Abstract
There is an immediate need for identification of new antifungal targets in opportunistic pathogenic fungi like Candida albicans. In the past, efforts have focused on synthesis of chitin and glucan, which confer mechanical strength and rigidity upon the cell wall. This paper describes the molecular analysis of CaMNT1, a gene involved in synthesis of mannoproteins, the third major class of macromolecule found in the cell wall. CaMNT1 encodes an α-1,2-mannosyl transferase, which adds the second mannose residue in a tri-mannose oligosaccharide structure which represents O-linked mannan in C. albicans. The deduced amino acid sequence suggests that CaMnt1p is a type II membrane protein residing in a medial Golgi compartment. The absence of CaMnt1p reduced the ability of C. albicans cells to adhere to each other, to human buccal epithelial cells, and to rat vaginal epithelial cells. Both heterozygous and homozygous Camnt1 null mutants of C. albicans showed strong attenuation of virulence in guinea pig and mouse models of systemic candidosis, which, in guinea pigs, could be attributed to a decreased ability to reach and/or adhere internal organs. Therefore, correct CaMnt1p-mediated O-linked mannosylation of proteins is critical for adhesion and virulence of C. albicans.
Candida albicans is an opportunistic fungal pathogen of humans that has been increasingly found to cause systemic infections in immuno-compromised patients (1). The efficacy of the limited number of antifungals that are available to treat these patients is further reduced owing to the increased incidence of drug resistance (2). Hence, there is an urgent need to identify new antifungal targets and compounds that inactivate them. Fungi are protected from various environmental stresses including potentially fungicidal compounds by their cell wall. Chitin, glucan, and mannan are the three main macromolecules in the cell wall of Saccharomyces cerevisiae and C. albicans (3). Of these, chitin and glucan provide the cell wall with its strength and rigidity and allow it to maintain shape despite disruptive forces like turgor and mechanical shearing (4). In addition these compounds seem to be absent from mammalian cells, and hence there has been great interest in the exploitation of their synthetic machinery as an antifungal target (5, 6).
The potential of mannan synthesis as an antifungal target has received less attention. Mannan consists of a large number of various hyper-mannosylated proteins (7) that are deposited mainly at the outside of the cell wall thereby covering the surface with a layer, protecting internal regions of the cell wall from large molecules like proteases (8). Many critical aspects of the interaction between the fungus and the host, such as adhesion, immuno-surveillance, and immuno-modulation, are mediated by host recognition and interaction with this mannan-rich surface of the cell wall (7, 9). Therefore, disturbance of the mannosylation process is likely to have a broad range of damaging effects on the fungus and its interaction with the host, as many mannoproteins involved in pathogenesis, including excreted virulence determinants, are likely to be affected.
Mannosylation of proteins can be subdivided into N (asparagine)-linked and O (serine/threonine)-linked glycosylation. N-linked glycosylation involves the addition of large oligosaccharides to the protein, and its importance is reflected by the conservation of the core structure in both fungi and mammalian cells (10). However, this functional conservation makes N-glycosylation a less specific and thereby less attractive antifungal target. O-linked glycosylation is fundamentally different in fungi and mammalian cells. Short chains of mannose are added in fungi (11), whereas mannosylated serine and/or threonine residues are rare in mammalian glycoproteins (12). In S. cerevisiae, O-linked oligosaccharides are up to five mannose residues long (13), and a number of genes have been shown to be involved in their synthesis. Attachment of the first mannose to the protein in the endoplasmic reticulum (ER) is mediated by Pmt proteins and has been shown to be an essential process in yeast (14). Addition of the second and third mannose residues can be attributed to Ktr3p and Mnt1p (15, 16), which are likely to be α-1,2-mannosyl transferases residing in a medial Golgi compartment (16, 17). Transfer of the terminal α-1,3-mannose residues is mediated by Mnn1p in a medial Golgi compartment (18, 19).
Absence of Mnt1p in S. cerevisiae resulted in the synthesis of truncated O-linked oligosaccharides. This interfered with the functioning and/or synthesis of cell wall compounds as indicated by the killer toxin K1-resistant phenotype of these strains (13, 20). MNT1 is a member of a gene family of nine putative mannosyl transferases (21) of which YUR1 KTR1 KTR3 and KTR3 have been analyzed further. Ktr1p, Ktr3p, and Kre2p/Mnt1p have overlapping roles in the collective addition of the second and third α-1,2-linked mannose residues of O-linked mannan (16), whereas YUR1 and KTR2 are not thought to be involved in O-glycosylation (22, 23). In addition, MNN6 is a member of the MNT1 family and is involved in mannophosphate transfer to N-linked oligosaccharides containing one or more α-1,2-linked mannobiose units (24). We set out to identify a similar gene family in C. albicans and assess its potential as an antifungal target. This report describes the characterization of CaMNT1, a member of this gene family in C. albicans, and the biochemical analysis of its function in vivo. We show that cells lacking CaMnt1p have a reduced ability to adhere to human buccal epithelial cells and to colonize epithelial cells in rat vaginal models of candidosis. In systemic animal models of candidosis, absence of CaMnt1p leads to decreased virulence both in mice and guinea pigs. Therefore, although CaMnt1p is not essential for viability, Mnt1p-mediated O-glycosylation of proteins of C. albicans is essential for normal host–fungus interactions.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions.
