Abstract
Macrophages are considered to be the mediators of resistance to extra-intestinal Salmonella infections. Nevertheless, the initial cellular response to Salmonella infections consists primarily of polymorphonuclear leukocytes (PMN). To determine whether PMN serve an important function for the infected host, we made mice neutropenic with the rat mAb to RB6–8C5 and infected them i.v. with ≈103 Salmonella dublin or an isogenic derivative that lacks the virulence plasmid (LD842). We infected BALB/c mice, which have a point mutation in the macrophage-expressed gene Nramp1 that makes them susceptible to Salmonella, and BALB/c.D2 congenic mice, which have the wild-type Nramp1 gene that makes them resistant to Salmonella. Both mouse strains were resistant to LD842, and neutropenia made only the BALB/c strain susceptible to this infection. Neutropenic congenic mice, however, were susceptible only to wild-type S. dublin (plasmid+). These results show a complex interplay between plasmid-virulence genes in Salmonella, host macrophages, and PMN. Mice with normal macrophages need PMN to defend against nontyphoid Salmonella that carry a virulence plasmid but not against Salmonella without virulence plasmids. Mice with a mutant Nramp1 gene need PMN to defend against all Salmonella, even those that lack virulence plasmids. These results, plus the evidence that PMN kill Salmonella efficiently in vitro, suggest that Salmonella have adapted to grow inside macrophages where they are relatively sheltered from PMN. The adaptations that allow Salmonella to survive in macrophages do not protect them from PMN.
Salmonella are facultative intracellular pathogens that cause infections in animals and humans (1). Salmonella cause a spectrum of diseases that includes gastroenteritis, septicemia, and enteric fever (2). A number of microbial factors influence the severity of enteric fever in experimental animals, including lipopolysaccharides (3), rpoS-controlled spv genes on the virulence plasmid (4, 5), and several phoP/phoQ-controlled chromosomal genes (6–8). Numerous genes found in pathogenicity island 1 (SPI1) are involved with epithelial cell invasion but not systemic infection (9) whereas genes in a second, newly described pathogenicity island (SPI2) are involved in systemic infections (10, 11). Although all pathogenic salmonella contain SPI 1 and 2, only a small number of salmonella serotypes carry spv genes on virulence plasmids (12, 13). However, the serotypes with virulence plasmids account for most of the systemic nontyphoid Salmonella infections in people (14, 15) and are the host-adapted pathogens of domestic animals (15, 16).
There are several host genes that contribute to resistance to Salmonella infections (17). The best characterized host resistance gene is Nramp1, which is polymorphic in mice by virtue of a single amino acid substitution. The mutation results in susceptibility to infection by Salmonella, bacillus Calmette–Guérin, and Leishmania donovani (18, 19). Nramp1 is expressed in macrophages, providing additional evidence that macrophages are crucial cells involved in resistance to Salmonella infection (19). Mice with mutant Nramp1 genes die from Salmonella infection within the first 7 days after i.p. injection of ≤101 Salmonella dublin whereas congenic mice without the Nramp1 mutation are not killed by >104 bacteria injected i.p. (20, 21). The mechanism of action of the Nramp1 protein is not known, but it is homologous to a family of transmembrane proteins that are ion transporters (22).
The importance of macrophages in Salmonella pathogenesis is shown by the fact that all macrophage-sensitive mutants of Salmonella typhimurium are avirulent in BALB/c mice (23). Perhaps because the macrophage is clearly so important in the pathogenesis of enteric fever, the role of neutrophils [polymorphonuclear leukocytes (PMN)] in resistance to that infection has not been given much attention. PMN are known to play a crucial role in resistance to a number of pyogenic bacteria, but they generally are not considered to be an important component of host resistance to facultatively intracellular bacteria. Recently, Conlon and North (24, 25) used antibodies to either CR3 or a neutrophil-specific membrane protein (RB6–8C5) to make mice neutropenic, and they found that neutropenic mice have increased susceptibility to several facultative intracellular pathogens including Listeria, Francisella, and Salmonella (24, 25). In this research we employed the rat mAb to RB6–8C5 to make mice neutropenic and to determine how neutropenia affects infection with Salmonella and how this relationship is altered by the Nramp1 allele of the host. As a result of these experiments, we propose an hypothesis to explain natural resistance to Salmonella infections.
METHODS
Mice and Infection.
