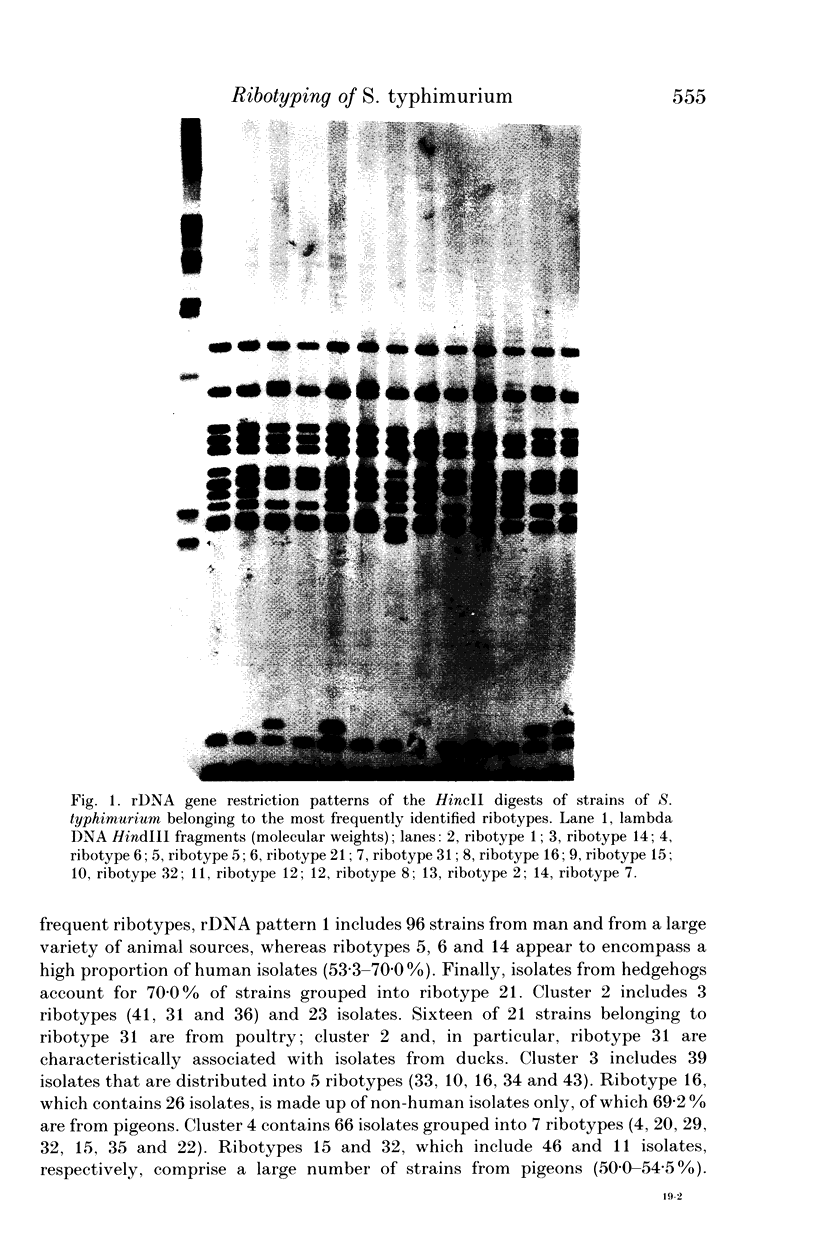
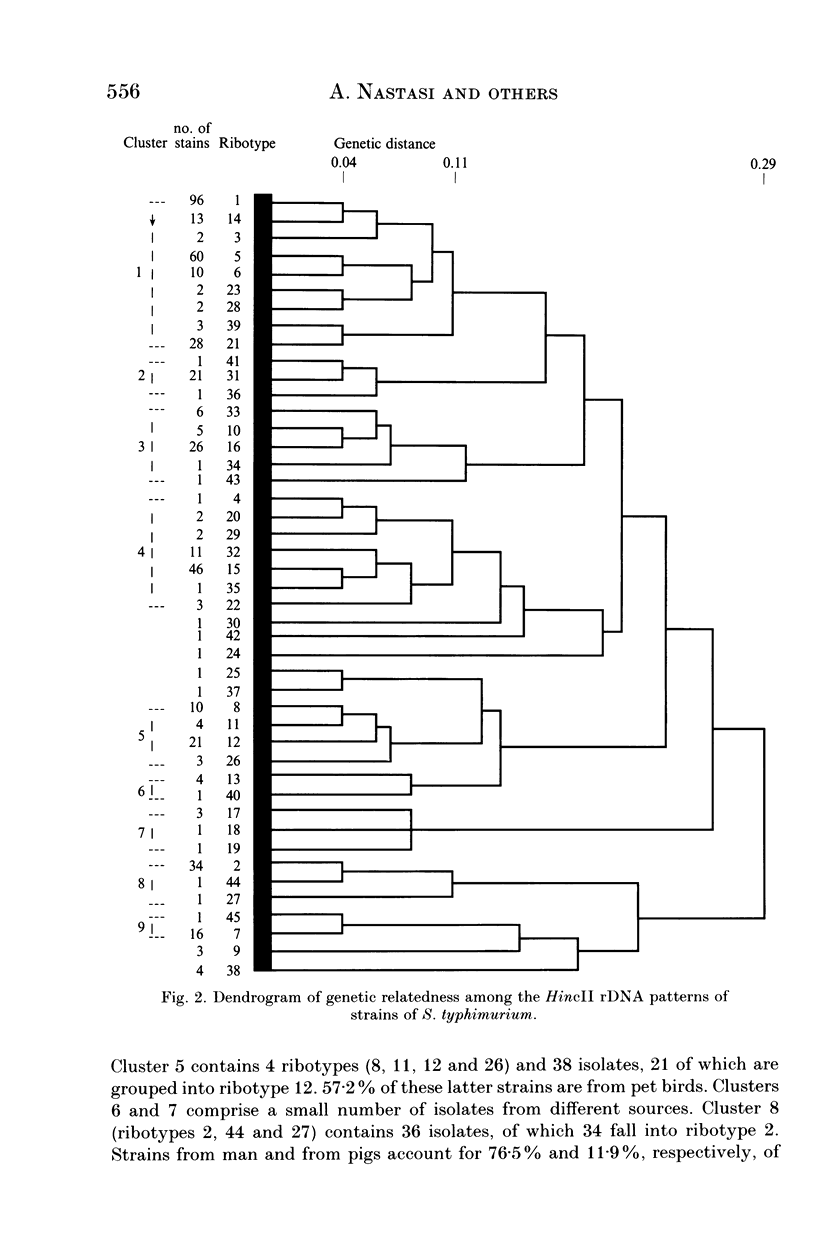
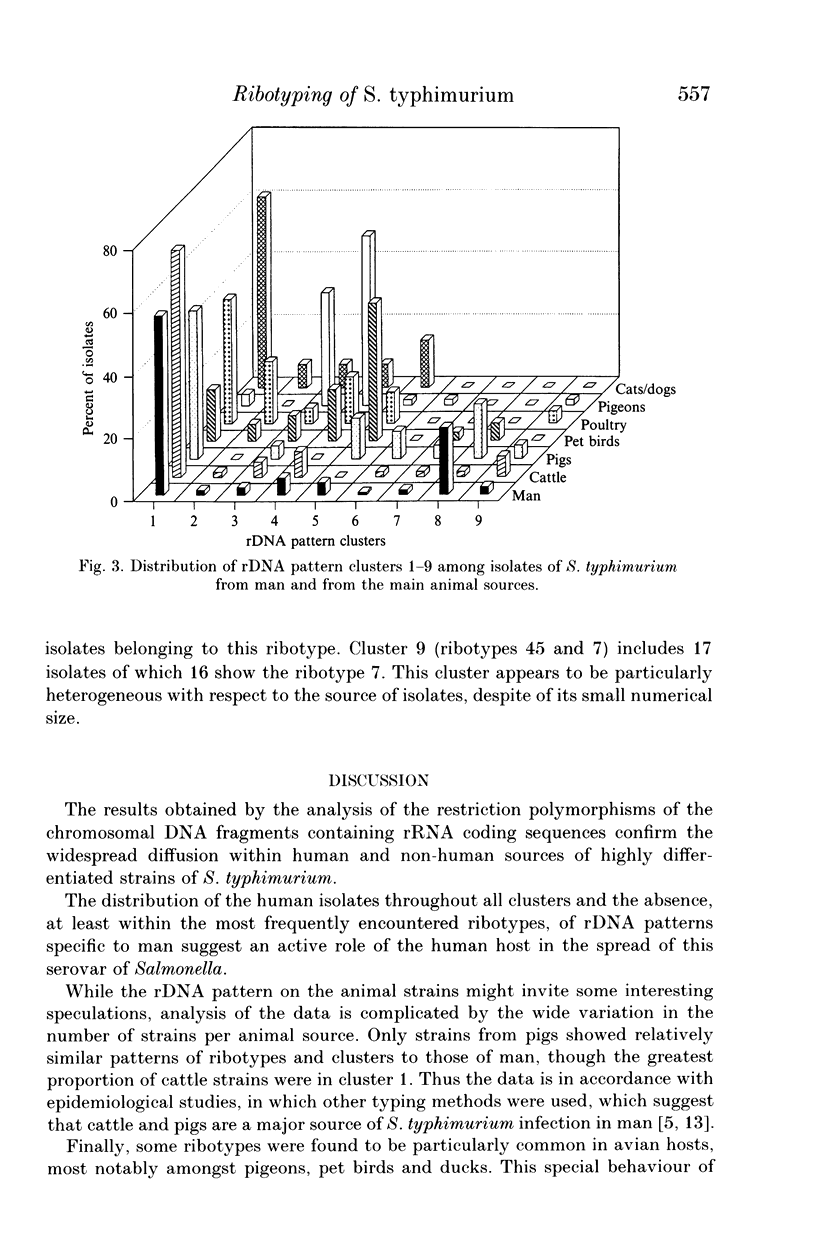
Abstract
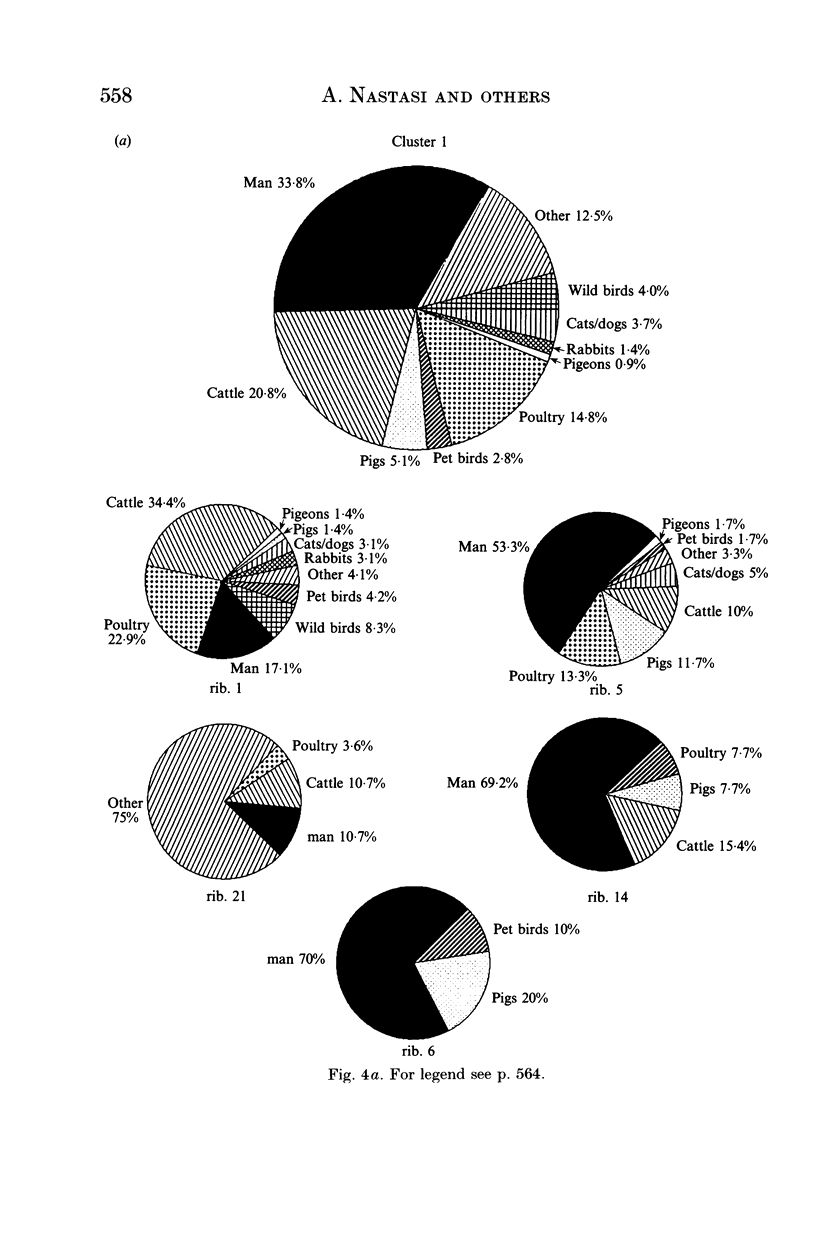
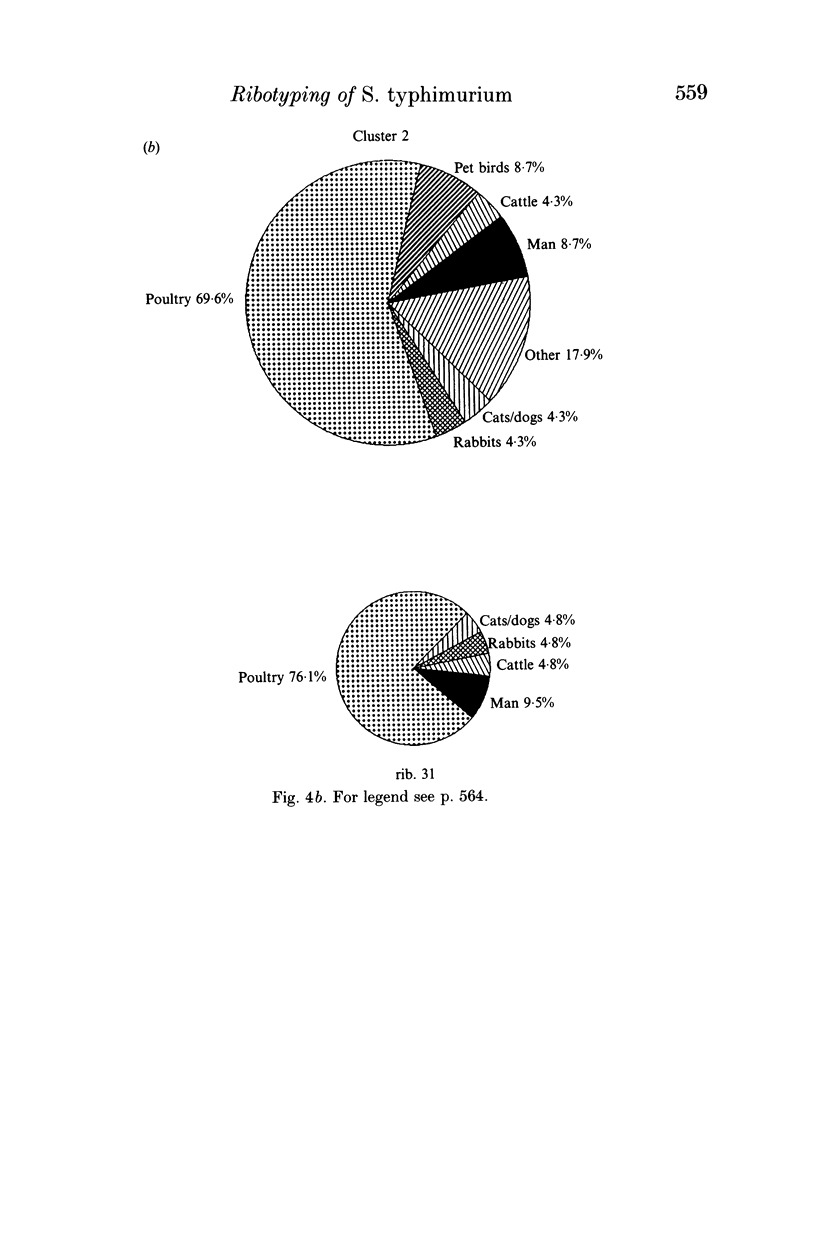
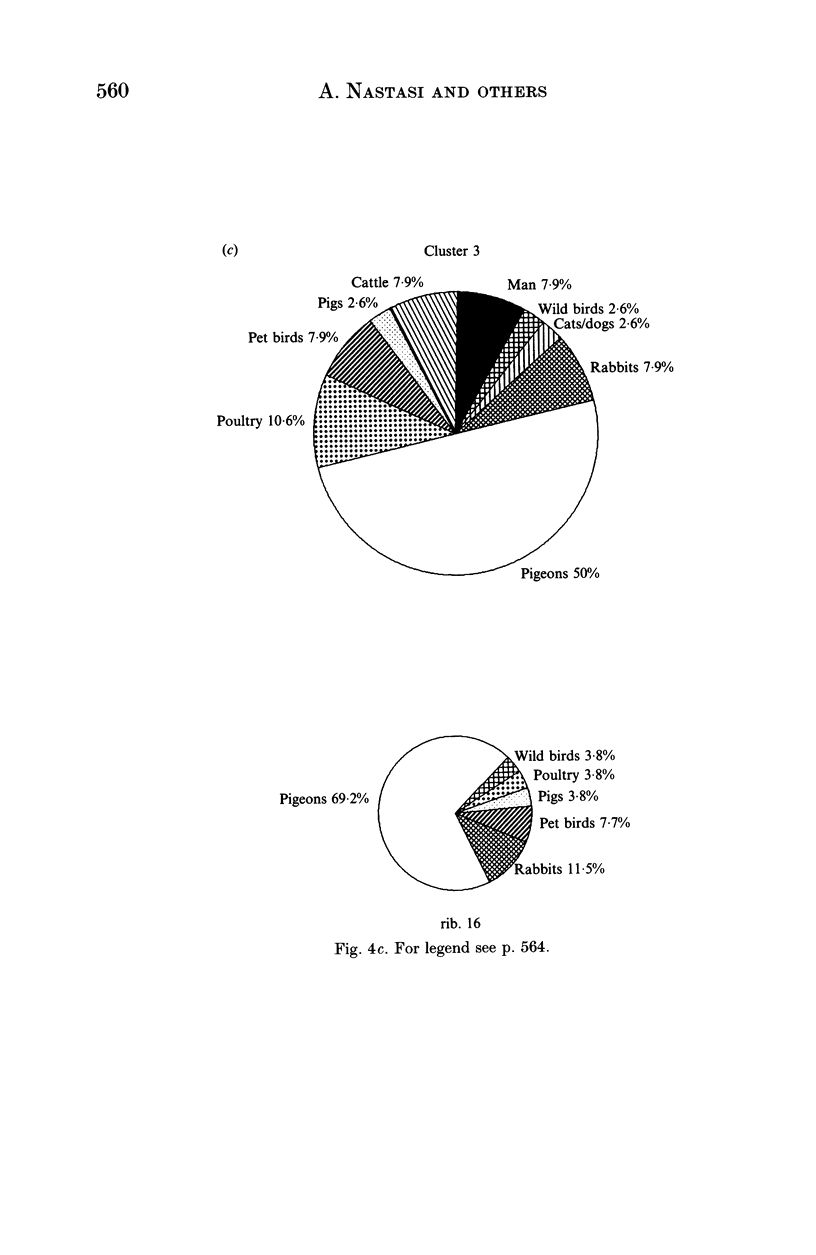
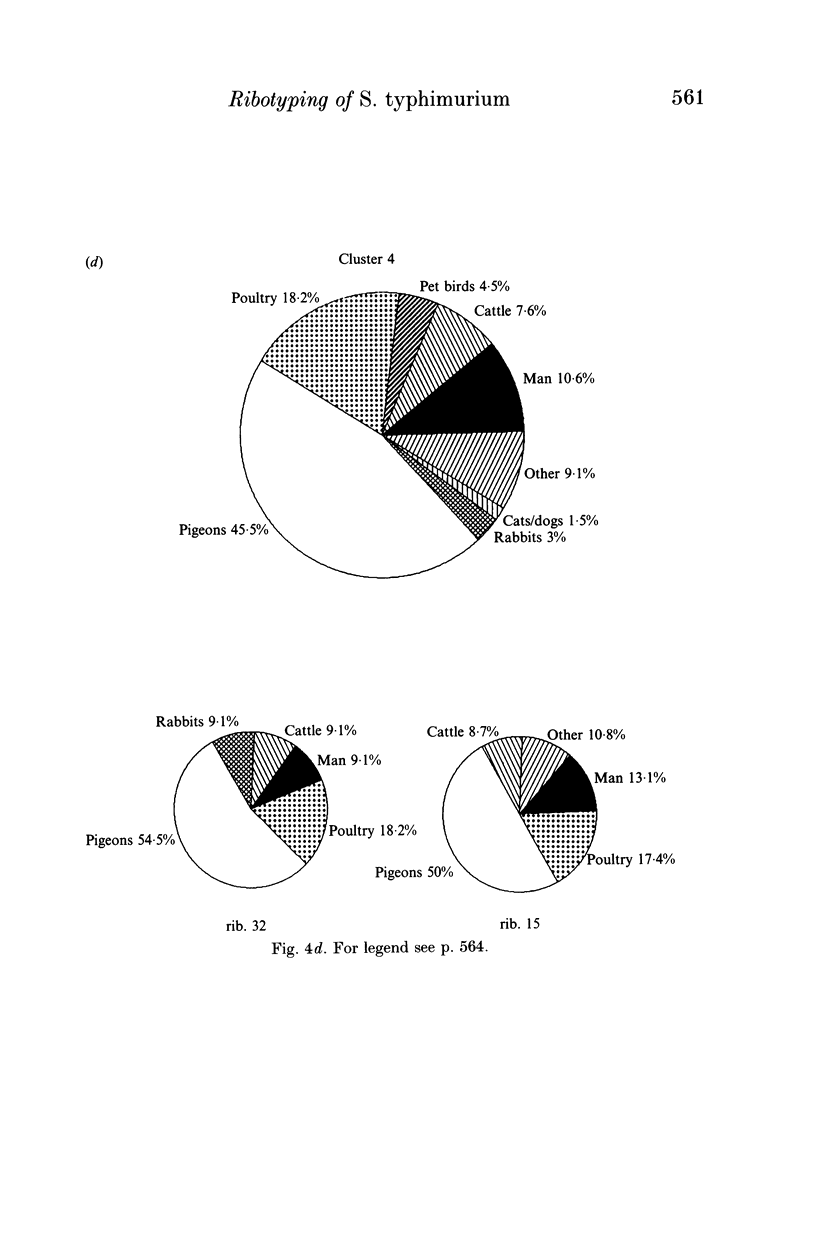
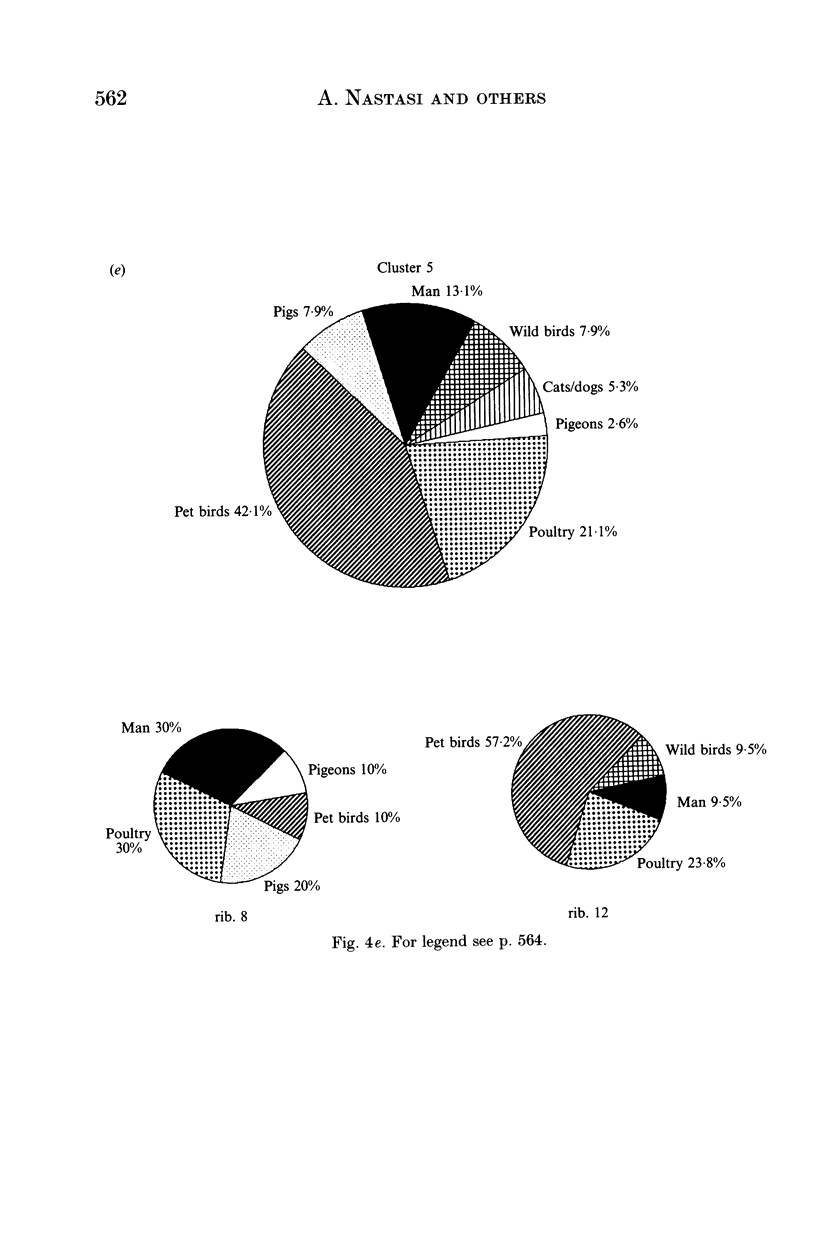
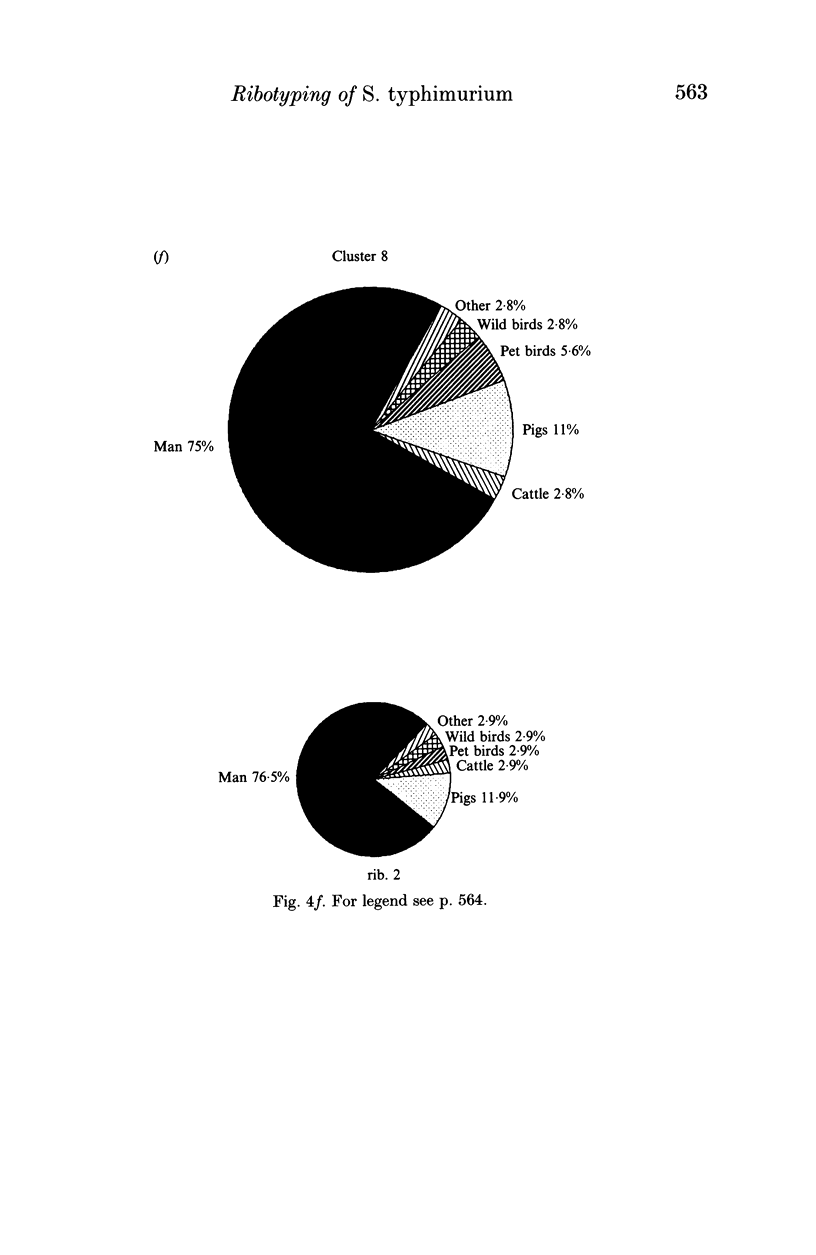
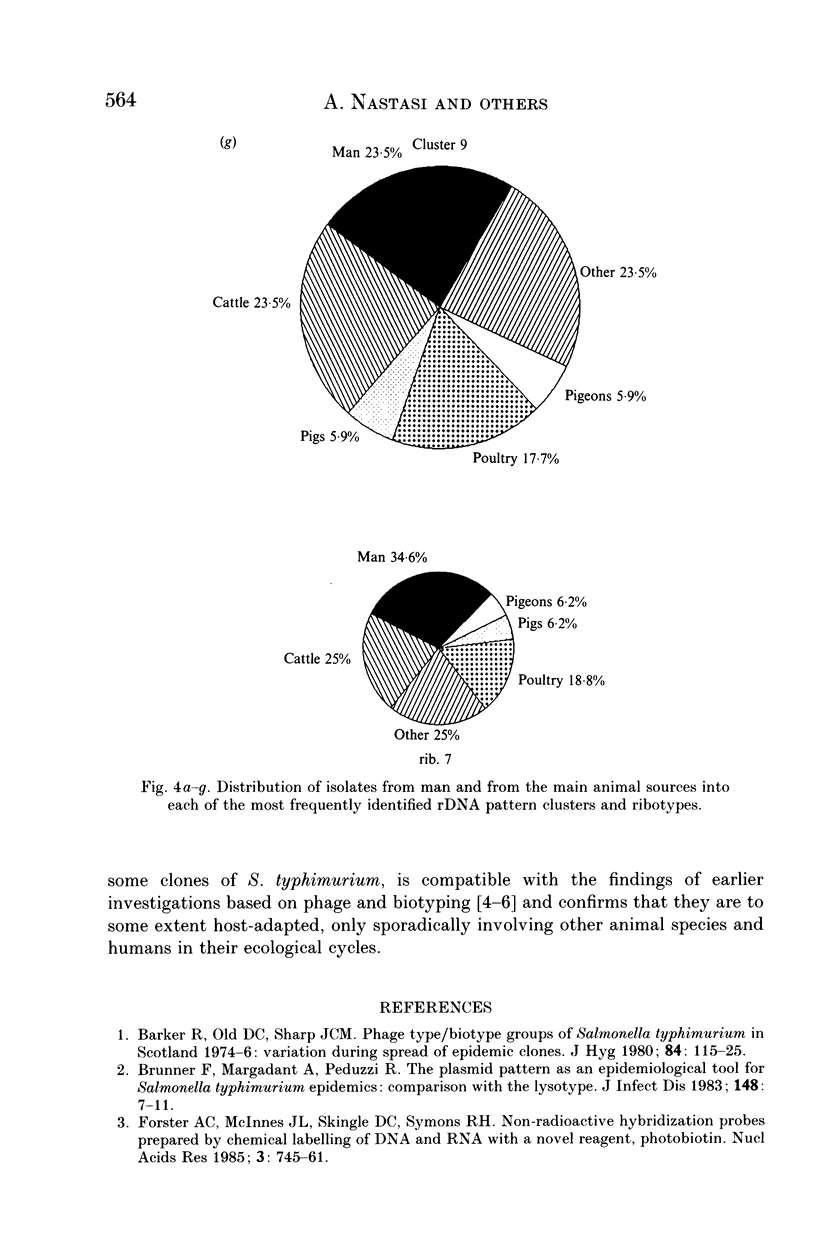
Salmonella typhimurium is the most frequently identified serovar of Salmonella