The following strains of C. albicans, except SGY243, were derived from strain SC5314: SGY243 (Δura3::ADE2/Δura3::ADE2) (25), SC5314 (clinical systemic isolate) (26), CAF2 (URA3/Δura3::λimm434) (27), CAI4 (Δura3::λimm434/Δura3::λimm434) (27), NGY21 (Δura3::λimm434/Δura3::λimm434 MNT1/Δmnt1::hisG-URA3-hisG), NGY22 (Δura3::λimm434/Δura3::λimm434 MNT1/Δmnt1::hisG), NGY22A as NGY22 but with the other MNT1 allele disrupted, NGY23 (Δura3::λimm434/Δura3::λimm434 Δmnt1::hisG-URA3-hisG/Δmnt1::hisG), and NGY24 (Δura3::λimm434/Δura3::λimm434 Δmnt1::hisG/Δmnt1hisG). Media (28) and conditions to induce germ tube growth (29–31) were standard as was the adherence assay (32). Escherichia coli XL1-Blue (Stratagene) was used in cloning experiments.
Cloning and Sequencing of CaMNT1.
Chromosomal DNA of C. albicans SGY243 was digested with EcoRI, and size-fractionated fragments of 2 kb were cloned in pUC18 (Stratagene). Colony hybridizations using a 1.9-kb EcoRI fragment encoding the ScMNT1 gene (15) as a probe resulted in isolation of pNG41, which was sequenced using the USB version 2 Sequenase kit (Amersham). To obtain the remaining part of CaMNT1, inverse PCR was used (33). Templates were prepared by digesting genomic DNA of C. albicans CAI4 with either EcoRI at position 452 (to obtain the distal part of the gene) or PstI at position 1074 (to obtain the proximal part), and circularized at 3 μg/μl. The use of primers p1 (5′-CTCCAATAGTAATCATAATCG-3′, position 701–681) and p2 (5′-ATAATGGTTGTCATTTCT-3′, position 924–943) resulted in a single 2-kb fragment encoding the distal, conserved part of CaMNT1, the sequence of which was identical to that of the clone. Owing to restriction fragment length polymorphism (data not shown), amplification of the possibly more variable, proximal region resulted in two products, a major product of 2.0 kb and a minor product of 2.4 kb. The resulting nucleotide sequence can therefore confidently be ascribed to one particular copy of CaMNT1. PCR products were sequenced using the SequiTherm Cycle Sequencing kit (Epicentre Technologies, Cambio, Cambridge).
Disruption of CaMNT1.
A 0.5-kb EcoRI–HindIII CaMNT1 fragment representing the C terminus of the predicted protein was cloned in pSK (Stratagene) to yield plasmid pBsub3. Plasmid pMB7 (27) was digested with BglII, and the overhangs were blunt-ended using Klenow. Subsequently, the hisG-URA3-hisG cassette was excised by digestion with HindIII and inserted in HindIII plus HincII-digested pBsub3 yielding plasmid pCW. A 1.4-kb fragment containing the distal region of CaMNT1 was amplified using PCR and plasmid pNG41 as a template with the pUC forward primer and CaMNT1 specific primer p3 (5′-GTGATGCTCGAGTTCATTCCATTGC-3′, position 1045–1072). The product was digested with XhoI and KpnI and cloned into pCW to yield plasmid pJW. Both alleles of CaMNT1 in C. albicans CAI4 were disrupted by twice transforming with PvuII-digested pJW (35) and subsequently regenerating the ura− derivative using fluoroorotic acid (FOA) (27). The resulting strains contained alleles with a 69-base deletion in the catalytic domain and an insertion at this site of either the hisG-Ura3-hisG cassette or, after FOA-selection, a resolved single copy of hisG.
Southern and Northern Blot Analyses.
Chromosomal DNA of C. albicans was isolated as described by Philippsen et al. (36). Conditions for Southern and Northern blot analyses have been described previously (37). For heterologous hybridization of chromosomal DNA of C. albicans with the ScMNT1 gene, the formamide concentration was lowered to 30%, and washes were performed with 2 × SSC, 1% SDS at 53°C.
Analyses of in Vitro and in Vivo Mannosylation.
Mannan was isolated as described by Ballou (18). Reductive β-elimination using [3H]NaBH4, purification, and Biogel-P4 chromatography were performed as described by Ferguson (38). Labeling of mannan using [3H]mannose, β-elimination, digestions using mannosidases, purification steps, high-performance thin-layer chromatography (HPTLC), and mannosyl transferases assays have been described previously (13).
Vaginal and Systemic Models of Candidosis.
In rat models of vaginal candidosis, groups of six young adult female Wistar rats (200 g) were pretreated by surgical removal of their ovaries and uterus and by subcutaneous injection of oestradiol undecylate to induce a state of pseudoestrus. Pseudoestrus was sustained by further subcutaneous injections after the animals were infected intravaginally with a suspension of C. albicans. At various time points, the vaginas of two randomly chosen animals were sampled with a swab that was thoroughly stirred in saline, after which serial dilutions of the suspensions were plated on Sabouraud glucose agar and incubated for 48 h at 37°C. Guinea pig and mouse models of candidosis have been described previously (39). The identity of rescued C. albicans cells was checked by fingerprinting of whole fungal cell DNA with the moderately repeated sequence Ca3 (40).