BALB/c mice were purchased from The Jackson Laboratory. Congenic BALB/c.DBA/2.Idh-Ib-Pep-3b mice (26) were bred in our laboratory from breeding pairs kindly supplied by B. Zwilling (Ohio State University). Male and female mice were used and mixed evenly in all experiments. Mice were kept under specific pathogen-free conditions and were allowed free access to food and water. Mice were infected via a tail vein by injecting bacteria suspended in 0.2 ml of normal saline. At time of death, organs were removed and processed for quantitative cultures as described (27). To do quantitative blood cultures, 50 μl of peripheral blood was collected from the tail in a heparinized tube and was diluted appropriately in sterile saline before being applied to trypticase soy agar plates.
mAb.
The rat hybridoma against RB6–8C5 was obtained from DNAX and was grown in a serum-free medium. The IgG2b antibody was purified on a protein G Sepharose column (Pharmacia LKB) and was adjusted to 0.9 mg/ml, as measured by radial immunodiffusion (The Binding Site, Birmingham, U. K.).
White Blood Cell Counts.
Peripheral blood was obtained from the tail in heparinized tubes. White blood cells were stained with methylene blue to facilitate counting in a Petrous Hauser chamber. Blood smears were prepared and stained with Diff-Quick stain (Baxter Diagnostics, McGraw Park, IL). To obtain a differential count, at least 100 nucleated blood cells from each mouse were examined at a magnification of ×1,000.
Bacteria.
S. dublin lane (a clinical isolate) and the isogenic plasmid-cured strain LD842 have been described (20). The purA mutant of S. dublin lane was a gift from B. Stocker (Stanford University). S. typhimurium 14028 was obtained from F. Heffron (Oregon Health Sciences University) and the isogenic phoP- mutant kindly was supplied by E. Groisman (Washington University). All bacteria were grown overnight in trypticase soy broth, were washed twice, and then were diluted for injection.
Isolation of Human Neutrophils (PMN).
Peripheral blood (30 ml) was collected in acid citrate dextrose solution from normal volunteers. Erythrocytes were sedimented with 6% dextran (molecular weight 500,000) 1:1 in saline. The supernatant was centrifuged at 140 × g for 10 min and the white blood cells were resuspended in PBS. Cells were underlayered beneath 7 ml of Histopaque-1077 (Sigma) and were centrifuged at 330 × g for 20 min at 10°C. The pellet was removed and resuspended in PBS; erythrocytes were lysed if necessary with iced sterile water followed by 0.6 M KCl.
In vitro phagocytosis was assayed by using PMN in Hanks balanced salt solution with CaCl2 (1M) 0.13% (vol/vol), glucose (1M) 0.56%, MgCl2 (1M) 0.05%, Hepes 0.25%, and 1% (vol/vol) of 10% gelatin in water. For opsonization, we added an equal volume of autologous normal human serum. Bacteria were added in a volume of 0.1 ml, and the reaction mixture of 1.0 ml was rotated at 37°C. Aliquots of 100 μl were removed at intervals, and PMN were lysed with cold water before the aliquots were diluted and plated on trypticase soy agar. After 30 min, 100 μl were removed and used to prepare a Giemsa-stained slide that was examined at ×1000 magnification to determine the percentage of PMN that contained bacteria.
Statistics.
Geometric means and standard deviations were calculated. The significance of differences between means was determined with the Mann-Whitney U test using spss version 7.5.1.
RESULTS
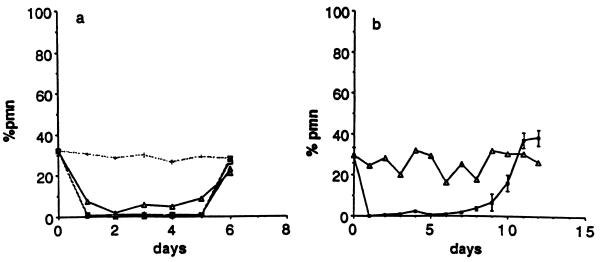
We injected several doses of anti-RB6–8C5 into mice to determine the optimal dose; all doses above 150 μg resulted in profound neutropenia that lasted for 5 days, but the higher doses did not extend the period of neutropenia (Fig. 1a). We were able to prolong the period of neutropenia to 7–8 days by injecting another 300 μg i.p. on day 4 (Fig. 1b). By day 9, the percentage of PMN rose to 7–10% in the mice given two doses of antibody, and by day 11 white blood cells were normal in all treated mice. None of the mice became ill spontaneously while they were neutropenic. In subsequent experiments, we used a regimen of 150 μg followed by 300 μg of antibody on day 4. To insure that anti-RB6–8C5 did not cause reticuloendothelial system blockade, we measured the clearance rate of 5 × 103 S. dublin during the first 120 min after the bacteria were injected i.v. into mice. There was no difference in the clearance of the bacteria by neutropenic and control mice, and by 120 min, there were <100 colony-forming units/ml in neutropenic and control mice (data not shown).
