Abstract
Although infection by primary HIV type 1 (HIV-1) isolates normally requires the functional interaction of the viral envelope protein with both CD4 and the CCR-5 coreceptor, a subset of such isolates also are able to use the distinct CCR-3 receptor. By analyzing the ability of a series of wild-type and chimeric HIV-1 envelope proteins to mediate CCR-3-dependent infection, we have determined that CCR-3 tropism maps to the V1 and V2 variable region of envelope. Although substitution of the V1/V2 region of a CCR-3 tropic envelope into the context of a CCR-5 tropic envelope is both necessary and sufficient to confer CCR-3 tropism, this same substitution has no phenotypic effect when inserted into a CXCR-4 tropic HIV-1 envelope context. However, this latter chimera acquires both CCR-3 and CCR-5 tropism when a CCR-5 tropic V3 loop sequence also is introduced. These data demonstrate that the V1/2 region of envelope can, like the V3 loop region, encode a particular coreceptor requirement and suggest that a functional envelope:CCR-3 interaction may depend on the cooperative interaction of CCR-3 with both the V1/V2 and the V3 region of envelope.
HIV type 1 (HIV-1) infection normally requires the functional interaction of the viral envelope glycoprotein with at least two cell surface molecules (reviewed in refs. 1–3). These are the CD4 primary receptor and a coreceptor, belonging to the chemokine receptor family of seven-membrane spanning receptors, that can vary depending on the identity of the particular HIV-1 isolate under study. The majority of patient HIV-1 isolates use CCR-5 as a coreceptor, and this CCR-5 tropism correlates with the ability of such isolates to infect primary macrophages (M-tropism) (4–7). In contrast, so-called “laboratory-adapted” isolates of HIV-1, as well as patient isolates able to induce syncitium formation in culture, generally use a distinct coreceptor molecule termed “CXCR-4” either instead of, or in addition to, CCR-5 (8–11).
In addition to CCR-5 and CXCR-4, a number of other chemokine receptors also have been reported to function as coreceptors for a subset of HIV-1 isolates (7, 9, 12–14). Of these, the most prevalent is probably CCR-3, a chemokine receptor that is expressed in vivo on eosinophils and basophils as well as on microglial cells and on a small percentage of lymphocytes of the T helper 2-cell type (15, 16). Although CCR-3 does not therefore contribute to HIV-1 infection of macrophages or the large majority of CD4+ lymphocytes, the presence of CCR-3 on microglial cells could be important in that these CD4+ myeloid cells serve as the major target for HIV-1 replication in the brain (17). Recently, direct evidence demonstrating that CCR-3 can serve as a functional coreceptor for microglial cell infection by HIV-1 has been presented (15). Because brain microglial cell infection may contribute significantly to the development of dementia in AIDS patients (17), the appearance of CCR-3 tropism could have a significant impact on the pathogenic potential of HIV-1. Indeed, it has been reported that the expansion of HIV-1 coreceptor usage to include CCR-3 can be observed in a significant proportion of patients displaying disease progression (12).
In this manuscript, we attempt to define which regions within the HIV-1 envelope glycoprotein confer CCR-3 tropism, and we report that CCR-3 tropism depends on specific sequences located within the variable V1/V2 region of envelope. Although CCR-3 tropism therefore is determined by a different region of envelope than CXCR-4/CCR-5 tropism, which largely maps to the V3 loop (7, 18–21), CCR-3 tropism does require the presence of a CCR-5 tropic V3 loop sequence in cis. These data, obtained by using several CCR-3 tropic and nontropic HIV-1 isolates, therefore identify a specific region of the HIV-1 envelope as the major determinant of CCR-3 tropism and hence support the hypothesis that CCR-3 tropism is likely to be a selected viral phenotype. In addition, these data demonstrate that two distinct regions of the HIV-1 envelope, i.e., the V1/V2 region and the V3 loop, can cooperate in mediating the interaction of envelope with a specific coreceptor molecule.
MATERIALS AND METHODS
Construction of Molecular Clones and Chimeras.
