antibody for Ich-1L.
DISCUSSION
We have observed the induction of KARP-1 mRNA and protein by DNA damage. To our knowledge, this is the first demonstration of a mammalian gene presumed to be involved in DNA DSB repair, which is inducible by DNA damage. Importantly, the induction of KARP-1 was mediated by the ATM and p53 genes, suggesting that it lies on a critical pathway for controlling the cellular response to x-irradiation.
KARP-1 Expression Is p53 Responsive, Whereas Ku86 Expression Is Not.
Under normal cellular conditions the Ku86 and KARP-1 genes are constitutively expressed, albeit the level of Ku86 expression is one to two orders of magnitude higher than KARP-1 expression (19). In the presence of elevated amounts of p53 protein, however, Ku86 transcription is relatively unaltered, whereas that of KARP-1 is sharply increased (Fig. 3). Thus, the p53 response element, which is approximately equidistant between the promoters of these two genes (Fig. 1), appears to only positively influence KARP-1 transcription. A thorough analysis of the KARP-1 and Ku86 promoters, and the transcription factors that bind to them, may shed some light on how p53 selectively augments KARP-1 transcription.
The Timing of KARP-1 Induction Implies That It Performs a Function Late in DNA DSB Repair.
Studies of asynchronously growing cells exposed to x-irradiation suggested that DSB repair was biphasic with the bulk of the repair being completed within 2 hr (“fast”) and the remainder being completed in the subsequent 24–48 hr (“slow”) (30, 31). Thus, the timing of KARP-1 protein induction, which is first observed 3–4 hr after x-irradiation (Fig. 5), suggests that KARP-1 performs a function during the slow phase of DNA DSB repair. Intriguingly, a requirement for DNA-PK during the slow phase of repair has also been reported (32). That function may involve the repair of specific or recalcitrant lesions. Alternatively, large complexes are required for the repair of DNA lesions (12). For example, a number of strand break repair proteins, including hMRE11/hRAD50 (33) and hRAD51/BRCA1 (34), are localized into independent discrete nuclear foci, which persist many hours after x-irradiation. If the formation of such complexes is inhibitory to subsequent metabolic processes (e.g., DNA replication) KARP-1/DNA-PK could be involved in their disassembly.
KARP-1 Regulation of DNA-PK Activity.
Our previous demonstration that KARP-1 regulated DNA-PK activity (19) coupled with the current observation that KARP-1 is induced following x-ray exposure (Fig. 3) suggests that DNA-PK activity should increase subsequent to x-irradiation. This hypothesis is inconsistent with a report which demonstrated that, in whole cell extracts derived from either AT or wild-type cells, DNA-PK phosphorylation activity was indistinguishable and not inducible following x-irradiation (18). It is conceivable, however, that these in vitro assays do not accurately reflect DNA-PK activity in vivo, and/or that the phosphorylation activity of DNA-PK is not required for DNA DSB repair. Alternatively, it has been shown that DNA-PKcs can be active in a Ku-independent manner (32, 35). Thus, in vivo, the entity described in vitro as “DNA-PK” may be a complex mixture of various Ku, KARP-1, and DNA-PKcs subunits that interact in tissue-specific or cell cycle-specific manners. KARP-1 may thus only regulate DNA-PK in the repair of specific lesions or in specific phases of the cell cycle (36).
KARP-1 Regulation of DNA DSB Repair.