Figure 1.
(a) The effects of various doses of mAb RB6–8C5 on circulating PMN are shown with all the solid lines. Open triangles, 100 μg; dark triangles, 150 μg; open squares, 200 μg; hatched squares, 300 μg; dotted line, control mice. (b) Maintenance of neutropenia. The effect of injecting 150 μg i.p. on day 0 and 300 μg i.p. 4 days later. Each point is the mean of 3 mice.
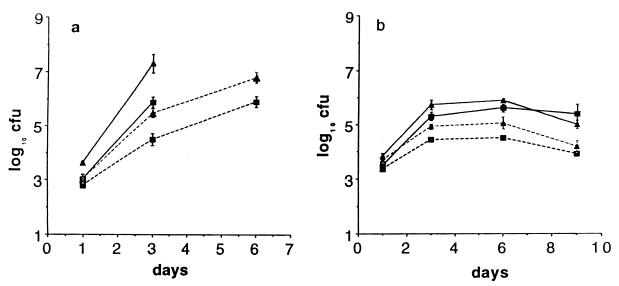
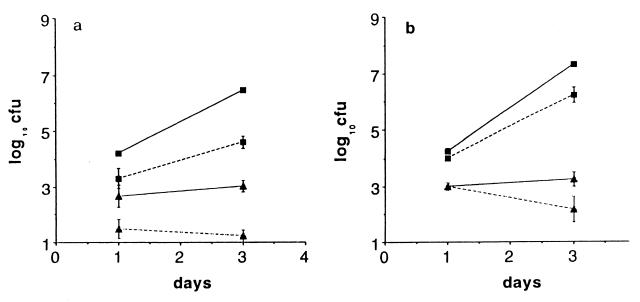
To determine the role of PMN in the systemic phase of S. dublin infection, we infected neutropenic, congenic BALB/c.D2 mice with 5 × 103 S. dublin lane i.v. One day after infection, there was almost no difference in the numbers of bacteria in the livers and spleens of the neutropenic and control congenic mice. However, between days 1 and 3 of infection, bacteria grew faster in neutropenic mice (Fig. 2a and Table 1) so that, on day 3, there were ≈10× more bacteria in the spleens and 100× more bacteria in the livers of neutropenic than of control mice. After day 3, the bacteria continued to grow rapidly in neutropenic mice, so that by day 6, no neutropenic mice were alive whereas four of four mice were alive in the control group (Fig. 2a). To determine whether the enhanced virulence of S. dublin lane in neutropenic mice depended on the virulence plasmid, we infected congenic mice with the same number of the isogenic, plasmid-cured strain LD842 (12). As shown in Fig. 2b, one day after infection, the numbers of LD842 in the liver and spleen of neutropenic and control mice were quite similar and were also nearly identical to what we found in the mice infected with the virulent lane strain. However, from day 1–3, the number of LD842 increased only ≈10-fold (Fig. 2b), and the rate of growth of LD842 was only slightly higher in neutropenic than in control mice (Table 1). After day 3, there was essentially no further net growth of LD842 in either the neutropenic or the control congenic mice, and all mice survived until day 9, the last day of the experiment. Thus, in BALB/c.D2 mice, which have a wild-type Nramp1, host cells other than PMN (presumably macrophages) controlled the growth of LD842, but those cells were not sufficient to control the growth of S. dublin lane, which has a virulence plasmid.
Figure 2.
Effect of neutropenia on infection with S. dublin (a) and LD842 (b) in BALB/c.D2 (ItyR) congenic mice. Mice were infected 1 day after being treated with anti-RB6–8C5. CFU in spleen (squares) and liver (triangles) at time points after i.v. injection of bacteria. In this and subsequent figures, the spleens are cfu per organ and the livers are cfu per gram of tissue. Solid lines are neutropenic mice, and dotted lines are control mice. Each point is the geometric mean ± SD of at least four mice. All of the neutropenic mice (4/4) died by day 5, so there are no values for cfu on day 6 for this group.