Mammalian expression plasmids encoding full length human CCR5 (pCMV5/CCR-5) and CXCR4 (pCMV5/CXCR-4) bearing an amino-terminal influenza hemaglutinin (HA) epitope tag have been described (21). A similar expression plasmid encoding human CCR-3 was generated by PCR amplification of a full length CCR-3 cDNA clone by using DNA primers that inserted a unique NcoI site coincident with the CCR-3 translation initiation codon (5′-CCATGG-3′) and a unique XhoI site immediately 3′ to the CCR-3 translation termination codon. After cleavage with NcoI and XhoI, this fragment was substituted into the pCMV5/CCR-5 expression plasmid in place of CCR-5. In this context, the CCR-3 ORF is expressed under the control of the cytomegalovirus (CMV) immediate early promoter and is linked to the 5′ leader of rat preproinsulin (22) and to sequences encoding an amino-terminal HA epitope tag.
The wild-type HIV-1 ADA, BaL, JR-FL, YU-2, and IIIB isolate env genes (18, 20, 23, 24) were expressed by using the pCR3.1 plasmid (Invitrogen) as described (25), and unique restriction sites were used to generate chimeras in this same vector context (Fig. 1). To simplify nomenclature, the following abbreviations are used for each of the cloned envs: ADA (A), BaL (B), JR-FL (J), YU-2 (Y), and IIIB (T). The chimeric IIIB/V3-BaL env gene has been described (18) and was derived by the precise replacement of the V3 loop of IIIB with the V3 loop of BaL. Each of the chimeric env genes was designed to substitute one or more of the five hypervariable regions of the gp120 component of the HIV-1 envelope, designated V1 to V5. For example, the A1–2/B envelope chimera contains ADA envelope sequences encoding hypervariable regions V1 and V2 substituted into the BaL envelope context. A total of 11 envelope chimeras was constructed (Fig. 1). For fusion assays, each wild-type or chimeric envelope was substituted in place of the env gene present in the previously described pIIIB proviral expression plasmid (18, 21).
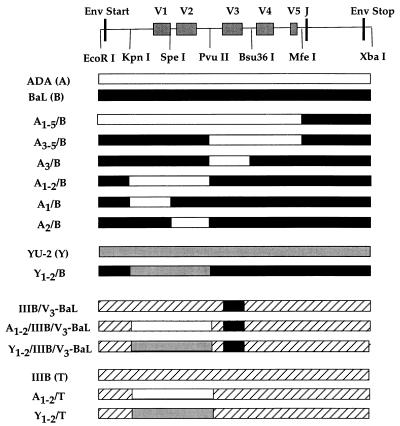
Figure 1.
Structure of envelope chimeras. Wild-type and chimeric envelope proteins were expressed either in the context of the pCR3.1 plasmid or in an HIV-1 proviral context. Chimeric env genes were constructed by using the indicated restriction enzyme sites in the gp120 coding region. By using coordinates based on the 504 amino acid (aa) ADA gp120 protein, we inserted the following ADA envelope sequences into the BaL envelope: A1–5/B, aa 1–473; A3–5/B, aa 284–473; A3/B, aa 284–359; A1–2/B, aa 42–284; A1/B, aa 42–145; and A2/B, aa 145–284. All other chimeras substituted equivalent env sequences with the exception of IIIB/V3-BaL, which has been described previously (18). J, junction between gp120 and gp41.
Virus Infection and Cell Fusion Assays for Coreceptor Function.
293T and COS cells were maintained as described (18, 21). Luciferase reporter viruses were prepared by lipofectamine-mediated transfection of a 35-mm culture of 293T cells by using 1 μg of the indicated env expression plasmid together with 1 μg of a second plasmid encoding an HIV-1 provirus lacking a functional env gene and bearing the luciferase indicator gene in place of the nef gene (pNL-Luc-E-R-) (12, 26). Two days after transfection, the virus containing supernatant media (3 ml total) was harvested and passed through a 0.2-μM filter, and 1 ml was used to infect each 35-mm target cell culture. Normally, target cells for infection were generated by the lipofectamine-mediated transfection of 293T cells (35-mm culture) with 50 ng of pCMV5/CD4, 50 ng of a pCMV5-based coreceptor expression plasmid, and 900 ng of the parental pCMV5 plasmid. Alternatively, target COS cells were transfected with 800 ng of pCMV5/CD4 and 800 ng of a pCMV5-based coreceptor expression plasmid. Infected target cells were lysed 4 days postinfection and then assayed for luciferase activity by using commercially available reagents (Promega).
