Abstract
Agrobacterium tumefaciens induces crown gall tumors on plants by transferring a nucleoprotein complex, the T-complex, from the bacterium to the plant cell. The T-complex consists of T-DNA, a single-stranded DNA segment of the tumor-inducing plasmid, VirD2, an endonuclease covalently bound to the 5′ end of the T-DNA, and perhaps VirE2, a single-stranded DNA binding protein. The yeast two-hybrid system was used to screen for proteins interacting with VirD2 and VirE2 to identify components in Arabidopsis thaliana that interact with the T-complex. Three VirD2- and two VirE2-interacting proteins were identified. Here we characterize the interactions of VirD2 with two isoforms of Arabidopsis cyclophilins identified by using this analysis. The VirD2 domain interacting with the cyclophilins is distinct from the endonuclease, omega, and the nuclear localization signal domains. The VirD2–cyclophilin interaction is disrupted in vitro by cyclosporin A, which also inhibits Agrobacterium-mediated transformation of Arabidopsis and tobacco. These data strongly suggest that host cyclophilins play a role in T-DNA transfer.
Molecular studies on host–pathogen interactions have revealed that many pathogenic bacteria exploit host cell functions and signal transduction pathways for their own benefit (1). Agrobacterium tumefaciens is a classic example of such a pathogen (2). This bacterium genetically engineers plants to create a favorable metabolic niche. It achieves this aim by transferring a piece of its DNA, the T-DNA, from its tumor-inducing (Ti) plasmid into a plant cell. The T-DNA is integrated into the plant genome, and the expression of its oncogenes leads to overproduction of plant growth hormones, resulting in tumor formation. The T-DNA also confers the ability to synthesize and excrete opines, a group of sugar and amino acid conjugates, that are used by the inciting bacteria as a carbon and nitrogen source. The opines give Agrobacterium cells a growth advantage because specialized enzymes needed for the uptake and metabolism of these compounds are encoded by the Ti plasmid (2). This natural “genetic colonization” event serves as a paradigm for studying other host–microbe interactions.
The molecular events that occur inside the bacterium during its interaction with a plant host have been studied intensively (2). In response to signal molecules released from wounded plants, a set of virulence (vir) genes on the Ti plasmid are induced. The vir genes comprise a large regulon composed of at least 10 (virA to virJ) operons. The expression of these genes leads to the production and transfer of a nucleoprotein complex, the T-complex, from the bacterium to the plant cell. The proteins encoded by the virD and virE operons are involved in the production of the T-complex. VirD2, an endonuclease, nicks at the T-DNA border sequences generating a single-stranded T-DNA molecule (T-strand) to which VirD2 covalently binds at the 5′ end (3, 4). VirE2 is a single-stranded DNA binding protein that may coat the T-strand and protect it from nuclease degradation (5). The T-strand, together with the two Vir proteins, VirD2 and VirE2, form the T-complex (6).
It is apparent that both VirD2 and VirE2 perform important functions in transferring and integrating the T-DNA once the T-complex is inside the plant cell. First, both VirD2 and VirE2 have nuclear localization signals (NLS) that guide the T-complex into the plant cell nucleus and are required for tumorigenesis (6). Second, VirE2 and VirD2 appear to preserve the integrity of the T-DNA and participate in T-DNA integration (7, 8). These data suggest that VirD2 and VirE2 may interact with plant cell machinery, such as intracellular nucleoprotein trafficking or DNA integration/recombination/repair apparatus, to facilitate transfer and integration of the T-DNA.
Despite extensive efforts to uncover plant factors involved in T-DNA transfer and integration, the nature of these factors remains elusive. This is due to the complexity of plant genetics and the process of Agrobacterium-mediated tumorigenesis. The yeast two-hybrid system provides a different approach for identifying interacting proteins (9, 10). Recently, it has been successfully used to study protein–protein interactions in Agrobacterium (11, 12) and to identify a plant NLS receptor for VirD2 (13). We used this technique to look for VirD2- and VirE2-interacting proteins involved in T-complex trafficking and targeting in Arabidopsis thaliana. Here we report the identification of a class of VirD2 interactors in Arabidopsis, the cyclophilins (CyPs), and characterize this interaction by using yeast two-hybrid and biochemical analyses.
MATERIALS AND METHODS
Plasmid Construction.