in Italy. This serovar is characterized by the widespread dissemination among human and non-human sources of phenotypically and genetically well-differentiated clones. In this study 457 strains of S. typhimurium isolated in Italy in the years 1982-91 from human and animal sources were submitted to characterization by the rDNA fingerprinting technique. Application of this typing method, after digestion of chromosomal DNA with HincII endonuclease, confirmed the greatest genetic differentiation of clones of S. typhimurium, allowing reliable identification of 45 rDNA patterns linked into 9 major clusters. rDNA pattern clusters or ribotypes specific to man were not recognized, whereas some rDNA patterns were characteristically related to ducks, pigeons and pet birds. The ribotyping results for isolates from animal hosts suggest that pig and cattle are the main source of human infection.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S., Ward L. R., De Saxe M. J., Old D. C., Barker R., Duguid J. P. Correlation of phaga type, biotype and source in strains of Salmonella typhimurium. J Hyg (Lond) 1978 Oct;81(2):203–217. doi: 10.1017/s0022172400025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R., Old D. C., Sharp J. C. Phage type/biotype groups of Salmonella typhimurium in Scotland 1974-6: variation during spread of epidemic clones. J Hyg (Lond) 1980 Feb;84(1):115–125. doi: 10.1017/s0022172400026607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F., Margadant