For fusion assays, indicator cells were generated by cotransfection of 293T cells (35-mm cultures) by using lipofectamine with 400 ng of pCMV5/CD4, 400 ng of pC5/HIV/SEAP, and 50 ng of a pCMV5-based coreceptor expression plasmid (21). Simultaneously, virus-producing cells were generated by transfection of 293T cells with 2 μg of the indicated proviral construct. At 48 h posttransfection, producer and indicator cells were harvested by trypsinization and equal numbers (5 × 104) were co-cultivated in 48-well plates. After 48 h, culture supernatants were harvested and SEAP activity was determined as described (21).
RESULTS
Previously, there has been significant controversy as to which HIV-1 isolates are able to use CCR-3 as a coreceptor with, for example, the JR-FL HIV-1 isolate being described as both unable (5, 6, 11) and able (12, 14, 15) to use CCR-3. It has been suggested that these discrepancies are due to the difficulty of expressing CCR-3 at significant levels in transfected or transduced cells (13). We previously described (21) pCMV5-based expression plasmids for human CCR-5 and CXCR-4 in which these ORFs are expressed under the control of the CMV immediate early promoter and linked, at the translation initiation codon, to the leader region of rat preproinsulin, which can facilitate efficient translation (22), and to an HA epitope tag. A similar pCMV5-based CCR-3 expression plasmid was constructed, and the ability of CCR-3 to function as a coreceptor for HIV-1 infection was determined.
To measure coreceptor activity, we used the previously described pNL-Luc-E-R- HIV-1 proviral expression plasmid (13, 26). pNL-Luc-E-R- contains a full length NL4–3 HIV-1 provirus in which the env gene has been inactivated by a frame-shift mutation and the luciferase indicator gene substituted in place of nef. Virions produced upon transfection of pNL-Luc-E-R- into 293T cells are not infectious but can be rescued by pseudotyping with an env gene expressed from a cotransfected plasmid. Infection of cells with these virions results in a single round of replication and produces readily detectable luciferase activity.
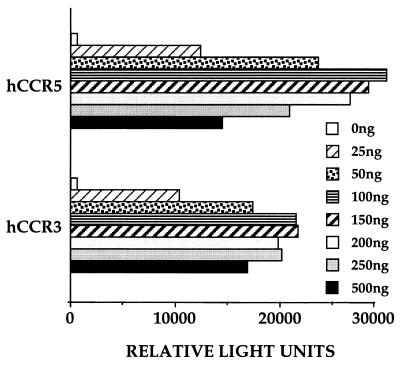
To examine the ability of CCR-3 to function as a coreceptor for HIV-1 infection, we transfected 293T cells with 50 ng of the pCMV5/CD4 expression plasmid and increasing levels of either pCMV5/CCR-5 or pCMV5/CCR-3. Total levels of transfected DNA were maintained at 1 μg/35-mm culture by supplementation with the parental pCMV5 vector, and the level of coreceptor expression therefore is predicted to be directly proportional to the level of the transfected coreceptor expression plasmid. At 48 h after transfection, these target cells were infected with a pNL-Luc-E-R- virus preparation pseudotyped with the HIV-1 isolate ADA envelope protein, which has been shown to use both CCR-5 and CCR-3 as a coreceptor (7, 15). As can be seen in Fig. 2, CCR-3 proved only marginally less effective than CCR-5 in mediating HIV-1 strain ADA envelope-dependent infection. Both coreceptors also gave a very similar dose response, with both CCR-3 and CCR-5 showing maximal infection at between 100 and 200 ng of transfected coreceptor expression plasmid. FACS analysis of 293T cells transfected with either pCMV5/CCR-5 or pCMV5/CCR-3, using a mAb directed against the HA epitope tag introduced at the amino terminus of both chemokine receptors, demonstrated that CCR-5 was expressed at a 2- to 3-fold higher level on the cell surface than was CCR-3 (data not shown). Based on these data, all subsequent experiments were performed by using 50 ng of coreceptor expression plasmid per transfection, which gave substantial but not maximal HIV-1 infection efficiency (Fig. 2).