RESULTS
Identification of CaMNT1 of C. albicans.
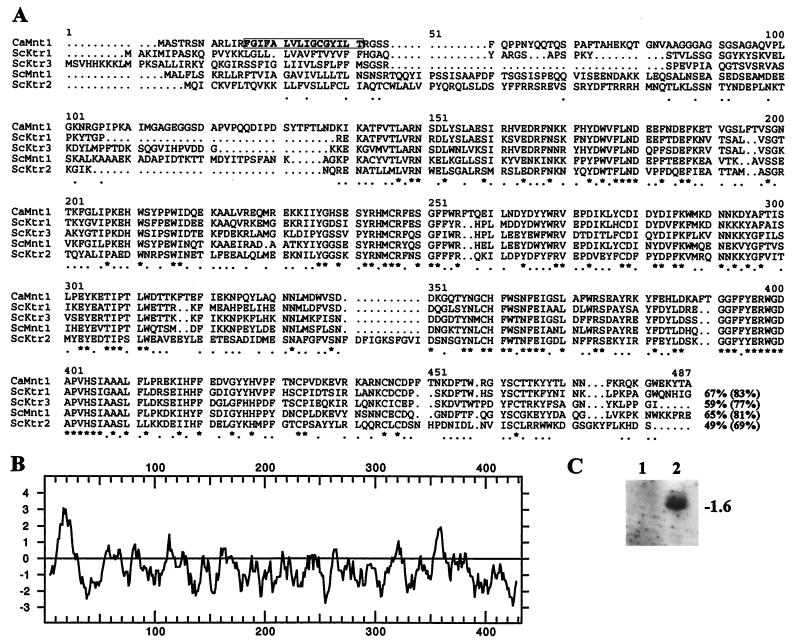
Southern blots of various restriction digests of C. albicans DNA were probed with radiolabeled DNA fragments containing the MNT1 gene of S. cerevisiae, and a 2-kb EcoRI fragment was found to hybridize (data not shown). A partial EcoRI library of C. albicans DNA was constructed from which a single hybridizing clone was isolated. The nucleotide sequence of the insert was found to contain 1023 bp of the 3′-end of an ORF that was highly homologous to the MNT1 sequence of S. cerevisiae. Inverse PCR was used to obtain the 5′-end of the ORF revealing the complete sequence of the CaMNT1 gene (Fig. 1A) of 1296 bp long, which corresponds to the transcript of 1.6 kb found in Northern analysis (Fig. 1C). The deduced amino acid sequence was 65% identical and 81% similar to that of Mnt1p of S. cerevisiae, with the highest homology located in the distal 300 amino acid residues where the catalytic domain resides (15). Hydrophobicity analysis suggested a 12-amino acid cytosolic loop followed by a single short membrane-spanning region of 16 amino acids flanked by two positively charged residues (Fig. 1B). This indicates that, like ScMnt1p, CaMnt1p is a type II membrane protein residing in a medial Golgi compartment (17). Overexpression of CaMnt1p in Pichia pastoris resulted in high mannosyl transferase activities, indicating that CaMNT1 encodes a mannosyl transferase rather than a regulator of expression of mannosyl transferases (L. Thomson, Y. Aoki, S. Yamazaki, E.T.B., and N.A.R.G., unpublished results). CaMNT1 was located on chromosome 3 by hybridization of Southern blots of electrophoretically separated chromosomes of C. albicans with radiolabeled DNA fragments containing part of the gene (B. B. Magee, unpublished observations; W. A. Fonzi, unpublished observation).
Figure 1.
(A) Analysis of the deduced amino acid sequence of CaMNT1 of C. albicans CAI4 and comparison with S. cerevisiae sequences ScMnt1p and ScKtr1–3p using Seqnet GCG program. Positions of complete identity are indicated with asterisks and by full stops where three or more residues are conserved between any of the five sequences. Values for percentage similarities (parentheses) and identities between CaMnt1p and the other genes are given. (B) Hydrophobicity analysis (34) with a window of nine amino acids indicated a single transmembrane spanning region (box in A). (C) Northern analysis of total RNA obtained from C. albicans strains CAI4 and NGY24, respectively, hybridized with radiolabeled CaMNT1 probe. rRNA bands visible on ethidium bromide-stained gels (not shown) were used both as a loading control and molecular weight markers.
Both Northern analysis and in vitro mannosyl transferase activities in microsomal fractions of C. albicans cells grown under various conditions revealed that CaMNT1 was expressed constitutively with somewhat higher levels of transcripts in early mid-exponential growth phase cells and 30 min after induction of hyphae.
Sequential Disruption of the CaMNT1 Gene.