The data presented in our earlier work (19) combined with this study suggest that the following pathway may exist in primate cells: x-irradiation > ATM > p53 > KARP-1 > DNA-PK > DNA DSB repair. It should be emphasized, however, that this pathway is specific for DNA-PK-dependent DSB repair. There are additional complexes (including the hRAD51, hRAD52, and BRCA2 proteins) capable of carrying out DNA DSB repair (12). Indeed, p53 may link these pathways together in a consistent fashion. Thus, p53 is required for induced expression of KARP-1 and, presumably, for DNA-PK activity. In addition, p53 has been shown to directly interact with and inhibit hRAD51 (37). Hence, a working hypothesis may be that the exposure of cells to x-irradiation induces p53, which simultaneously causes a G1 cell cycle arrest, activates nonhomologous DSB repair via DNA-PK, and inhibits homologous repair. After nonhomologous repair has taken place, p53 levels recede, inactivating DNA-PK and activating RAD51-dependent pathways in either S phase (BRCA2 complex) or G2 (RAD52 complex). In cell lines defective for p53, the DNA-PK pathway would be impaired, but this would be compensated for by the enhancement of the RAD51-dependent pathways. This model is consistent with the observation that ATM−/− and p53−/− cell lines are proficient for DNA DSB repair (1, 7).
Acknowledgments
We would like to thank Drs. A.-K. Bielinsky, A. Landy, J. Sedivy, and Z. Han for their comments and helpful discussions. E.A.H. is a Leukemia Society of America Scholar. This work was supported in part by National Institutes of Health Grant AI35763 to E.A.H.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AT, ataxia telangiectasia; ATM, AT mutated; DNA-PK, DNA-dependent protein kinase; DSB, double-strand break; RT, reverse transcription.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF039597).
References
- 1.Lavin M F, Shiloh Y. Curr Opin Immunol. 1996;8:459–464. doi: 10.1016/s0952-7915(96)80030-6. [DOI] [PubMed] [Google Scholar]
- 2.Meyn M S. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- 3.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle P A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 4.Jackson S P. Curr Biol. 1995;5:1210–1212. doi: 10.1016/s0960-9822(95)00238-7. [DOI] [PubMed] [Google Scholar]
- 5.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Forance A J J. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 6.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 7.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Baltimore D. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 9.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 10.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson E A. Am J Hum Genet. 1997;61:795–800. doi: 10.1086/514895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weaver D T, Alt F W. Nature (London) 1997;388:428–429. doi: 10.1038/41225. [DOI] [PubMed] [Google Scholar]
- 14.Jeggo P A. Mutat Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 15.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 17.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jongmans W, Artuso M, Vuillaume M, Bresil H, Jackson S P, Hall J. Oncogene. 1996;13:1133–1138. [PubMed] [Google Scholar]
- 19.Myung K, He D M, Lee S E, Hendrickson E A. EMBO J. 1997;16:3172–3184. doi: 10.1093/emboj/16.11.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Z, Chatterjee D, Early J, Pantazis P, Hendrickson E A, Wyche J H. Cancer Res. 1996;56:1621–1628. [PubMed] [Google Scholar]
- 21.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 22.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Yaseen N R, Hogan P G, Rao A, Sharma S. J Biol Chem. 1995;270:20653–20659. doi: 10.1074/jbc.270.35.20653. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickson E A, Qin X-Q, Bump E A, Schatz D G, Oettinger M, Weaver D T. Proc Natl Acad Sci USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Schwedes J F, Parks D, Mann K, Tegtmeyer P. Mol Cell Biol. 1995;15:2157–2165. doi: 10.1128/mcb.15.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zauberman A, Lupo A, Oren M. Oncogene. 1995;10:2361–2366. [PubMed] [Google Scholar]
- 27.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Nature (London) 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 28.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo P A. Mutat Res. 1990;239:1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- 31.Nevaldine B, Longo J A, King G A, Vilenchik M, Sagerman R H, Hahn P J. Radiat Res. 1993;133:370–374. [PubMed] [Google Scholar]
- 32.Evans J W, Liu X F, Kirchgessner C U, Brown J M. Radiat Res. 1996;145:39–46. [PubMed] [Google Scholar]
- 33.Maser R S, Monsen K J, Nelms B E, Petrini J H J. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 35.Yaneva M, Kowalewski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S E, Mitchell R A, Cheng A, Hendrickson E A. Mol Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturzbecher H-W, Donzelmann B, Henning W, Knippschild U, Buchhop S. EMBO J. 1996;8:1992–2002. [PMC free article] [PubMed] [Google Scholar]