Table 1.
Effect of neutropenia on the rate of growth of Salmonella in the spleens of susceptible and resistant mice (days 1–3)
| Mice | Pathogen | Neutropenia | Rate of growth* (log10) |
|---|---|---|---|
| Congenic | S. dublin | − | 0.83 |
| Congenic | S. dublin | + | 1.44 |
| Congenic | LD842 | − | 0.53 |
| Congenic | LD842 | + | 0.92 |
| BALB/c | LD842 | − | 0.65 |
| BALB/c | LD842 | + | 1.61 |
Slope [(y − a)/x], days 1–3 after i.v. infection with 2–5 × 103 cfu.
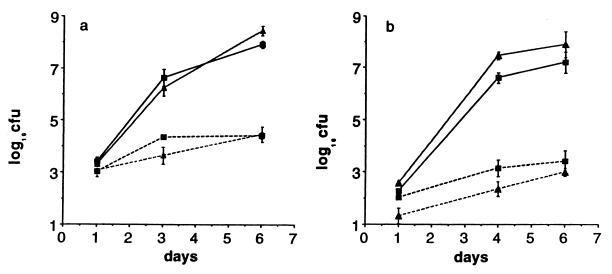
We then asked whether PMN would play an even larger role in mice with a mutant Nramp1 gene. We have shown (20) that BALB/c mice succumb to S. dublin lane infection but can survive an i.p. infection with up to 105 LD842. We injected 5 × 103 colony-forming units (cfu) of LD842 i.v. into neutropenic and control BALB/c mice and, to our surprise, the bacteria grew rapidly in neutropenic BALB/c mice, reaching over 106 cfu/spleen by day 3 and killing two of the four remaining mice before day 6 (Fig. 3a). In control BALB/c mice, there was only a small increase in bacterial numbers from day 1 to 6, which is consistent with what we have reported for oral and i.p. infections with LD842 in BALB/c mice (20). We repeated the experiment, injecting only 90 cfu of LD842 into neutropenic mice; again, the bacteria grew rapidly and killed half of the mice by day 6 (Fig. 3b). Thus, in mice with a defective Nramp1 gene, nearly all of the host defense against LD842 actually depends on PMN.
Figure 3.
Effect of neutropenia on infection with LD842 in BALB/c mice. Each point is the mean (±SD) of four mice. Two different inocula are shown: (a) 5 × 103 and (b) 9 × 101. In the 5 × 103 group, two of four mice died before day 6, and the last point is the mean of only two mice. See legend for Fig. 2.
These results implied that during the course of a Salmonella infection, macrophages from congenic but not BALB/c mice acquire the ability to inhibit or kill the bacteria. We attempted to confirm this by removing peritoneal macrophages from both mouse strains on the third day after i.v. infection with LD842. After adhering the cells to plastic overnight, we added either opsonized (normal human serum) S. dublin lane or LD842. There was equal uptake of the Salmonella by both sets of macrophages, and there was no significant difference in bacterial counts after 4 and 18 hr of incubation (data not shown).
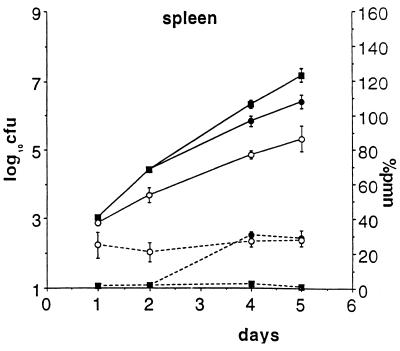
In the in vivo experiments described above, there was only a small difference between the numbers of cfu in control and neutropenic mice on day 1 after i.v. infection, but the differences increased with time. This suggested that PMN act primarily after the Salmonella have been taken up by their target cells, the tissue macrophages, and, probably, PMN act on bacteria that are released by dying macrophages. To see whether recovery from neutropenia PMN during the course of infection would slow the growth of salmonella, we delayed the injection of S. dublin an additional 2 days after the treatment with anti-RB6–8C5, which was long enough for the mice to recover from neutropenia before they died. By day 4 after infection, the mice were no longer neutropenic, and, coincident with that, the rate of bacterial growth in the spleen (Fig. 4) and liver (data not shown) slowed. From day 4 to 6, the rate of growth of S. dublin in the recovered mice slowed while bacteria continued to multiply rapidly in the neutropenic controls. The difference between the slopes (rate of growth) in recovered and neutropenic mice was significant (Mann–Whitney U test, 2-tailed; P = 0.05). This difference shows that PMN play a role in reducing bacterial growth even after the infection is fully established presumably because Salmonella are released by dying macrophages throughout the course of infection.
