Abstract
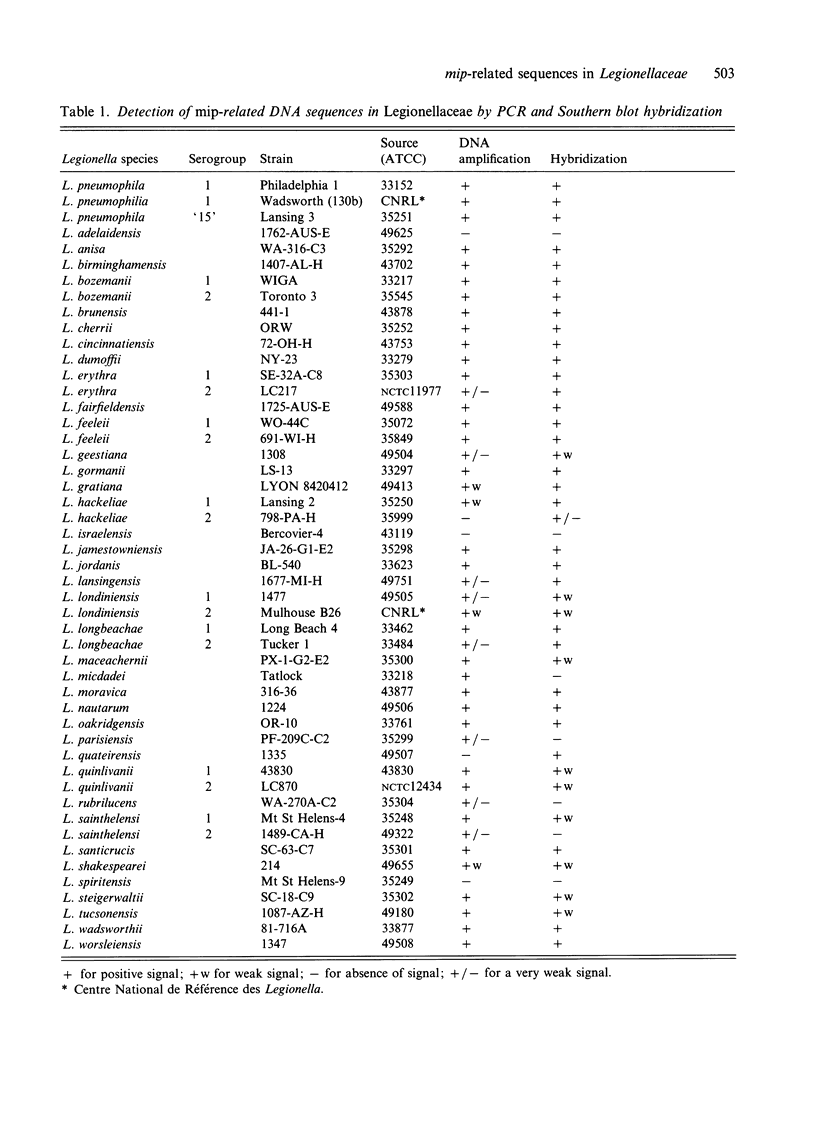
The macrophage infectivity potentiator gene (mip) from Legionella pneumophila is a major virulence factor of the species. Thus, mip-detection by amplification has been proposed to assess the presence of L. pneumophila in clinical and environmental samples. The distribution of mip-related sequences within the Legionellaceae was studied by DNA amplification using mip-specific primers followed by Southern blot hybridization with an internal probe. Thirty-nine species (48 serogroups) of Legionellaceae were screened in this attempt. Using this approach, sequences related to mip were observed in 89% of the tested species including the most recently described L. fairfieldensis, L. lansingensis and L. shakespearei. In several cases, cloning and sequencing of the amplified products confirmed the high levels of similarity between the sequence found in non-pneumophila species with that of the L. pneumophila mip gene. This confirms previous reports that mip related genes are widespread among Legionellaceae and therefore specific detection of the species L. pneumophila cannot be based on mip-targeted amplification.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bangsborg J. M., Cianciotto N. P., Hindersson P. Nucleotide sequence analysis of the Legionella micdadei mip gene, encoding a 30-kilodalton analog of the Legionella pneumophila Mip protein. Infect Immun. 1991 Oct;59(10):3836–3840. doi: 10.1128/iai.59.10.3836-3840.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto N. P., Bangsborg J. M., Eisenstein B. I., Engleberg N. C. Identification of mip-like genes in the genus Legionella. Infect Immun. 1990 Sep;58(9):2912–2918. doi: 10.1128/iai.58.9.2912-2918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto N. P., Eisenstein B. I., Mody C. H., Engleberg N. C. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990 Jul;162(1):121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- Cianciotto N. P., Eisenstein B. I., Mody C. H., Toews G. B., Engleberg N. C. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989 Apr;57(4):1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto N. P., O'Connell W., Dasch G. A., Mallavia L. P. Detection of mip-like sequences and Mip-related proteins within the family Rickettsiaceae. Curr Microbiol. 1995 Mar;30(3):149–153. doi: 10.1007/BF00296200. [DOI] [PubMed] [Google Scholar]
- Cianciotto N. P., Stamos J. K., Kamp D. W. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr Microbiol. 1995 Apr;30(4):247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- Edelstein P. H. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981 Sep;14(3):298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg N. C., Carter C., Weber D. R., Cianciotto N. P., Eisenstein B. I. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun. 1989 Apr;57(4):1263–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Barbaree J. M., Sanden G. N., Morrill W. E. Virulence of a Legionella anisa strain associated with Pontiac fever: an evaluation using protozoan, cell culture, and guinea pig models. Infect Immun. 1990 Sep;58(9):3139–3142. doi: 10.1128/iai.58.9.3139-3142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Barbaree J. M., Shotts E. B., Jr, Feeley J. C., Morrill W. E., Sanden G. N., Dykstra M. J. Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect Immun. 1986 Sep;53(3):553–559. doi: 10.1128/iai.53.3.553-559.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulhac B., Nowicki M., Bornstein N., Meunier O., Prevost G., Piemont Y., Fleurette J., Monteil H. Detection of Legionella spp. in bronchoalveolar lavage fluids by DNA amplification. J Clin Microbiol. 1992 Apr;30(4):920–924. doi: 10.1128/jcm.30.4.920-924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig B., Rahfeld J., Schmidt B., Mann K., Wintermeyer E., Fischer G., Hacker J. Characterization of Mip proteins of Legionella pneumophila. FEMS Microbiol Lett. 1994 May 1;118(1-2):23–30. doi: 10.1111/j.1574-6968.1994.tb06798.x. [DOI] [PubMed] [Google Scholar]
- Rowbotham T. J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980 Dec;33(12):1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Surgot M., Bornstein N., Paucod J. C., Marmet D., Isoard P., Fleurette J. Relationship between free amoeba and Legionella: studies in vitro and in vivo. Zentralbl Bakteriol. 1990 Mar;272(3):265–275. doi: 10.1016/s0934-8840(11)80027-7. [DOI] [PubMed] [Google Scholar]