All recombinant DNA methods followed standard protocols (14). To make in-frame fusions of VirD2 and VirE2 to the DNA-binding protein LexA, the full-length virD2 and virE2 genes of pTiA6 from A. tumefaciens strain A348 (4, 15) were amplified with specific primers by using the high-fidelity Vent DNA Polymerase (New England Biolabs) and standard PCR techniques. Restriction sites were introduced into the primers for virD2 (EcoRI in the upstream primer and XhoI in the downstream primer) and virE2 (BamHI and NdeI upstream and XhoI downstream) for convenient cloning into pEG202 and other vectors (10). The fusion junctions were verified by DNA sequencing. The two plasmids, designated pEG202D2 and pEG202E2, were used for the yeast two-hybrid screen (10). The virD2 and virE2 genes were also subcloned into pJG4-5 (10) to construct pJG-D2 and pJG-E2.
To overexpress VirD2 and VirE2, the EcoRI/XhoI fragment of pEG202D2 containing virD2 and the NdeI/XhoI fragment of pEG202E2 containing virE2 were subcloned into vector pET-28(a)+ (Novagen) to make in-frame fusions to the histidine (His) tag. The plasmids, designated pET-D2 and pET-E2, were introduced into Escherichia coli strain BL21(DE3) (Pharmacia Biotech) for expression.
Plasmid pGEX-5X-1 (Pharmacia Biotech) was used to make glutathione S-transferase (GST) fusions to CyPs. EcoRI/XhoI fragments containing the cDNA library inserts (in pJG4-5) from clones pD2-130 (Roc1), pD2-342 (CypA), and pD2-294 (Roc4) encoding different isoforms of CyPs were ligated into pGEX-5X-1 to make in-frame fusions to GST. These plasmids, called pGST-Roc1, pGST-CypA, and pGST-Roc4, were transformed into E. coli strain BL21 for expression. The same set of cDNA inserts was also subcloned into pEG202 for the yeast two-hybrid assay, and these plasmids were named pEG-Roc1, pEG-CypA, and pEG-Roc4.
The Yeast Two-Hybrid Screen.
The interaction trap/two-hybrid system was provided by R. Brent (Massachusetts General Hospital). The basic protocols and all the strains and plasmids used in the assay are as described (10). A cDNA library (in pJG4-5) of A. thaliana ecotype C24 seedlings was supplied by H. Zhang and H. Goodman (Massachusetts General Hospital). This cDNA library was amplified in E. coli and then used to screen for proteins interacting with either VirD2 or VirE2. A series of recommended specificity tests (10) were carried out to verify the interactions identified among the yeast colonies passing the initial library screen. Proteins were designated as true interactors only if they interacted with VirD2 or VirE2 and not with the control protein bicoid (encoded by pRFHM-1). The library plasmids encoding these proteins were transformed into E. coli, purified, analyzed by restriction digestion, and grouped according to their restriction profiles. The cDNA insert of one representative from each group was then sequenced, and compared with the database by using the blast program (16).
Protein Affinity Purification and in Vitro Protein Binding Assays.
Expression and purification of GST fusion proteins and affinity purification of proteins binding to GST fusion proteins were performed as described (14) with some minor modifications. Isopropyl β-d-thiogalactoside (IPTG)-induced E. coli BL21 cells carrying pGEX-5X-1 (GST vector control), pGST-Roc1, pGST-CypA, pGST-Roc4, pET-D2, or pET-E2 were collected and resuspended in bead binding buffer (14). Bacteria were lysed by sonication, and the sonicate was centrifuged twice for 15 min at 27,000 × g to remove insoluble materials. Binding reactions used equal volumes of crude bacterial lysates in a final volume of 500 μl with 20 μl of glutathione-agarose bead slurry (G beads; Pharmacia Biotech). Reactions were incubated at 4°C for 1–2 hr with gentle shaking. The G beads were pelleted by centrifugation and washed five times in 1 ml of bead binding buffer. The G beads with the bound proteins were then resuspended in 25 μl of 1× SDS sample buffer (14) and boiled for 5 min before being subjected to SDS/PAGE and Western blot analysis. SDS (0.05 or 0.1%), 2-mercaptoethanol (0.1 or 1%), NaCl (0.5 or 1 M), or cyclosporin A (CsA, Sigma; 0.5, 5, 10, or 50 μM) were added to the bead binding buffer during incubation to examine their effect on CyP–VirD2 interactions. The effect of CsA on CyP–VirD2 complexes was also examined by allowing the CyPs and VirD2 to bind for 1 hr in the absence of CsA and then treating the binding mixture with CsA for an additional 15 min to 1 hr.