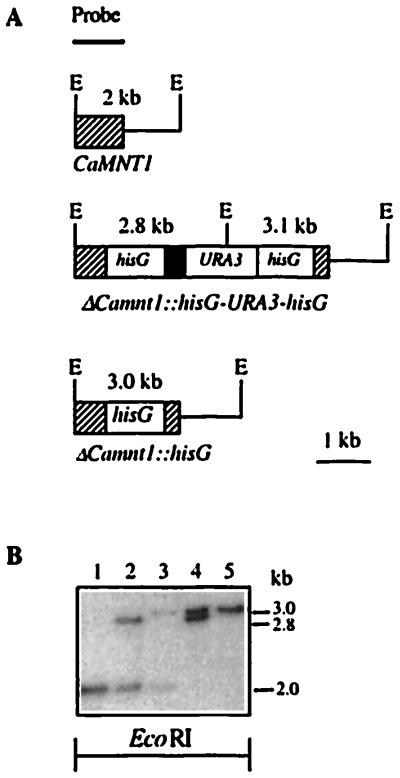
The “ura-blaster” technique (25) was used to disrupt both copies of the CaMNT1 gene into the uridine-requiring C. albicans strain CAI4 with disruption plasmid pJW. Chromosomal DNA of transformants was digested with EcoRI and analyzed by Southern hybridization using part of CaMNT1 as a probe. In addition to the 2.0-kb band also observed in the wild type, transformants displayed a 2.8-kb band, confirming disruption of one CaMNT1 allele (Fig. 2). Following selection on FOA medium, the loss of one copy of hisG and the URA3 gene increased the length of the 2.8-kb band to 3.0 kb (Fig. 2). The second allele of CaMNT1 was disrupted similarly and led to removal of all CaMNT1 alleles, as confirmed by the absence of CaMNT1 transcript in Northern analysis (Fig. 1C), indicating that CAI4 is diploid for this locus. Parental strain CAI4 and homozygous Camnt1 disruptant NGY24 displayed similar growth rates in YPD at 30°C (0.25 h−1 and 0.23 h−1, respectively), similar amounts of biomass in stationary phase, and were fully competent to form germ tubes in 20% serum, Spider medium and in 4 mM N-acetylglucosamine-containing medium.
Figure 2.
Sequential disruption of CaMNT1 using the ura-blaster technique. (A) Schematic representation of the construction of the CaMNT1, ΔCamnt1::hisG-URA3-hisG and the ΔCamnt1::hisG alleles. The probe used for Southern analysis was a 500-bp EcoRI–HindIII fragment of CaMNT1. E, EcoRI. (B) Southern analysis of EcoRI-digested chromosomal DNA obtained from Camnt1 mutant strains at various stages of construction of a homozygous Camnt1 disruptant strain. Lanes: 1, parental strain C. albicans CAI4; 2, primary transformant NGY21; 3, post-FOA progeny of NGY21, NGY22; 4, secondary transformant NGY23; 5, post-FOA progeny of NGY23, NGY24.
CaMNT1 Encodes an α-1,2-Mannosyl Transferase.
To determine whether CaMNT1 encodes a mannosyl transferase, specific mannose transfer rates from GDP-mannose to methyl α-d-mannoside were determined in mixed membrane fractions isolated from wild-type strain CAI4 and both the heterozygous and homozygous disruptant strains, NGY22 and NGY24, respectively. NGY24 contained only 25% of the activity observed in CAI4. Activities in NGY22 were intermediate, indicating a gene dosage effect (Table 1).
Table 1.
Specific in vitro mannosyl transferase activities in pmol mannose transferred/mg protein measured in mixed membrane fractions isolated from parental C. albicans strains and the derived heterozygous (NGY22 and NGY22A) and homozygous (NGY24) Camnt1 disruptant strains grown in YPD
| Strain | Activity (x ± SD, n = 8) |
|---|---|
| SC5314 | 3856 ± 356 |
| CAI4 | 3715 ± 402 |
| NGY22 | 2516 ± 368 |
| NGY22A | 2602 ± 245* |
| NGY24 | 950 ± 446 |
*Average based on seven replicates.
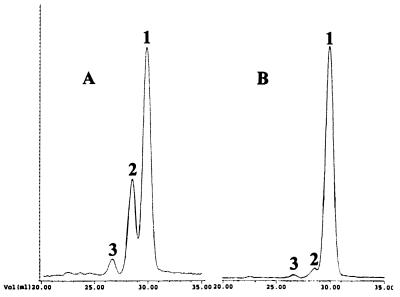
It was conceivable that this large reduction of in vitro mannosyl transferase activity was reflected in altered mannan structures. Therefore, total cell wall mannan was isolated from both C. albicans strains, CAI4 and NGY24. The carbohydrate/protein ratios of both mannan preparations were found to be unchanged at 200 A495/mg protein. Because 88% of cell wall carbohydrates are N-linked (41), this indicated that absence of CaMnt1p does not lead to severe truncation of the large N-linked oligosaccharides. Therefore, O-linked oligosaccharides were isolated by radioactive, reductive β-elimination and separated chromatographically using a Biogel-P4 column. Three peaks were observed in the isolate from CAI4, corresponding to Man1 (65%), Man2 (30%), and Man3 (5%) oligosaccharides (Fig. 3A). In contrast, O-linked mannan from disruptant strain NGY24 yielded one main peak at Man1 (95%) and two minor peaks at Man2 (3%) and Man3 (2%) (Fig. 3B). This indicates that CaMnt1p is responsible for adding the second mannose to O-linked oligosaccharides.
Figure 3.
Biogel-P4 chromatogram of β-eliminated and reductively labeled mannan isolated from C. albicans CAI4 (A) and the Camnt1 disruptant strain NGY24 (B). Nonlabeled partially hydrolyzed dextrane was used as internal standard (not shown). 1, Man1; 2, Man2; 3, Man3.