Figure 4.
Effect of recovery of PMN during infection with S. dublin. Congenic mice were infected 2 days after treatment with RB6–8C5, and so recovered from neutropenia by day 4 after infection. Both the percentage of circulating PMN (dotted lines) and cfu per spleen (solid lines) are shown. Dark squares are mice that were kept neutropenic throughout the experiment, dark circles are mice that were allowed to recover from neutropenia, and open circles are normal controls. Note that the reduction in the rate of bacterial growth coincides with the recovery from neutropenia.
It has been claimed that Salmonella actually multiply extracellularly in murine livers (28), where they would be vulnerable to PMN. To try to clarify whether Salmonella were primarily inside macrophages or were extracellular, we did two experiments. In one, we infected mice with phoP-, a macrophage-sensitive mutant of S. typhimurium, or purA-, a macrophage-sensitive mutant of S. dublin lane; both of these mutants do not survive in macrophages and are avirulent in BALB/c mice (29, 30). As expected, neither mutant grew in control mice from day 1 to 3, but neither did they grow in the spleens of neutropenic mice, even though both mutants carry virulence plasmids (Fig. 5). In contrast, the parent salmonella grew 2–3 logs in two days in neutropenic mice. To be certain that the mutants were not also hypersensitive to killing by PMN, we mixed them with human PMN and normal serum, and they were killed at the same rate as wild-type Salmonella (data not shown). Thus, two different macrophage-sensitive mutants were unable to grow in the liver and spleen of neutropenic BALB/c mice, which implies that the salmonella were inside macrophages, not growing extracellularly in neutropenic mice.
Figure 5.
Effect of neutropenia on growth of S. typhimurium phoP- (a) and S. dublin purA- (b) in the spleens of BALB/c mice. Solid lines are neutropenic mice, and dotted lines are infected control mice. Triangles show mice infected with the mutants, and squares show mice that were infected with wild-type bacteria. Each time point is the geometric mean (±SD) from three mice. See legend for Fig. 2.
In another experiment, we infected neutropenic mice with S. dublin lane, and, 2 hr later, we treated them with the long acting aminoglycoside antibiotic, amikacin. Three hr later, we killed the mice and cultured their spleens and livers; there was no reduction in cfu in either organ compared with untreated mice (Table 2). However, when we treated mice in the same way with ciprofloxacin, an antibiotic that enters into cells (31), there was a significant (>2 logs) drop in bacterial counts (Table 2). This is further evidence that the S. dublin were inside cells; they were protected from amikacin but susceptible to ciprofloxacin.
Table 2.
Lack of effect of amikacin on S. dublin after i.v. infection
| Treatment | cfu
|
|
|---|---|---|
| Spleen | Liver | |
| None | 2.63 ± 0.2* | 3.50 ± 0.1 |
| Amikacin | 2.56 ± 0.1† | 3.33 ± 0.2† |
| Ciprofloxacin | 1.05 ± 0.2‡ | 1.15 ± 0.5‡ |
log; geometric mean of three mice ± SEM.
Not significant compared to untreated.
P = <0.01 compared to untreated.
These in vivo experiments showed a profound effect of neutropenia on the course of Salmonella infection, which implies that PMN kill Salmonella. We tested this hypothesis in vitro using human PMN because they are easier to obtain in large amounts than mouse PMN. S. dublin was killed rapidly by human PMN, regardless of whether or not the bacteria carry a virulence plasmid. In the presence of 50% normal human serum as an opsonin, 90% of the inoculum was killed within the first 30 min. Without serum, there was neither phagocytosis nor killing (data not shown). With lesser percentages of human serum, we had less phagocytosis, hence less killing (not shown). We repeated the bacterial killing experiment with murine peritoneal exudate cells obtained from mice 3 hr after treatment with casein and fresh mouse serum (32). The mouse cells (≈75% PMN) were equally bactericidal for S. dublin as human PMN (not shown).