Protein Gels and Western Blot Analysis.
Protein analysis with SDS/PAGE followed standard protocols (14). Gels were either stained with Coomassie blue or processed for Western blot analysis. Protein transfer to polyvinylidene fluoride membranes (Millipore) was carried out on the Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad). The ECL Western blotting analysis system (Amersham) was used for detection. Polyclonal antibodies against VirD2 and VirE2 proteins were raised in rabbits (5, 17).
Deletion Analysis of VirD2 by Using the Yeast Two-Hybrid System.
A series of VirD2 deletions were made to define the domain in VirD2 that interacts with the CyPs (Roc1 and CypA). Primers complementary to specific regions of the virD2 coding region were used to amplify the desired fragments by PCR. The PCR products were then cloned into pEG202 to make in-frame fusions to LexA that were verified by DNA sequencing. The VirD2 deletions encoded by these plasmids (pEG202D2Δ1-4 and pEG202D2ΔNLS) were analyzed to determine if they could interact with the CyPs by using yeast two-hybrid analysis (10).
Arabidopsis Transformation.
Transformation of Arabidopsis (ecotype NO) roots was performed as described (18) by using A. tumefaciens strain EHA101 (pJR301) (ref. 19; J. Rout and E.W.N., unpublished data). This strain contains a binary vector (pJR301) carrying a CaMV 35S promoter-driven β-glucuronidase (GUS) gene construct that contains a plant intron in its coding region. Roots from 2-week-old seedlings were precultured in callus-inducing medium (CIM) (18) with various concentrations of CsA (0, 1, 5, or 10 μM) for 3 days. About 50 mg of roots (fresh weight) were used for each treatment. The precultured roots were then infected with EHA101 (pJR301) grown overnight in AB medium (20) and diluted 10-fold in CIM. Root explants and bacterial cells were cocultivated in CIM medium in the presence or absence of CsA for 2 days prior to staining for GUS activity. Histochemical staining with 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc; Jersey Lab Supply, Livingston, NJ) of Arabidopsis roots was conducted as described (21). Blue GUS-positive spots were counted by using a dissecting microscope.
Tobacco Transformation.
Tobacco BY-2 (Nicotiana tabacum L. cv. Bright Yellow 2) cells were grown in Modified Linsmaier–Skoog medium (LS, pH 5.5) (22) supplemented with 1 μM 2,4-dichlorophenoxyacetic acid (2,4-D). A. tumefaciens LBA4404 (pBI121) (23, 24) cells from an overnight culture were resuspended to OD600 = 1.0 in IBPO4 medium (25) supplemented with 100 μM acetosyringone (AS) and incubated at 25°C for 2 hr. Three days following subculture (5% weekly), 4 ml of BY-2 cells were cocultured with or without 10 μM CsA in the presence of 100 μl of bacteria and 100 μM AS. After 2 days, the cells were rinsed, resuspended in LS medium containing 2,4-D and 500 μg/ml carbenicillin, and grown for 8 days prior to staining for GUS activity. Histochemical staining was performed as described (21). GUS-positive cells were scored under the microscope as intact cells whose cytoplasm stained blue without showing any coloration of the cell wall. Treatment effects were analyzed by ANOVA using sas (version 6.12 for UNIX, 1996, SAS Institute, Cary, NC). Mean comparisons were made after analysis of variance by using Tukey’s Studentized Test (α = 0.05).
RESULTS
Identification of Arabidopsis Proteins That Interact with VirD2 and VirE2 by Using the Yeast Two-Hybrid System.