Figure 2.
Effect of coreceptor expression levels on ADA infection efficiency. Target 293T cells were transfected with 50 ng of pCMV5/CD4 and increasing levels of either pCMV5/CCR-5 or pCMV5/CCR-3. Total transfected DNA was maintained at 1 μg by addition of the parental pCMV5 expression plasmid. At 48 h after transfection, target cells were infected with the pNL-Luc-E-R- HIV-1 indicator virus pseudotyped with the HIV-1 strain ADA envelope protein. The level of luciferase activity resulting from productive pNL-Luc-E-R- infection was determined 4 days later and is given as relative light units measured by using a luminometer.
CCR-3 Coreceptor Tropism Maps to the V1/V2 Region of Envelope.
We next wished to identify an M-tropic HIV-1 isolate that uses CCR-5 effectively yet is unable to use CCR-3. As shown in Fig. 3, the ADA, YU-2, JR-FL, and BaL envelopes all mediate efficient infection of cells bearing CCR-5 and CD4. However, although the ADA, YU-2, and JR-FL envelope also permit the efficient infection of CCR-3+ target cells, BaL is essentially inactive with CCR-3.
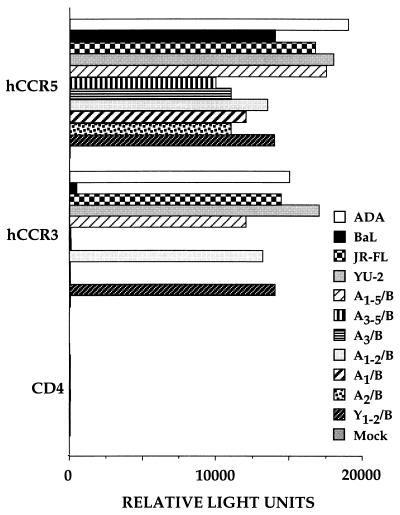
Figure 3.
CCR-3 tropism maps to the V1 and V2 hypervariable regions of gp120. The pNL-Luc-E-R- HIV-1 indicator construct was pseudotyped with the indicated wild-type and chimeric HIV-1 envelope proteins and the resultant virions used to infect 293T cells expressing CD4 alone or together with the CCR-5 or CCR-3 chemokine receptor. The resultant luciferase activity is indicated in relative light units.
To identify the region of envelope involved in mediating CCR-3 tropism, we next constructed a set of six chimeras between ADA and BaL, in which different regions of the ADA env gene and in particular different hypervariable regions, were substituted into the BaL env context (Fig. 1). As shown in Fig. 3, substitution of essentially the entire ADA gp120 coding region into BaL, in clone A1–5/B, conferred full CCR-3 tropism. In contrast, insertion of the ADA V3, V4, and V5 regions (A3–5/B) or of the ADA V3 loop region only (A3/B) failed to confer CCR-3 tropism on the BaL envelope, although infection via CCR-5 remained efficient. However, substitution of the combined V1/V2 region of ADA into BaL, in A1–2/B, did permit the efficient infection of CCR-3+ cells. Both V1 and V2 proved critical for this phenotype, in that BaL-based chimeras containing only the V1 (A1/B) or the V2 (A2/B) region of the ADA envelope proved unable to infect CCR-3+ cells, although these chimeras again remained fully functional on CCR-5+ cells (Fig. 3). To determine whether the ability of the V1/V2 region of env to confer CCR-3 tropism was unique to ADA, we prepared a similar chimera in which the BaL V1/V2 region was replaced by sequences derived from the CCR-3 tropic YU-2 isolate (Fig. 1). As shown in Fig. 3, the resultant Y1–2/B chimera, like the A1–2/B chimera, again displayed a marked tropism for CCR-3+ cells.
Functional Interaction of the HIV-1 Envelope with CCR-3 Requires a CCR-5 Tropic V3 Loop.