DISCUSSION
Recently, we found that intestinal epithelial cells make interleukin 8 and other chemokines in response to invasion by Salmonella, which implies that PMN play a role of the host defense against Salmonella (33, 34), and Conlan and North (24) published their observation that neutropenia exacerbates S. typhimurium infection in mice. We now show the deleterious effect of neutropenia on mice infected with S. dublin, but we have found that the importance of PMN depends on the host’s Nramp1 allele and on whether S. dublin has its virulence plasmid. To summarize our findings, neutropenia severely compromised the ability of congenic mice (normal Nramp1 allele) to control the growth of S. dublin lane, (plasmid +) (Fig. 2a). However, if the S. dublin did not have a virulence plasmid (LD842), then PMN were not required to suppress bacterial multiplication (Fig. 2b), indicating that normal macrophages are sufficient to deal with nontyphoid Salmonella that lack a virulence plasmid but that they are not sufficient to defend against S. dublin with a virulence plasmid. In contrast, neutropenic BALB/c mice were unable to control the growth of LD842 (Fig. 3), revealing the profound functional impairment that results from the Nramp1 mutation. Thus, by making mice neutropenic before we infected them with S. dublin lane, we not only confirmed the important role of PMN in this systemic infection, but we revealed the nearly total inability of BALB/c macrophages to limit the growth of nontyphoid Salmonella in vivo, let alone to kill those bacteria.
Our results also suggest that tissue macrophages in nonimmune mice cannot effectively inhibit the growth of Salmonella that carry a virulence plasmid. Others have shown (35) that the virulence plasmid enhances the growth rate of Salmonella in the liver and spleen. The current experiments show that the virulence plasmid has a much greater effect on the ability of S. dublin to grow in reticuloendothelial system organs than previously believed. In congenic mice, S. dublin lane grew nearly twice as rapidly as LD842 between day 1 and 3 after infection, but the most dramatic difference between the two strains was seen after day 3 of infection. There was no net growth of LD842 in neutropenic congenic mice after day 3 whereas S. dublin lane multiplied nearly as fast in neutropenic mice in the last 3 days as it did in the first 3 days after infection, and three of four mice died before day 6 (Fig. 2). This implies that macrophages became activated in vivo after a few days of infection because they acquired the ability to prevent further growth of LD842 even in the absence of PMN. However, the same macrophages could not prevent the growth of S. dublin (plasmid+) in the absence of PMN. In other words, the virulence plasmid functions to negate the antimicrobial activity of “activated” macrophages. In contrast, macrophage activation did not seem to occur in mice with a mutant Nramp1 because LD842 grew exponentially in neutropenic BALB/c mice until they died. Although BALB/c mice appear to be resistant to LD842, resistance was entirely caused by PMN.
Using cultured peritoneal macrophages, we were not able to confirm that macrophages from BALB/c.D2 congenics had more acquired antimicrobial activity than BALB/c macrophages for LD842. Our assumption was that, by using macrophages from infected mice, the macrophages would have been activated in vivo. [Others have shown (36, 37) that interferon-γ and tumor necrosis factor-α are required for resistance to salmonella in vivo (36, 37), but there may be other mediators of macrophage activation.] It is possible that activating signals act only locally at sites of infection, so that the peritoneal macrophages were not actually activated cells because i.v.-injected salmonella do not enter the peritoneum (J.F., unpublished observation). It is also possible that bacteria grown overnight in trypticase soy broth were not susceptible to macrophage killing. We know that salmonella change their phenotype in vivo, including altering their lipopolysaccharide (38).
In vitro, PMN were very efficient at killing Salmonella. The bactericidal effect of PMN against Salmonella has been reported by several groups (39–42). The significance of those observations has not been appreciated by most investigators, although some have speculated that the invasion of host cells was a mechanism used by salmonella to escape from PMN (43). The increased severity of Salmonella infection in neutropenic mice confirms that PMN kill Salmonella in vivo as well as they do in vitro. In contrast, unactivated macrophages have limited bactericidal activity against virulent Salmonella in vitro (44), and in vivo Salmonella grow quite well in reticuloendothelial system organs (27, 45).