A series of control experiments were performed to ensure that pEG202D2 and pEG202E2 were suitable for library screening by using the yeast two-hybrid system (10). When transformed into yeast strain EGY48 containing two reporter genes (the lexA operator LEU2 in the chromosome and the lexA operator lacZ in pSH18-34), neither pEG202D2 nor pEG202E2 activated transcription of the lacZ reporter gene. Western blot analyses by using VirD2 or VirE2 antibodies showed that the fusion proteins, LexA–VirD2 (encoded by pEG202D2) and LexA–VirE2 (encoded by pEG202E2), matched the predicted molecular sizes (data not shown). A repression assay (10) indicated that both fusion proteins entered the yeast nucleus and bound to the lexA operators. This finding was not surprising because both VirD2 and VirE2 contain NLSs (6). Interestingly, LexA–VirD2 repressed reporter gene expression in the repression assay more strongly than LexA–VirE2, suggesting that LexA–VirD2 can enter the yeast nucleus more efficiently than LexA–VirE2. EGY48 cannot grow in the absence of leucine unless the lexA operator LEU2 gene is expressed (10). EGY48 carrying pEG202E2 cannot grow on leucine-free medium, whereas EGY48(pEG202D2) formed small colonies after long (4-day) incubations. However, the background level is low and can be tolerated. We conclude that both LexA–VirE2 and LexA–VirD2 are good candidates for interaction screens of cDNA libraries.
The two plasmids, pEG202D2 and pEG202E2, were used to screen a cDNA library of A. thaliana C24 seedlings. After the recommended specificity tests eliminated the false positive clones (10), three VirD2- and two VirE2-interacting proteins of Arabidopsis were identified. The three VirD2-interacting proteins (DIP) were designated DIP1, DIP2, and DIP3, and the two VirE2-interacting proteins (EIP) were called EIP1 and EIP2. Based on their protein sequences and their similarities to other proteins in the database, they represented both cytoplasmic and nuclear factors (data not shown). However, no clone encoding AtKAPα, an Arabidopsis NLS binding protein recently shown to interact with VirD2 by using yeast two-hybrid analysis (13), was isolated in our screen. In this report we characterize the interactions between VirD2 and one class of Arabidopsis proteins, the CyPs (DIP1). The other interactors identified will be the subject of future reports.
CyPs Specifically Interact with VirD2 in the Yeast Two-Hybrid Assay.
DIP1 represents a family of highly conserved proteins, the CyPs (26). It consists of three isoforms of Arabidopsis CyPs, one cytosolic (Roc1; ref. 27), one chloroplast (Roc4; ref. 27), and the other unknown (a protein belonging to the CyP A class and referred to here as CypA; ref. 28) (Table 1). VirD2 interacted most strongly with CypA in the most stringent yeast reporter strain, EGY191 (10), whereas VirD2-Roc1 and VirD2-Roc4 showed only weak interactions (data not shown). Roc4, a chloroplast CyP, is unlikely to be involved in T-complex trafficking based on its cellular location. However, the fact that Roc4 was identified during two-hybrid analysis supports the contention that the CyPs are true VirD2 interactors because the C terminus of Roc4 shares significant amino acid sequence similarity with other CyPs (27). CyPs did not interact with the control protein bicoid (encoded by pRFHM-1) or VirE2 (Table 1), and they did not activate the reporter genes (LEU2 and lacZ) by themselves. In addition, when the position of the interacting pairs in the yeast two-hybrid assay was switched (CyPs-encoding cDNAs cloned in pEG202 and virD2 cloned in pJG4-5), CyPs and VirD2 still interacted (data not shown), strongly indicating that the CyP–VirD2 interactions are specific.
Table 1.
Arabidopsis CyPs interact with VirD2 in the yeast two-hybrid assay
| cDNA clone (in pJG4-5) | Protein encoded | GenBank match (accession no. and ref.) | Interaction with LexA fusion to
|
||
|---|---|---|---|---|---|
| VirD2 (pEG202D2) | VirE2 (pEG202E2) | Bicoid (pRFHM1) | |||
| pJG4-5 | Vector only | − | − | − | |
| pD2-130 | Roc1 | L14844 (27) | + | − | − |
| pD2-294 | Roc4 | L14845 (27) | + | − | − |
| pD2-342 | CypA/Cyp1 | U07276 (28) | + | − | − |
The two-hybrid assay was done in yeast strain EGY48, which contains two reporter genes (LEU2 and lacZ) (10). A positive interaction was scored if EGY48 cells carrying a combination of pEG202 and pJG4-5 plasmids stained blue on X-Gal medium and grew in leucine-free medium in the presence of galactose but not glucose.
CyPs Interact with VirD2 in Vitro.
