Abstract
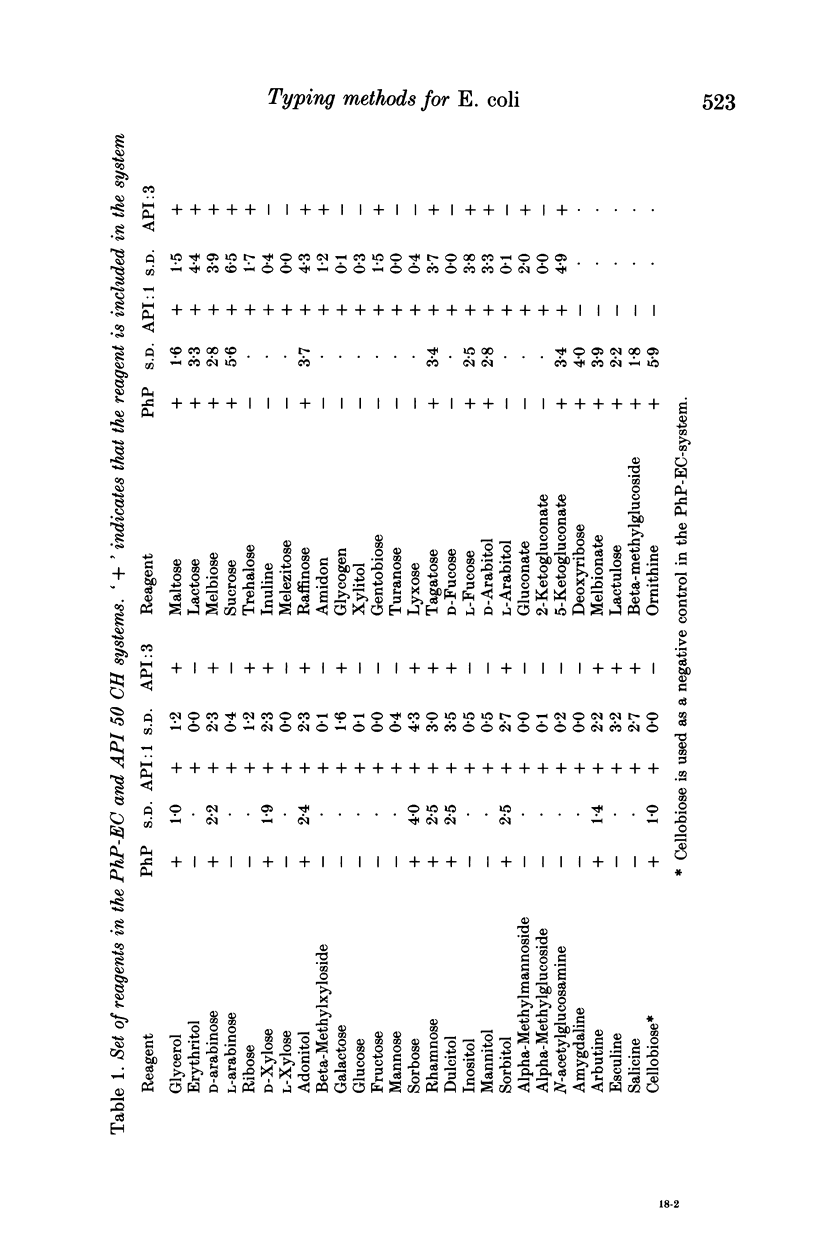
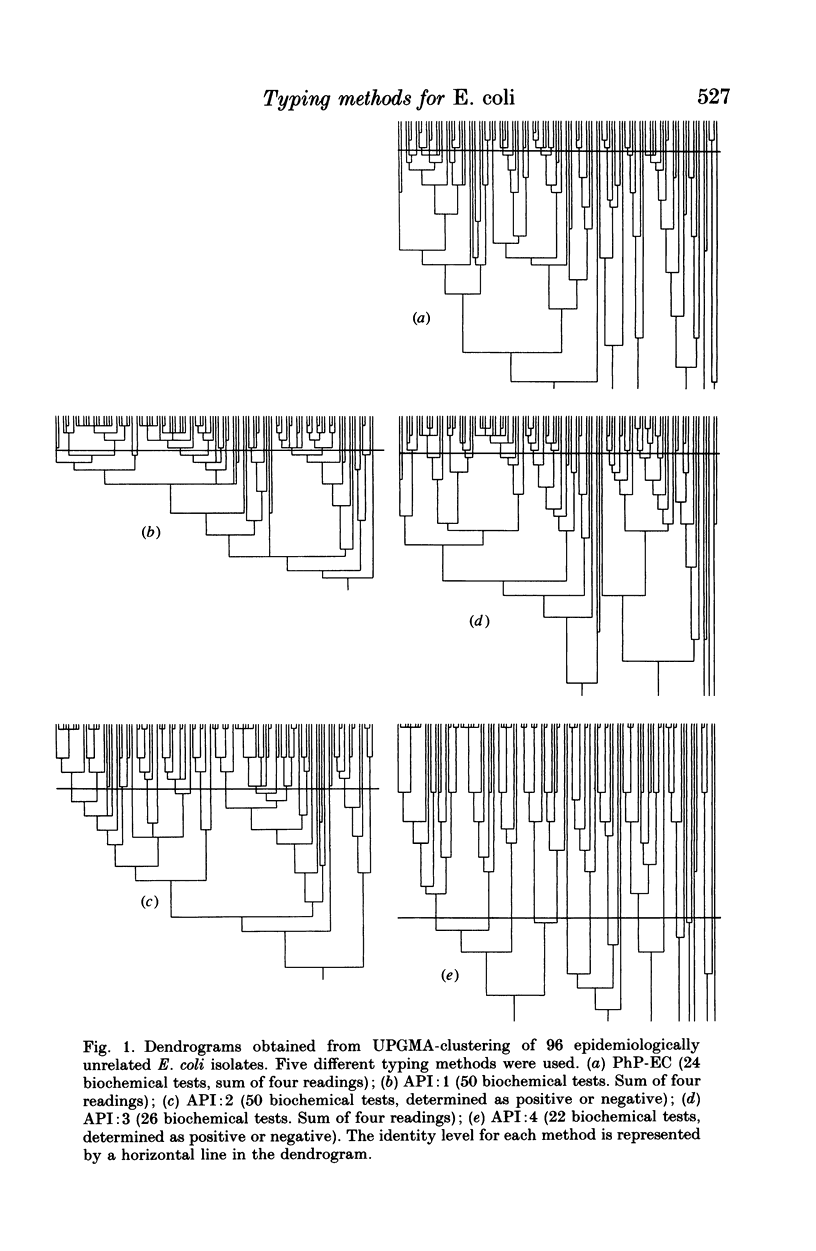
Reproducible and discriminating typing methods are required for epidemiological investigations. Numerical typing systems analyse patterns obtained in various ways by calculating similarity coefficients between isolates. In the present study, various measures of the efficiency of a numerical typing system are quantified. These include reproducibility, accuracy, and discrimination power. Three different numerical typing methods for Escherichia coli were compared using these measures: (a) Biotyping with API 50 CH system, (b) Biochemical fingerprinting with the API 50 CH system and (c) Biochemical fingerprinting with the PhP-EC system. Biotyping qualitatively measures the results of a set of biochemical reactions as + or -. Biochemical fingerprinting also uses biochemical reactions, but the tests are scored quantitatively by measuring the kinetics and intensity of each reaction. It was found that biotyping yielded poor reproducibility. When biochemical fingerprinting analysis was used with the API 50 CHE system the reproducibility and the discrimination was good. The PhP-EC system for biochemical fingerprinting showed equal reproducibility but was superior to the API 50 CH system with regard to discrimination power.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Mercer A., Kusecek B., Pohl A., Heuzenroeder M., Aaronson W., Sutton A., Silver R. P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983 Jan;39(1):315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Pluschke G. Clonal analysis of descent and virulence among selected Escherichia coli. Annu Rev Microbiol. 1986;40:185–210. doi: 10.1146/annurev.mi.40.100186.001153. [DOI] [PubMed] [Google Scholar]
- Brauner A., Boeufgras J. M., Jacobson S. H., Kaijser B., Källenius G., Svenson S. B., Wretlind B. The use of biochemical markers, serotype and fimbriation in the detection of Escherichia coli clones. J Gen Microbiol. 1987 Oct;133(10):2825–2834. doi: 10.1099/00221287-133-10-2825. [DOI] [PubMed] [Google Scholar]
- Brauner A., Hylander B., Wretlind B., Ostenson C. G., Kühn I. Escherichia coli populations from diabetic and non-diabetic patients with bacteraemia and faecal samples from healthy subjects--a comparative study. Epidemiol Infect. 1990 Dec;105(3):533–540. doi: 10.1017/s0950268800048159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicmanec J. F., Evans A. T. Classification of urinary tract infections by biotype identification of the pathogens. J Urol. 1980 Jul;124(1):68–69. doi: 10.1016/s0022-5347(17)55300-8. [DOI] [PubMed] [Google Scholar]