Based on direct observation of bacteria in hepatocytes, Conlan (24) suggested that they were an important site of bacterial multiplication in neutropenic mice. Our results do not exclude the possibility that hepatocytes are infected by Salmonella, but they suggest that macrophages are the functionally important site of bacterial growth in neutropenic mice. First, within hours after i.v. injection, the Salmonella were protected from the bactericidal effect of amikacin but were killed by ciprofloxacin, which is consistent with the bacteria being in an intracellular location at the start of the infection. Second, the Nramp1 gene had a tremendous effect on the course of infection in neutropenic mice, which, because the Nramp1 gene is expressed in macrophages and not hepatocytes (18), implies that the bacteria were in the former. Finally, macrophage-sensitive mutants of Salmonella did not become virulent in neutropenic mice. Because those bacteria are not impaired in their growth in cells other than macrophages, their failure to grow more rapidly in neutropenic mice again suggests they were inside macrophages in vivo. However, because the rate of growth of bacteria was slightly faster in the livers than in the spleens of neutropenic mice, they may be multiplying inside hepatocytes as well as in Kupffer cells.
Our conclusion that Salmonella are primarily intracellular in phagocytes in vivo is corroborated in a recent report by Richter-Dahlfors et al. (46). They infected BALB/c mice with ≈100 cfu of S. typhimurium (all mice died by day 6), and made thick sections of the liver that were examined by confocal microscopy. With immunostaining, they co-localized Salmonella and CD18 positive cells and concluded that the CD18(+) cells were mostly macrophages. Even though they did not find S. typhimurium associated with PMN, their findings are consistent with our results. Because salmonella multiply inside macrophages but PMN kill salmonella quickly, one would expect to see many bacteria inside macrophages and very few in PMN at any time point.
Based on our results, we would like to propose the following hypothesis to explain the resistance of mice to systemic Salmonella infections during the pre-immune phase of infection. We propose that Salmonella initially are ingested by macrophages in the liver and spleen, but, if the salmonella contain a virulence plasmid, they can grow inside macrophages. We assume that the growing salmonella kill the macrophages, and, when released, the bacteria are vulnerable to phagocytosis and killing by PMN; virulence plasmids do not protect Salmonella from killing by PMN. If infected macrophages die by apoptosis (47, 48) in vivo, as they do in vitro, this would be a host defense mechanism rather than a virulence factor for the Salmonella. Thus, rather than using a capsule to avoid phagocytosis by PMN, nontyphoid Salmonella have adapted to invade “hospitable” macrophages to avoid PMN. The adaptations that enable Salmonella to grow in macrophages are very complex, and mutations that affect that adaptation make the organism avirulent, as they are then subject to attack by both kinds of phagocytes. Thus, PMN are the primary host defense against nontyphoid Salmonella that carry virulence plasmids, and macrophage invasion is the principal bacterial strategy.
Acknowledgments
We thank R. Haubrich and the University of California at San Diego Treatment Center Biostatistical Unit for help with statistical analysis. This work was supported by National Institutes of Health Grant DK35108.
ABBREVIATIONS
- PMN
polymorphonuclear leukocytes
- cfu
colony-forming units
References
- 1.Sanders E, Brachman P S, Friedman E A, Goldsby J, McCall C E. Am J Epidemiol. 1965;81:370–384. doi: 10.1093/oxfordjournals.aje.a120523. [DOI] [PubMed] [Google Scholar]
- 2.Rubin R H, Weinstein L. Salmonellosis: Microbiologic, Pathologic, and Clinical Features. New York: Stratton Intercontinental Medical Book Corporation; 1977. [Google Scholar]
- 3.Roantree R J. In: Microbial Toxins. Weinbaum G, Kadis S, Ajl S J, editors. Vol. 5. New York: Academic; 1971. pp. 1–37. [Google Scholar]
- 4.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coynault C, Robbe-Saule V, Norel F. Mol Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 6.Groisman E A, Chiao E, Lipps C J, Heffron F. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller S I, Kukral A M, Mekalanos J J. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alpuche-Aranda C M, Racoosin E L, Swanson J A, Miller S I. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo C M, Galan J E. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochman H, Soncini F C, Solomon F, Groisman E A. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shea J E, Hensel M, Gleeson C, Holden D W. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiney, D. G., Fang, F. C., Krause, M., Libby, S., Buchmeier, N. A. & Fierer, J. (1995) Clin. Infect. Dis. 21 Suppl. 2, S146–S151. [DOI] [PubMed]