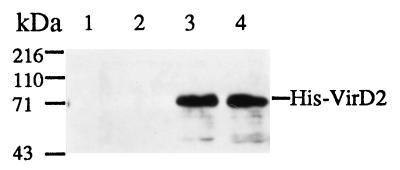
The GST fusion system is widely used to demonstrate protein–protein interactions in vitro because of the high solubility of GST fusion proteins and the ease of its single step purification scheme by using G-beads (14). To verify the specificity of the interactions between VirD2 and the three Arabidopsis CyP isoforms, the cDNA inserts of each CyP were subcloned into pGEX-5X-1 to make GST–CyP fusions. Roc4 was highly insoluble when fused to GST, probably because of its hydrophobic chloroplast transit peptide, and its interaction with VirD2 was not studied further. Western blot analysis (Fig. 1) reveals that His-tagged VirD2 protein specifically bound to GST–Roc1 and GST–CypA (lanes 3 and 4), but not to G-beads alone (lane 1) or GST (lane 2). The interaction of GST–Roc1 and GST–CypA with VirD2 was so efficient that the VirD2 band can be easily seen after Coomassie blue staining of the protein gel (data not shown). The His-tagged VirE2 protein did not bind to the GST–CyP fusions (data not shown), indicating that the CyP interaction is specific for VirD2. GST-Roc1 and GST–CypA can also extract VirD2 protein specifically from a total bacterial lysate of induced A. tumefaciens A348 cells in the binding assay (data not shown), further supporting our contention that the VirD2–CyP interaction is specific.
Figure 1.
Specific binding of VirD2 to GST–CyP fusions in vitro. An equal amount of total bacterial lysate expressing His-tagged VirD2 was incubated with buffer alone (lane 1) or with an equal volume of total bacterial lysate expressing GST (lane 2), GST–Roc1 (lane 3), or GST–CypA (lane 4). GST or GST fusion proteins were affinity purified with G-beads and subjected to SDS/PAGE and Western blot analysis.
CyPs Interact Strongly with VirD2.
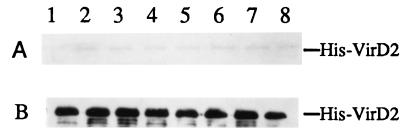
To test the strength of the interaction between CyPs and VirD2, the effect of salt, detergent, and 2-mercaptoethanol (2-ME) on the interaction was examined. As shown in Fig. 2, 0.5 or 1 M NaCl and 0.1% SDS had little, if any, effect on the interaction between His–VirD2 and GST–Roc1 or GST–CypA when they were present in the binding buffer. Similarly, 0.1 or 1% 2-ME did not affect the CyP–VirD2 interaction (data not shown). These tests suggest a strong interaction between VirD2 and the two CyP isoforms, Roc1 and CypA, and indicate that intermolecular disulfide bonds are not involved.
Figure 2.
Specific binding of VirD2 and GST–CyP fusions in the presence of detergent and high salt. An equal amount of total bacterial lysate expressing His-tagged VirD2 was incubated with an equal volume of total bacterial lysate expressing GST–Roc1 (lanes 1–4) or GST–CypA (lanes 5–8). Lanes 1 and 5, no additional salt or detergent added to the binding buffer; lanes 2 and 6, 0.5 M NaCl; lanes 3 and 7, 1 M NaCl; and lanes 4 and 8, 0.1% SDS. GST fusion proteins were affinity purified with G-beads and subjected to SDS/PAGE and Western blot analysis. (A) Coomassie blue stained SDS/PAGE. (B) Immunoblot with rabbit anti-VirD2 antibody.
CsA Inhibits the VirD2–CyP Interaction in Vitro.
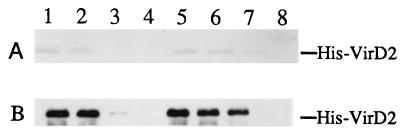
CyPs were first identified as proteins that interacted with the immunosuppressive drug CsA (26). CsA can disrupt protein–CyP interactions in vitro as shown for human CypA and the capsid protein of HIV-1 (29, 30). Different isoforms of CyPs also appear to have different affinities for CsA (26). To determine whether CsA would affect the Roc1–VirD2 or CypA–VirD2 interaction, various concentrations of CsA were included during or after the binding between GST–CyPs and His–VirD2 in the G-bead binding assay. In the presence of 5 μM CsA, His–VirD2 and GST–Roc1 did not associate (Fig. 3, lane 3). Interestingly, the His–VirD2–GST–CypA interaction was less sensitive to CsA, and more than 10 μM CsA was needed to abolish the binding between GST–CypA and His–VirD2 (Fig. 3, lane 8). This result is consistent with the yeast two-hybrid data which suggest that the VirD2–CypA interaction is stronger than the VirD2–Roc1 association. If CsA was added after VirD2 and GST–Roc1 or GST–CypA were first mixed and allowed to bind, VirD2 and CyP interactions were still disrupted, and the CsA concentrations needed for this disruption were similar to those required for inhibition when CsA was included during the binding (data not shown). These data suggest either that CsA and VirD2 compete for the same binding site on CyPs or that CsAs binding causes a conformational change in CyPs such that CyPs can no longer interact with VirD2.