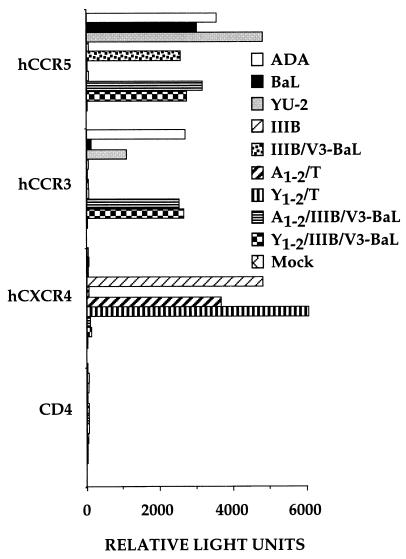
As noted above, the majority of primary HIV-1 isolates use CCR-5 as a coreceptor whereas laboratory-adapted HIV-1 isolates, as well as a subset of primary isolates that induce efficient syncytium formation in culture, generally use CXCR-4 (1–3). The tropism of HIV-1 isolates for CCR-5 vs. CXCR-4 largely is controlled by sequences located within the short V3 hypervariable loop (7, 18–20, 24). Thus, the precise substitution of the V3 loop of the CCR-5 tropic BaL isolate into the context of the CXCR-4 tropic IIIB isolate envelope protein, to give the IIIB/V3-BaL chimera, not only confers CCR-5 tropism on the IIIB envelope but also entirely blocks CXCR-4 tropism (18, 21). This result is confirmed in Fig. 4, which shows that the IIIB envelope permits infection of CXCR-4+ but not CCR-5+ COS cells, whereas the IIIB/V3-BaL env chimera facilitates infection of CCR-5+ but not of CXCR-4+ cells. Of interest, neither the IIIB envelope nor the IIIB/V3-BaL envelope was able to mediate infection of cells expressing CCR-3.
Figure 4.
Effect of V3 loop sequences on CCR-3 tropism. This experiment was performed as in Fig. 3, except that COS cells, which are CXCR-4-negative, were used as target cells for infection with the pseudotyped pNL-Luc-E-R- virions in place of 293T cells, which express CXCR-4.
To examine whether insertion of the V1/V2 region of either ADA or YU-2 would confer CCR-3 tropism on either the IIIB or the IIIB/V3-BaL envelope, we generated the appropriate envelope chimeras (Fig. 1) and tested their ability to infect COS cells expressing CCR-5, CCR-3, or CXCR-4. In the case of IIIB/V3-BaL, insertion of either the ADA V1/V2 region, in A1–2/IIIB/V3-BaL, or the YU-2 region, in Y1–2/IIIB/V3-BaL, proved fully sufficient to confer CCR-3 tropism (Fig. 4). In contrast, substitution of these same regions into the CXCR-4 tropic IIIB envelope had no detectable phenotypic effect. In particular, both the A1–2/T and the Y1–2/T chimera failed to display any ability to use CCR-3 yet fully retained their ability to mediate infection of cells bearing CXCR-4 (Fig. 4).
CCR-3-Mediated Cell Fusion.
The experiments presented thus far have measured the ability of different chemokine receptors to mediate HIV-1 infection. Alternately, it is also possible to measure the ability of CD4 and chemokine receptors to mediate fusion with cells expressing HIV-1 envelope proteins, a process that is critical for syncytium formation in infected cultures. We previously described a quantitative fusion assay in which cells expressing the HIV-1 envelope and Tat proteins are mixed with cells expressing CD4 and a candidate coreceptor and containing an indicator construct consisting of the HIV-1 long terminal repeat linked to the SEAP indicator gene (21). Fusion between these cells results in the Tat-mediated activation of the HIV-1 long terminal repeat promoter and, hence, in enhanced SEAP expression.