- 13.Roudier C, Krause M, Fierer J, Guiney D G. Infect Immun. 1990;58:1180–1185. doi: 10.1128/iai.58.5.1180-1185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fierer J, Krause M, Tauxe R, Guiney D. J Infect Dis. 1992;166:639–642. doi: 10.1093/infdis/166.3.639. [DOI] [PubMed] [Google Scholar]
- 15.Helmuth R, Stephan R, Bunge C, Hoog B, Steinbeck A, Bulling E. Infect Immun. 1985;48:175–182. doi: 10.1128/iai.48.1.175-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodward M J, McLaren I, Wray C. J Gen Microbiol. 1989;135:503–511. doi: 10.1099/00221287-135-3-503. [DOI] [PubMed] [Google Scholar]
- 17.O’Brien A D, Rosenstreich D L. J Immunol. 1983;131:2613–2615. [PubMed] [Google Scholar]
- 18.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 19.Govoni G, Vidal S, Cellier M, Lepage P, Malo D, Gros P. Genomics. 1995;27:9–19. doi: 10.1006/geno.1995.1002. [DOI] [PubMed] [Google Scholar]
- 20.Chikami G K, Fierer J, Guiney D G. Infect Immun. 1985;50:420–424. doi: 10.1128/iai.50.2.420-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckmann L, Fierer J, Kagnoff M F. J Immunol. 1996;156:2894–2900. [PubMed] [Google Scholar]
- 22.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields P I, Swanson R V, Haidaris C G, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conlan J W, North R J. Infect Immun. 1992;60:5164–5171. doi: 10.1128/iai.60.12.5164-5171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conlan J W. Infect Immun. 1997;65:630–635. doi: 10.1128/iai.65.2.630-635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter M, O’Brien A D, Skamene E, Gros P, Forget A, Kongshavn P A L, Wax J S. Infect Immun. 1983;40:1234–1235. doi: 10.1128/iai.40.3.1234-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan E J, Fierer J, Chikami G, Guiney D. J Infect Dis. 1987;155:1254–1259. doi: 10.1093/infdis/155.6.1254. [DOI] [PubMed] [Google Scholar]
- 28.Hsu H S. Microbiol Rev. 1989;53:390–409. doi: 10.1128/mr.53.4.390-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groisman E A, Saier M H., Jr Trends Biochem Sci. 1990;15:30–33. doi: 10.1016/0968-0004(90)90128-x. [DOI] [PubMed] [Google Scholar]
- 30.Sigwart D F, Stocker B A D, Clements J D. Infect Immun. 1989;57:1858–1861. doi: 10.1128/iai.57.6.1858-1861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Easmon C S F, Crane J P. J Clin Pathol. 1985;38:442–444. doi: 10.1136/jcp.38.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf D, Robb L, Dunn A R, Mifsud S, Di Rago L. Blood. 1996;88:3755–3764. [PubMed] [Google Scholar]
- 33.Eckmann L, Kagnoff M F, Fierer J. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung H C, Eckmann L, Yang S-K, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff M F. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gulig P A, Doyle T J. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauciel C, Espinasse-Maes F. Infect Immun. 1992;60:450–454. doi: 10.1128/iai.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastroeni P, Skepper J N, Hormaeche C E. Infect Immun. 1995;63:3674–3682. doi: 10.1128/iai.63.9.3674-3682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 39.Steigbigel R T, Lambert L H, Remington J S. J Clin Invest. 1974;53:131–142. doi: 10.1172/JCI107531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandell G L. Infect Immun. 1974;9:337–341. doi: 10.1128/iai.9.2.337-341.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papp-Szabo E, Firtel M, Josephy P D. Infect Immun. 1994;62:2662–2668. doi: 10.1128/iai.62.7.2662-2668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiloh M U, Ruan J, Nathan C. Infect Immun. 1997;65:3193–3198. doi: 10.1128/iai.65.8.3193-3198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller S I, Hohmann E L, Pegues D A. In: Principles and Practice of Infectious Diseases. Mandell G L, Bennett J E, Dolin R, editors. New York: Churchill Livingstone; 1995. pp. 2013–2033. [Google Scholar]
- 44.Buchmeier N A, Heffron F. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langermans J A M, van der Hulst M E B, Nibbering P H, van Furth R. J Immunol. 1990;144:4340–4346. [PubMed] [Google Scholar]
- 46.Richter-Dahlfors A, Buchan A M J, Finlay B B. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L M, Kaniga K, Galan J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 48.Monack D M, Raupach B, Hromockyj A E, Falkow S. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]