Figure 3.
CsA inhibits binding between VirD2 and GST–CyP fusions. An equal amount of total bacterial lysate expressing His-tagged VirD2 was incubated with an equal volume of total bacterial lysate expressing GST–Roc1 (lanes 1–4) or GST–CypA (lanes 5–8). Lanes 1 and 5, no CsA added; lanes 2 and 6, 0.5 μM CsA; lanes 3 and 7, 5 μM CsA; and lanes 4 and 8, 50 μM CsA. GST fusion proteins were affinity-purified with G-beads and subjected to SDS/PAGE and Western blot analysis. (A) Coomassie blue stained SDS/PAGE. (B) Immunoblot with rabbit anti-VirD2 antibody.
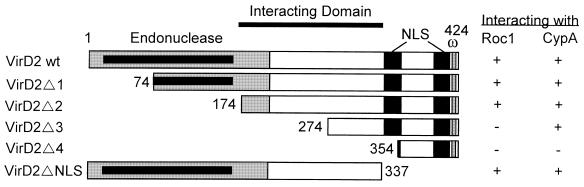
CyPs Interact with a Novel Domain of VirD2.
To delineate the domain in VirD2 which interacts with the CyPs, a series of deletion mutants of VirD2 were generated, fused to LexA in pEG202, and then tested by using yeast two-hybrid analysis to determine whether they interacted with Roc1 or CypA. As depicted in Fig. 4, a central domain of VirD2, from amino acid residue 174 to 337, was required for its interaction with Roc1. A smaller, overlapping region of VirD2 (residues 274–337) was involved in its interaction with CypA. Interestingly, the CypA–VirD2 interaction was stronger than that of Roc1–VirD2, and less sensitive to CsA inhibition (Fig. 3). Neither the NLSs (6) nor the omega domain in VirD2 (31) were required for VirD2–CyP interactions (Fig. 4).
Figure 4.
Delineating the CyP-interacting domain of VirD2. A series of deletions of the virD2 gene were generated by PCR amplification, subcloned into pEG202, and subjected to yeast two-hybrid analysis with cDNA clones of Roc1 and CypA in pJG4-5. A positive interaction was scored if EGY48 cells carrying a combination of pEG202 and pJG4-5 plasmids stained blue on X-Gal medium and grew in leucine-free medium in the presence of galactose but not glucose. Boxes represent the VirD2 region present in pEG202. The CyP interacting domain is indicated by the bar. The endonuclease (4), NLSs (6), and omega domains (31) are indicated. The rectangle within the endonuclease domain denotes critical residues essential to endonuclease activity (8, 52). Results of the yeast two-hybrid analysis are shown to the right of the figure.
CsA Inhibits Agrobacterium-Mediated Transformation of Plants.
Because CsA inhibited the interaction between VirD2 and CyPs in vitro, we tested whether CsA can inhibit Agrobacterium-mediated transformation of A. thaliana and N. tabacum. As shown in Table 2, concentrations of 5 and 10 μM CsA decreased the transformation frequency in Arabidopsis to about 20% of that without CsA treatment. The same concentrations of CsA had no effect on the growth of Agrobacterium (data not shown). The concentration of CsA needed for this inhibition was consistent with that needed to disrupt the VirD2–CyP interaction in vitro (Fig. 3). When CsA (up to 10 μM) was present during the 3-day preculture but not cocultivation with Agrobacterium, no inhibition of transformation was observed, indicating that the inhibition by CsA is reversible and that CsA is not highly toxic to Arabidopsis roots (data not shown). CsA at the same concentrations also reduced Agrobacterium-mediated transformation of N. tabacum suspension cultures suggesting that CyPs represent a common mechanism for T-complex trafficking in plants (Table 2).