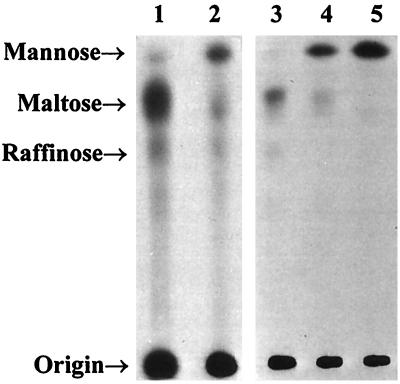
To determine the linkage between the first and second mannose residue, mannan from strains CAI4 and NGY24 was isolated that was radiolabeled by incorporation of [3H]mannose. O-linked oligosaccharides were removed by β-elimination without reduction and separated by HPTLC (Fig. 4). Using this technique, most of the radiolabeled O-linked oligosaccharides in CAI4 were found to be two mannose residues long, whereas in NGY24 most O-linked oligosaccharides were only one mannose long. This confirmed that CaMnt1p is responsible for adding the second mannose. In a separate experiment, oligosaccharides obtained from the wild-type strain were digested with jack bean α-mannosidase and α-1,2-mannosidase of Aspergillus satoi. Both enzymes digested Man2 and Man3 oligosaccharides and reduced them to Man1. Hence, both mannose residues are α-1,2-linked, and CaMnt1p is thus an α-1,2-mannosyl transferase.
Figure 4.
Autoradiogram of an HPTLC chromatogram of β-eliminated [3H]mannose-labeled mannan isolated from C. albicans CAI4 (lane 1) and the derived homozygous disruptant strain, NGY24 (lane 2). In a separate experiment, β-eliminated mannan isolated from C. albicans CAI4 was left undigested (lane 3) or digested with jack bean α-mannosidase (lane 5) or α-1,2-mannosidase of A. satoi (lane 4) prior to chromatography. Mannose, maltose, and raffinose were used as standards for Man1, Man2, and Man3, respectively, and visualized using sulfuric acid in ethanol spray (data not shown).
Absence of CaMNT1 Reduces Adherence.
Mannoproteins are important for adhesion of C. albicans to a number of surfaces (42). Therefore, the ability of both the wild-type and homozygous disruptant C. albicans strains, CAI4 and NGY24 respectively, to adhere to human buccal epithelial cells was tested. Growth of the fungus in high concentrations of galactose stimulates its adherence (43) and increased the number of CAI4 cells (x ± S.D, n = 3) adhering per epithelial cell 6-fold (from 2.1 ± 0.1 on glucose to 13.9 ± 0.3 on galactose). The ability of NGY24 to adhere when grown in galactose was only 0.7 ± 0.1, 5% of that of CAI4. Cells of NGY24 grown on glucose, adhered at a frequency of 0.5 ± 0.2, 75% reduced compared with that of CAI4 and were quite similar to adhesion of cells of NGY24 when grown on galactose.
Adherence to vaginal epithelial cells was tested in a rat model of vaginal candidosis. Approximately 106 and 107 colony-forming units (CFUs) were injected intravaginally in ovarectimized rats, and the number of fungal cells was counted in vaginal lavages taken at various times after infection (data not shown). Irrespective of the dosage administered, 99.9% of cells of the wild-type strain SC5314 were lost after 1 week, but a significant number was able to adhere and colonize vaginal epithelial cells as indicated by the presence of 40 to 200 fungal CFUs in the lavages. Similarly, the number of cells of the prototrophic, heterozygous, and homozygous disruptant strains NGY21 and NGY23, respectively, dropped quickly, but in contrast to infection with wild-type cells, complete clearance of CFUs from lavage samples had occurred after 3 weeks. No difference in clearance rate of NGY21 and NGY23 was observed.
Virulence Studies in Systemic Animal Models.
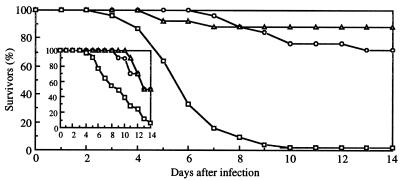
To test whether CaMnt1p influences virulence in systemic animal models, cells of the prototrophic wild-type, heterozygous, and homozygous disruptant strains, SC5314, NGY21, and NGY23, respectively, were injected intravenously into the lateral tail vein of immuno-competent mice and guinea pigs. At a dosage at which 50% of the guinea pigs died after 6 days owing to infection with cells of SC5314, infection with NGY21 and NGY23 showed survival after 14 days of 70 and 90%, respectively (Fig. 5). Similarly, a dosage of cells of SC5314 resulted in death of 50% of the mice after 8 days, but usage of either NGY21 and NGY23 delayed death of 50% of the mice until 14 days.
Figure 5.
Survival of Pirbright guinea pigs (500 g) intravenously infected with 40000 CFU/g of C. albicans SC5314 (□) and prototrophic heterozygous and homozygous Camnt1 disruptant strains, NGY21 (○) and NGY23 (▵), respectively. Survival of Swiss white mice (25 g) intravenously infected with the same strains at 8000 CFU/g is shown in the inset.