- Crichton P. B., Old D. C. Biotyping of Escherichia coli. J Med Microbiol. 1979 Nov;12(4):473–486. doi: 10.1099/00222615-12-4-473. [DOI] [PubMed] [Google Scholar]
- Gargan R., Brumfitt W., Hamilton-Miller J. M. A concise biotyping system for differentiating strains of Escherichia coli. J Clin Pathol. 1982 Dec;35(12):1366–1369. doi: 10.1136/jcp.35.12.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout-DeLasalle F., Higgins R. Biotyping of clinical isolates of Escherichia coli of animal origin, using the Analytab API 20E system. Can J Vet Res. 1986 Jul;50(3):418–421. [PMC free article] [PubMed] [Google Scholar]
- Hunter P. R., Gaston M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988 Nov;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn I., Franklin A., Söderlind O., Möllby R. Phenotypic variations among enterotoxinogenic Escherichia coli from Swedish piglets with diarrhoea. Med Microbiol Immunol. 1985;174(3):119–130. doi: 10.1007/BF02298122. [DOI] [PubMed] [Google Scholar]
- Kühn I., Möllby R. Phenotypic variations among enterotoxigenic O-groups of Escherichia coli from various human populations. Med Microbiol Immunol. 1986;175(1):15–26. doi: 10.1007/BF02123125. [DOI] [PubMed] [Google Scholar]
- Kühn I., Tullus K., Möllby R. Colonization and persistence of Escherichia coli phenotypes in the intestines of children aged 0 to 18 months. Infection. 1986 Jan-Feb;14(1):7–12. doi: 10.1007/BF01644802. [DOI] [PubMed] [Google Scholar]
- Okerman L., Devriese L. A. Biotypes of enteropathogenic Escherichia coli strains from rabbits. J Clin Microbiol. 1985 Dec;22(6):955–958. doi: 10.1128/jcm.22.6.955-958.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old D. C., Barker R. M. Numerical index of the discriminatory ability of biotyping for strains of Salmonella typhimurium and Salmonella paratyphi B. Epidemiol Infect. 1989 Dec;103(3):435–443. doi: 10.1017/s0950268800030831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orskov I., Orskov F., Jann B., Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977 Sep;41(3):667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