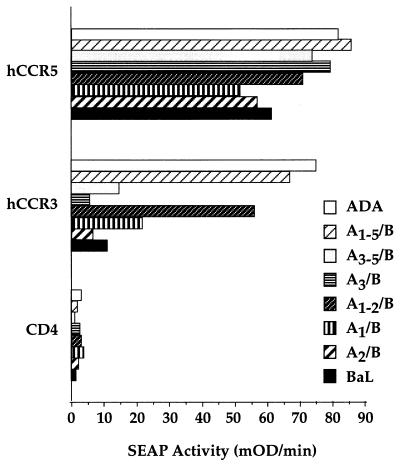
In Fig. 5, we have used this assay to measure the ability of wild-type and chimeric HIV-1 Env proteins to mediate fusion with CCR-3-expressing cells. As may be seen, this assay reproduces the observation that the ADA, but not the BaL, Env protein can interact functionally with CCR-3. Similarly, although both the A1–5/B and the A1–2/B chimeras displayed CCR-3 tropism in this fusion assay, the A3–5/B and A3/B chimeras again failed to use the CCR-3 molecule effectively. As in the case of the earlier infection data (Fig. 2), the individual substitution of either the V1 (A1/B) or the V2 (A2/B) loop of ADA into BaL again failed to confer the full CCR-3 tropism noted upon insertion of both V1 and V2, although there is the suggestion of a low degree of CCR-3 utilization with the A1/B chimera that was not observed in the infection assay (Fig. 5). Overall, these data confirm the finding that V1 and V2 together regulate CCR-3 tropism and demonstrate that this tropism can be measured effectively by using either infection or fusion assays for the HIV-1 envelope/co-receptor interaction.
Figure 5.
CCR-3-dependent fusion of HIV-1 envelope-expressing cells. The ability of coreceptors, acting in concert with CD4, to mediate fusion with cells expressing the indicated wild-type and mutant HIV-1 envelope proteins was determined as described in the text.
DISCUSSION
Although the importance of CCR-5 and CXCR-4 as coreceptors for HIV-1 is well established, it has remained unclear whether other chemokine receptors also can serve as coreceptors for HIV-1 infection in vivo (1–3). The significance of CCR-3 as a relevant coreceptor for HIV-1 has been particularly controversial, in that some early reports failed to observe HIV-1 envelope-mediated fusion or infection of cells transfected with a CCR-3 expression plasmid (5, 6, 11). It is important to note, however, that these early reports also failed to see any activity with envelope proteins from HIV-1 isolates, such as ADA and JR-FL, that have been shown subsequently to display a clear tropism for CCR-3 (7, 12, 14–16), thus leading to the suggestion that these earlier negative data may largely have reflected poor levels of cell surface CCR-3 expression (13). A subsequent report documenting CCR-3-dependent infection of primary microglial cells by several HIV-1 isolates (15) suggests that CCR-3 tropism is a real phenomenon and potentially of considerable significance in the development of HIV-1 pathogenesis. It is also of interest to note that human CCR-3 displays only ≈50% total amino acid sequence homology to human CCR-5, with a very low ≈24% sequence identity in the four receptor extracellular domains yet is effectively used by a number of HIV-1 isolates (7, 12, 14–16). In contrast, the murine CCR-5 receptor displays ≈82% total sequence identity and ≈79% identity over the four extracellular domains yet is not used by any HIV-1 isolate tested thus far (21, 27, 28). This discrepancy strongly suggests that CCR-3 tropism is likely to be a selected, rather than a fortuitous, viral phenotype.
By using both infection and fusion assays for coreceptor function, we have attempted to identify which regions within the HIV-1 envelope permit a functional interaction with CCR-3. Our starting points for this work were three envelope proteins, from the ADA, YU-2, and JR-FL isolates, that are able to interact functionally with both CCR-5 and CCR-3; one envelope, derived from the IIIB laboratory isolate, able to interact only with CXCR-4, and finally two envelopes, one from the BaL isolate and the second a synthetic chimera (IIIB/V3-BaL), containing the BaL V3 loop in an otherwise IIIB context, that can use only CCR-5. By generating envelope chimeras, we were able to demonstrate that insertion of either the ADA or the YU-2 V1/V2 envelope region into either the BaL or IIIB/V3-BaL envelope context was both necessary and sufficient to confer full CCR-3 tropism (Figs. 3–5). The fact that this same result was obtained with two distinct CCR-3 tropic “donor” env genes, i.e., ADA and YU-2, and with two equally distinct “recipient” env genes, i.e., BaL and IIIB/V3-BaL, suggests that the sequence of the V1/V2 domain of env genes present in M-tropic HIV-1 isolates is likely to be the major determinant of CCR-3 tropism. However, inspection of this region in the five viruses tested here does not reveal an obvious motif that correlates with CCR-3 tropism.