Internal organs were analyzed for colonization by C. albicans. In guinea pigs, it was found that infection with disruptant strains resulted in higher numbers of non-colonized skins, livers, and kidneys (Table 2). The number of C. albicans cells recovered from positive tissues, however, was found to be unchanged. This indicates that attenuated virulence is the result of reduced ability to reach and adhere to various organs. However, once the organ had been successfully colonized, invasion and proliferation was apparently comparable with that observed with cells of the wild-type strain. In mice, at most a very modest increase of non-colonized kidneys was found after infection with disruptants strains, whereas 50% of mice survived. Again, the number of C. albicans cells recovered from positive kidneys had not changed. This is similar to the phenotype of avirulent Cachs3 disruptant strains, which were fully able to colonize kidneys but did not kill the mice (44).
Table 2.
Colonization of various sites (in log CFU/g ± 1.0) in guinea pigs (dosage 40,000 CFU/g) and mice (dosage 8,000 CFU/g) after i.v. infection with C. albicans SC5314 and derived prototrophic heterozygous and homozygous Camnt1 disruptant strains NGY21 and NGY23, respectively
| Strain | n | Survivors, % | Negative organs, %
|
Colonization organs
|
||||
|---|---|---|---|---|---|---|---|---|
| Liver | Kidney | Skin | Liver | Kidney | Skin | |||
| Guinea pig | ||||||||
| SC5314 | 45 | 2 | 36 | 0 | 7 | 3.5 | 5.4 | 4.2 |
| NGY21 | 25 | 72 | 76 | 60 | 32 | 3.5 | 3.9 | 3.8 |
| NGY23 | 25 | 88 | 88 | 88 | 60 | 3.4 | 4.8 | 3.6 |
| Mouse | ||||||||
| SC5314 | 36 | 9 | 60* | 3 | ND | 3.2* | 6.0 | ND |
| NGY21 | 10 | 50 | 60 | 0 | ND | 3.6 | 6.3 | ND |
| NGY23 | 10 | 50 | 100 | 20 | ND | <1 | 5.6 | ND |
All animals were analyzed after death during the experiment or after death at the final 14th day of the experiment. n, number of animals; ND, not determined.
*n = 10.
DISCUSSION
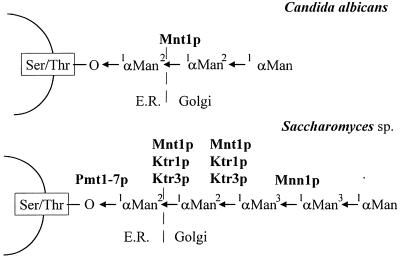
The CaMNT1 gene was isolated and cloned using heterologous hybridization with MNT1 of S. cerevisiae and was therefore expected to yield the functional homologue of this gene in C. albicans. Although the deduced amino acid sequences of CaMnt1p and ScMnt1p are similar and both seem to encode α-1,2-mannosyl transferases located in medial Golgi compartments, their roles in synthesis of O-linked glycosylation seem to be distinct. ScMnt1p participates in adding the second and third mannose sugars in the oligosaccharide, whereas CaMnt1p adds the second (Fig. 6). We cannot at this stage formally exclude the possibility that the CaMnt1p also adds the third mannose sugar to the O-linked mannobiose. However, our data imply that CaMnt1p may not be the true, biochemical homologue of ScMnt1p. The complete sequence of the S. cerevisiae genome has revealed a total of nine members of the ScMNT1 gene family (21). To date, we have identified at least five members in the Candida MNT1 gene family, which are presently under analysis (E. T. B., C. W., L. Thomson, Y. Aoki, S. Yamazaki, M. Arisawa, and N.A.R.G., unpublished results). The predicted ScKtr1p and ScKtr3p proteins are more homologous to CaMnt1p than ScMnt1p and have recently been shown to be collectively responsible, along with ScMnt1p, for addition of the second and third mannose residues in S. cerevisiae (16) (Fig. 6). Although we have no evidence as yet for this high level of redundancy in this glycosylation step in C. albicans, O-linked Man2 and Man3 were still detected at 20% of the level present in wild-type cells (Fig. 3B), and 25% of specific in vitro mannosyl transferase activity was still present in the homozygous disruptant strain (Table 1). This implies that other mannosyltransferases may contribute to the addition of the second mannose sugar.
Figure 6.
Structure of O-linked mannan and gene products involved in O-linked glycosylation in C. albicans and S. cerevisiae as described in the text.
Despite this significant residual mannosyl transferase activity in vitro and in vivo, the absence of CaMnt1p led to strong attenuation of virulence, which seems to be caused by a decreased ability to adhere. Absence of CaMnt1p reduced the adhesion of C. albicans to human buccal epithelial cells and to rat vaginal epithelial cells and reduced colonization of endothelial cells of liver and kidney and the skin of guinea pigs. However, as the glycosylation defect will influence virtually all mannoproteins, disruption of CaMNT1 is very likely to have pleiotropic effects. This is illustrated by the decreased virulence in mouse models of systemic candidosis, which could not be attributed to decreased adherence to internal organs. In this case, 50% of mice were alive after 14 days despite 80% of animals having their kidneys fully colonized (Table 2). Similar findings have been reported with avirulent Cachs3 null mutants of C. albicans, where colonization of the kidneys seemed normal but progress of the infection and subsequent death was for unknown reasons prevented (44).