Table 2.
Cyclosporin A inhibits Agrobacterium- meditated transformation
| Concentration of CsA | Arabidopsis, no. of GUS-positive spots | Tobacco BY-2, % transformation |
|---|---|---|
| 0 μM CsA, no bacteria | 0 | 0 |
| 0 μM CsA | 18.3 ± 0.6 | 20 ± 3.1 |
| 1 μM CsA | 10.0 ± 1.9 | ND |
| 5 μM CsA | 4.0 ± 0.8 | ND |
| 10 μM CsA | 3.3 ± 0.2 | 2.9 ± 1.1 |
| 10 μM CsA* | 4.3 ± 0.2 | ND |
Transformation of Arabidopsis and tobacco BY-2 suspension cultures is described in Materials and Methods. For Arabidopsis, both precultivation and cocultivation were performed in the presence of listed CsA concentrations unless otherwise noted. The number of GUS staining spots is the average of three experiments, followed by standard error. For tobacco, percent transformation shown is the mean of six repetitions per treatment followed by the standard error for a single representative experiment. ND, not determined.
*CsA was present only during cocultivation.
DISCUSSION
Upon exiting Agrobacterium, the T-complex enters the plant cell environment where it faces the formidable task of finding its way into the plant cell nucleus and integrating the T-DNA into the plant genome. Because the DNA between the T-DNA borders can be replaced by any random DNA and still be transferred, the protein components of the T-complex, VirD2 and VirE2, must specify the targeting of the T-complex (6). It seems reasonable to assume that in the complex process of T-complex transfer, VirD2 and VirE2 must interact with multiple host proteins. Many animal pathogens utilize host cell pathways during movement and pathogenesis (1), and it is likely that Agrobacterium exploits similar mechanisms in plants. Considering the broad range of eukaryotes to which Agrobacterium can transfer its T-DNA, it is likely that many of the proteins with which the T-complex interacts will belong to common cellular pathways involved in nucleoprotein uptake, trafficking, nuclear import, and DNA recombination and integration. In this context, VirD2 has recently been shown to interact with a plant NLS receptor (13).
Currently, little is known about the plant proteins involved in the intracellular transport of the T-complex or in the integration of the T-DNA. In this report, we describe the identification of a class of VirD2-interacting plant proteins, the CyPs. To date, eight distinct CyP isoforms have been identified in Arabidopsis (27, 28, 32, 33). We have identified three of these CyPs as VirD2 interactors (Table 1), but the possibility that VirD2 also interacts with other CyPs cannot be excluded. However, it is not clear whether this interaction with more than one CyP is biologically significant or whether it occurs because of similarities in the CyP binding domains. The fact that we isolated Roc1 and CypA repeatedly suggest that, unless the other CyPs are not highly expressed or the cDNA library used was not representative, VirD2 does not interact with other CyPs. Among the different isoforms of CyPs present in a plant cell (27, 28, 32, 33), the cytosolic and nuclear ones are most relevant to T-DNA transfer. Our analysis suggests that the VirD2-CyP interaction is strong and highly specific (Figs. 1 and 2). The VirD2 domain interacting with CyPs was identified and found to be distinct from the NLSs (6) and the endonuclease and omega domains (Fig. 4; ref. 31). CsA, which disrupts the VirD2–CyP interaction in vitro (Fig. 3), also inhibits Agrobacterium-mediated transformation of plants (Table 2). This finding strongly suggests that one or more CyPs play a role in T-complex transfer. Future studies will determine if CsA inhibits both transient and stable transformation.
CyPs constitute a large class of highly conserved proteins found in a diverse range of bacteria, yeast, plants, and animals (26, 34, 35). They exist in multiple isoforms that are located in different cellular compartments. The ubiquitous nature of CyPs suggests that they belong to common T-complex trafficking pathways predicted to exist in the various hosts to which Agrobacterium can transfer its T-DNA. Our data support this contention because CsA inhibited transformation of both A. thaliana and N. tabacum. Although the exact cellular function of the CyPs remains to be defined, evidence suggests that they can aid protein folding and serve as molecular chaperones (26, 36). CyPs were first identified in mammalian systems based on their ability to bind CsA (37). CsA blocks the peptidyl-prolyl isomerase (PPIase) activity of CyPs (38, 39), although the physiological role of this activity is unclear. CyPs are also components of intracellular signaling pathways (26, 34). In mammalian systems, CsA interaction with CyPs facilitates their binding to and inhibition of calcineurin, leading to immunosuppression (40). In contrast, CsA was found to disrupt VirD2 interactions with CypA and Roc1 as well as the interaction of the HIV-1 Gag structural protein with host CyPA (29, 30). The biological role of host CyPs is more intriguing in a number of host–pathogen interactions (41). CyPs are specific targets for the CsA-mediated attenuation of a variety of parasitic infections, including leishmaniasis, malaria and toxoplasmosis (42). Interestingly, human CyPA promotes HIV-1 infectivity by forming a stable complex with the capsid protein in HIV-1 virions and promoting viral uncoating (30). These data suggest that CyPs, and cellular pathways linked to CyPs, are frequently exploited by pathogens.