A., Peduzzi R., Piffaretti J. C. The plasmid pattern as an epidemiologic tool for Salmonella typhimurium epidemics: comparison with the lysotype. J Infect Dis. 1983 Jul;148(1):7–11. doi: 10.1093/infdis/148.1.7. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Holmberg S. D., Wachsmuth I. K., Hickman-Brenner F. W., Cohen M. L. Comparison of plasmid profile analysis, phage typing, and antimicrobial susceptibility testing in characterizing Salmonella typhimurium isolates from outbreaks. J Clin Microbiol. 1984 Feb;19(2):100–104. doi: 10.1128/jcm.19.2.100-104.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblavi S., Grimont F., Grimont P. A. Clonal diversity of Vibrio cholerae O1 evidenced by rRNA gene restriction patterns. Res Microbiol. 1990 Jul-Aug;141(6):645–657. doi: 10.1016/0923-2508(90)90059-y. [DOI] [PubMed] [Google Scholar]
- Nastasi A., Mammina C., Villafrate M. R. rDNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi infections. Epidemiol Infect. 1991 Dec;107(3):565–576. doi: 10.1017/s0950268800049268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Thomson-Carter F. M., Carter P. E., Pennington T. H. Differentiation of staphylococcal species and strains by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989 Jul;135(7):2093–2097. doi: 10.1099/00221287-135-7-2093. [DOI] [PubMed] [Google Scholar]
- Wauters G. Carriage of Yersinia enterocolitica serotype 3 by pigs as a source of human infection. Contrib Microbiol Immunol. 1979;5:249–252. [PubMed] [Google Scholar]
- van Leeuwen W. J., Guinée P. A. Frequency distribution of S. typhi-murium phage types in various countries. Zentralbl Bakteriol Orig A. 1975;230(3):320–335. [PubMed] [Google Scholar]
- van Leeuwen W. J., Pruys D., Guinée P. A. Phage typing of S. typhi murium in the Netherlands. 2. Frequency distributions of S. typhi murium phage types in the Netherlands during 1971 and 1972. Zentralbl Bakteriol Orig A. 1974 Feb;226(2):201–206. [PubMed] [Google Scholar]