Adhesion of C. albicans is thought to be mediated by a number of Candida-host cell recognition systems, each of which may or may not be specific for the type of host cell. Both the protein and carbohydrate moieties of mannoproteins have been implicated in the adhesion of cells to epithelial and endothelial cells (7). Adhesion and colonization of the Camnt1 null mutant to a number of different host cells (human buccal epithelial cells, rat vaginal cells, cells of liver, skin, and kidney of guinea pigs) was reduced. This suggests that either a single Candida binding protein(s) involved in interactions with all host tissues has been changed or that a spectrum of Candida cell wall proteins, each of which is specific for adhesion to a specific host cell type, must have been influenced. O-linked glycosylation has a profound influence on protein structure, which is mainly the result of interaction between peptide and the first amino acid-linked sugar (45). Because the Camnt1 disruption results in truncated, but still one mannose long, O-linked oligosaccharides, changes in the overall protein structure are unlikely. In contrast, disruptions in the Pmt-catalyzed addition of the first mannose unit is likely to lead to marked alterations in the tertiary structure of cell wall proteins and hence marked nonspecific pleiotropic effects that are unrelated to specific mannan–host surface interactions. Our results suggest that the O-linked carbohydrate component of cell wall mannoproteins is indeed critically important for the adhesion process and possibly other host recognition phenomena.
A desirable control for gene knock-out experiments is to restore a functional allele in the homozygous null strain. Re-introduction of CaMNT1 would have to be chromosomal to ensure stability under nonselective conditions and at a site different from the native locus, where both level and timing of expression matches that of the wild-type gene during the various stages of an establishing Candida infection. This would require a detailed understanding of the in vitro and, more importantly, in vivo activity of the promoter and the target locus, and further controls would be necessary to ensure that re-introduction did not alter the genetic background in a way that altered virulence. Because both the CaMNT1 promoter and a suitable target locus remain to be analyzed, it is hard to anticipate the virulence properties of a strain resulting from re-introduction of CaMNT1.
Despite disruption of CaMNT1, residual mannosyl transferase activity was present that enabled some mannoproteins to be properly glycosylated, thereby permitting some C. albicans cells to adhere and cause infection inefficiently. To inactivate the remaining mannosyl transferase activity, which could result in an avirulent or perhaps even a nonviable strain, we are currently analyzing other MNT1-like genes in Candida. In other fungi, there is evidence that O-linked mannosylation may also involve Mnt1p homologs (46–49). In mammalian cells, however, occurrence of mannose in O-linked oligosaccharides is rare, and in these cases mannose is directly linked to the serine or threonine in the protein (50). Furthermore, in mammalian cells protein glycosylation occurs in the Golgi and cytosol by transfer of mannose from NDP-mannose (51), whereas in fungi mannose is transferred from dolichol-P-mannose in the ER by Pmt proteins (10). To date, no mammalian homologs of the MNT1 family have been found. This report presents the first evidence of the importance of these apparently fungal-specific proteins for virulence of C. albicans. If multiple mutations in Candida MNT1-like genes prove to be lethal, these proteins would be seen as potential broad spectrum targets for future generations of antifungal drugs.
Acknowledgments
We thank Drs. A. Häusler and P. W. Robbins for advice in the initial stages of this project, Frans van Gerven for technical assistance with the animal experiments, Drs. W. A. Fonzi and B. B. Magee for chromosomal mapping of CaMNT1, Dr. J. Harthill and Prof. M. A. J. Ferguson (University of Dundee, UK) for help in analysis of mannan structures, and Dr. L. J. Douglas for advice with adherence assays. This work was supported by Wellcome Trust Grant 039643/2/93/z/1.27, a BBSRC studentship (to C.W.), and personal fellowships from the Royal Society of Edinburgh/Caledonian Research Foundation and Royal Society/Leverhulme Trust (to N.A.R.G.).
ABBREVIATIONS
- FOA
fluoroorotic acid
- HPTLC
high-performance thin-layer chromatography
- CFU
colony-forming unit
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. X99619).