An interesting phenomenon, reported in Fig. 4, is that insertion of the ADA or YU-2 V1/V2 region into the IIIB envelope neither conferred CCR-3 tropism nor detectably inhibited the functional interaction of the IIIB envelope with CXCR-4. This finding suggests that the V1/V2 region of envelope only can confer CCR-3 tropism when the V3 loop is tropic for CCR-5, as is indeed seen in the otherwise identical IIIB/V3-BaL chimera. Therefore, in this instance, CCR-3 tropism presumably reflects the cooperative interaction of the V1/V2 domain and of the V3 loop with the CCR-3 coreceptor. This finding further emphasizes the complexity of the envelope:co-receptor interaction, which previously has been shown to involve at least three of the four extracellular domains of the coreceptor (21, 27–29).
Although we are unaware of any previous attempt to map CCR-3 tropism in the HIV-1 envelope in detail, Choe et al. (7) did examine the CCR-3 tropism of a small number of ADA/IIIB or YU-2/IIIB chimeric envelope proteins. In agreement with our observations (Fig. 4), they reported that substitution of the V1/V2 domain of the CCR-3 tropic YU-2 envelope into IIIB neither conferred CCR-3 tropism nor inhibited CXCR-4 tropism. However, Choe et al. (7) also reported that insertion of the ADA V3 loop into IIIB conferred not only CCR-5 but also CCR-3 tropism. In our hands, the ADA V3 loop, which differs by only one amino acid from the BaL V3 loop, did not confer any detectable CCR-3 tropism when substituted into either the BaL envelope (Fig. 3) or the IIIB envelope (data not shown), although it did confer CCR-5 tropism in the latter case. The reason for this discrepancy is not known currently.
Although CCR-5 vs. CXCR-4 tropism is regulated largely by the envelope V3 loop, there has been a number of reports documenting the ability of the V1/V2 region to modulate HIV-1 cell tropism and replication potential (30–33), although others have suggested only a minor role for the V1/V2 domain (34). Our data documenting a major role for V1/V2 in regulating the interaction with CCR-3 support these previous findings and suggest that V1/V2 is indeed likely to play a significant role in modulating the envelope:coreceptor interaction.
Acknowledgments
We thank Nathaniel Landau for the pNL-Luc-E-R- indicator plasmid, Robert Doms for the human CCR-3 cDNA and the JR-FL env gene, and Lee Ratner for the ADA env gene. The YU-2 env gene was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and was donated by Beatrice Hahn and George Shaw. This research was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, Research Center on AIDS and HIV Infection and by a grant from the National Institute of Allergy and Infectious Diseases (1R01AI42538) to B.R.C. T.M.R. is supported by the Duke Interdisciplinary Research Training Program in AIDS grant from the National Institute of Allergy and Infectious Diseases (2T32AI07392).
ABBREVIATIONS
- HIV-1
HIV type 1
- M-tropism
macrophage tropism
- CMV
cytomegalovirus
References
- 1.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 2.Doms R W, Peiper S C. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Cullen B R. Frontiers Biosci. 1998;3:44–58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 4.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 5.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, et al. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 11.Zhang L, Huang Y, He T, Cao Y, Ho D D. Nature (London) 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 12.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, et al. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 16.Sallusto F, Mackay C R, Lanzavecchia A. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 17.Sharpless N, Gilbert D, Bandercam B, Zhou J M, Verdin E, Ronnett G, Friedman E, Dubois-Dalcq M. Virology. 1992;191:813–825. doi: 10.1016/0042-6822(92)90257-p. [DOI] [PubMed] [Google Scholar]
- 18.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 19.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westervelt P, Gendelman H E, Ratner L. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen B R. DNA. 1988;7:645–650. doi: 10.1089/dna.1988.7.645. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. Nature (London) 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 25.Doranz B J, Lu Z-H, Rucker J, Zhang T-Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, et al. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 27.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 28.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, et al. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 30.Groenink M, Fouchier R A M, Broersen S, Baker C H, Koot M, van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 31.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N, Zhu T, Ho D D. J Virol. 1995;69:2708–2715. doi: 10.1128/jvi.69.4.2708-2715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]