References
- 1.Beck-Sagué C M, Jarvis W R. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 2.Odds F C. Int J Antimicrob Agents. 1996;6:145–147. doi: 10.1016/0924-8579(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 3.Cabib E, Roberts R, Bowers B. Annu Rev Biochem. 1982;51:763–793. doi: 10.1146/annurev.bi.51.070182.003555. [DOI] [PubMed] [Google Scholar]
- 4.McLellan W L, Jr, McDaniel L E, Lampen J O. J Bacteriol. 1970;102:261–270. doi: 10.1128/jb.102.1.261-270.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hector R F, Schaller K. Antimicrob Agents Chemother. 1992;36:1284–1289. doi: 10.1128/aac.36.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz M B, Abruzzo G, Flattery A, Bartizal K, Marrinan J A, Li W, Milligan J, Nollstadt K, Douglas C M. Infect Immun. 1996;64:3244–3251. doi: 10.1128/iai.64.8.3244-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderone R A. Trends Microbiol. 1993;1:55–58. doi: 10.1016/0966-842x(93)90033-n. [DOI] [PubMed] [Google Scholar]
- 8.Zlotnik H, Fernandez M P, Bowers B, Cabib E. J Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutler J E. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 10.Lehle L, Tanner W. In: Glycoproteins. Montreuil J, Schachter H, Vliegenthart J F G, editors. Amsterdam: Elsevier; 1995. pp. 475–509. [Google Scholar]
- 11.Herscovics A, Orlean P. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 12.Hounsell E F, Davies M J, Renouf D V. Glycoconj J. 1996;13:19–26. doi: 10.1007/BF01049675. [DOI] [PubMed] [Google Scholar]
- 13.Häusler A, Ballou L, Ballou C E, Robbins P W. Proc Natl Acad Sci USA. 1992;89:6846–6850. doi: 10.1073/pnas.89.15.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentzsch M, Tanner W. EMBO J. 1997;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 15.Häusler A, Robbins P W. Glycobiology. 1992;2:77–84. doi: 10.1093/glycob/2.1.77. [DOI] [PubMed] [Google Scholar]
- 16.Lussier M, Sdicu A, Bussereau F, Jacquet M, Bussey H. J Biol Chem. 1997;272:15527–15531. doi: 10.1074/jbc.272.24.15527. [DOI] [PubMed] [Google Scholar]
- 17.Chapman R E, Munro S. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballou C E. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- 19.Yip C L, Welch S K, Klebl F, Gilbert T, Seidel P, Grant F J, O’Hara P J, MacKay V L. Proc Natl Acad Sci USA. 1994;91:2723–2727. doi: 10.1073/pnas.91.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill K, Boone C, Goebl M, Puccia R, Sdicu A M, Bussey H. Genetics. 1992;130:273–283. doi: 10.1093/genetics/130.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lussier M, Sdicu A, Winnett E, Vo D H, Sheraton J, Düsterhöft A, Storms R K, Bussey H. Yeast. 1997;13:267–274. doi: 10.1002/(SICI)1097-0061(19970315)13:3<267::AID-YEA72>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Romero P A, Lussier M, Sdicu A M, Bussey H, Herscovics A. Biochem J. 1997;321:289–295. doi: 10.1042/bj3210289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lussier M, Sdicu A-M, Camirand A, Bussey H. J Biol Chem. 1996;271:11001–11008. doi: 10.1074/jbc.271.18.11001. [DOI] [PubMed] [Google Scholar]
- 24.Wang X-H, Nakayama K, Shimma Y, Tanaka T. J Biol Chem. 1997;272:18117–18124. doi: 10.1074/jbc.272.29.18117. [DOI] [PubMed] [Google Scholar]
- 25.Kelly R, Miller S M, Kurtz M B, Kirsch D R. Mol Cell Biol. 1987;7:199–207. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillum A M, Tsay E Y H, Kirsch D R. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 27.Fonzi W A, Irwin M Y. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 29.Gow N A R, Gooday G W. J Gen Microbiol. 1982;128:2195–2198. doi: 10.1099/00221287-128-9-2195. [DOI] [PubMed] [Google Scholar]
- 30.Mattia E, Carruba G, Angiolella L, Cassone A. J Bacteriol. 1982;152:555–562. doi: 10.1128/jb.152.2.555-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Köhler J, Fink G R. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 32.Douglas L J, Houston J G, McCourtie J. FEMS Microbiol Lett. 1981;12:241–243. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 14.12. [Google Scholar]
- 34.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 35.Kurtz M B, Cortelyou M W, Kirsch D R. Mol Cell Biol. 1986;6:142–149. doi: 10.1128/mcb.6.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippsen P, Stotz A, Scherf C. Methods Enzymol. 1991;194:169–181. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 37.Hube B, Monod M, Schofield D A, Brown A J P, Gow N A R. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 38.Ferguson M A J. In: Glycobiology: A Practical Approach. Fukuda M, Kobata A, editors. Oxford: Oxford University Press; 1993. pp. 349–383. [Google Scholar]
- 39.Sanglard D, Hube B, Monod M, Odds F C, Gow N A R. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soll D R, Galask R, Schmid J, Hanna C, Mac K, Morrow B. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elorza M V, Marcilla A, Sentandreu R. J Gen Microbiol. 1988;134:2393–2403. doi: 10.1099/00221287-134-8-2393. [DOI] [PubMed] [Google Scholar]
- 42.Fukazawa Y, Kagaya K. J Med Vet Mycol. 1997;35:87–99. doi: 10.1080/02681219780000971. [DOI] [PubMed] [Google Scholar]
- 43.McCourtie J, Douglas L J. Infect Immun. 1981;32:1234–1241. doi: 10.1128/iai.32.3.1234-1241.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulawa C E, Miller D W, Henry L K, Becker J M. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jentoft N. Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 46.Ballou C E, Ballou L, Ball G. Proc Natl Acad Sci USA. 1994;91:9327–9331. doi: 10.1073/pnas.91.20.9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutzbach J S, Ankel H. J Biol Chem. 1971;246:2187–2194. [PubMed] [Google Scholar]
- 48.White C W, Jacobson E S. Can J Microbiol. 1993;39:129–133. doi: 10.1139/m93-019. [DOI] [PubMed] [Google Scholar]
- 49.Gunnarsson A, Svensson B, Nilsson B, Svensson S. Eur J Biochem. 1984;145:463–467. doi: 10.1111/j.1432-1033.1984.tb08578.x. [DOI] [PubMed] [Google Scholar]
- 50.Hart G W. Curr Opin Cell Biol. 1992;4:1017–1023. doi: 10.1016/0955-0674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- 51.Roth J, Wang Y, Eckhardt A E, Hill R L. Proc Natl Acad Sci USA. 1994;91:8935–8939. doi: 10.1073/pnas.91.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]