How might CyPs be involved in T-complex trafficking? Because the exact cellular function of various CyP isoforms is poorly defined, it is difficult to predict at which stage the CyPs function in T-DNA transfer. However, we are examining several possibilities. VirD2 has been viewed as the pilot protein for T-complex transport (6). When VirD2, along with the T-complex, is transferred from the bacterium into the plant cell, it may change conformation to fulfill its function(s) in the new environment. This conformational change may allow VirD2 to conceal the endonuclease domain, which may only be needed in the bacterium, or to expose signal domains (omega, NLS, and others) that are required for its function in plants. In this respect, CyPs have PPIase activity (26) that might aid VirD2 in folding into a different conformation inside a plant cell. CsA, the drug which inhibits the PPIase activity of CyPs (38, 39), also disrupts VirD2–CyP interactions (Fig. 3). It is also possible that CyP binding is required to maintain VirD2 in a functional and transfer-competent state because some CyPs exhibit chaperone activities (36, 43). Previous studies suggest that CyPs may play a chaperone role during stress responses (44). Some yeast CyPs are heat-inducible and required for optimal growth and metabolism at high temperatures (44, 45), although disruption of all yeast CyPs did not reduce viability (46). CyP mRNA levels in plants are also induced by abiotic stresses, including wounding (47). Interestingly, wounding is required for Agrobacterium to induce tumors.
The VirD2–CyP interaction may be important for T-complex transfer both in the cytoplasm and in the nucleus. Studies in mammalian systems have indicated that some CyPs, including those considered to be cytosolic, are distributed in the cytoplasm as well as in the nucleus (48, 49). The VirD2-interacting Roc1 is postulated to be a cytosolic isoform, although no evidence is available to support this view (27). The cellular location of CypA, the other VirD2-interacting CyP, is still unknown (28, 32). It is possible that both Roc1 and CypA, like their mammalian homologs, are present in both the cytosol and the nucleus of Arabidopsis. CyPs also function as DNA binding proteins (50) and have been implicated in the regulation of apoptosis because of their nuclease activity, which is independent of their PPIase activity (49). Integration of the T-DNA involves illegitimate recombination in plants which usually includes small deletions of the plant DNA at target sites (51), suggesting that a nuclease function is needed for T-DNA integration into the plant genome. It is tempting to speculate that CyPs play a role in T-DNA integration, although our data do not allow us to distinguish between transient or stable transformation events.
In summary, our studies on the VirD2–CyP interaction strongly suggest that Agrobacterium recruits and utilizes one or more plant CyPs during pathogenesis. We are currently determining the stage at which CyPs function for T-complex trafficking and integration and whether the PPIase activity of CyPs is required for their function.
Acknowledgments
We thank Dr. R. Brent for the yeast two-hybrid system, Drs. H. Goodman and H. Zhang for the cDNA library of Arabidopsis, Drs. S. Doty and M. Yanofsky for reviewing the manuscript, Lin Lee and Yuching Chen for technical assistance, and members of our laboratories for discussions. This research was supported by National Institutes of Health Grant GM32618 (to E.W.N.) and National Science Foundation Grant MCB-9723735 (to E.W.N. & L.C.). T.M. was supported by a Howard Hughes Undergraduate Summer Research Fellowship.
ABBREVIATIONS
- CyP
cyclophilin
- CsA
cyclosporin A
- GUS
β-glucuronidase
- NLS
nuclear localization signal
- vir
virulence
- Ti
tumor-inducing
- T-DNA
transferred DNA
- T-strand
single-stranded copy of the T-DNA
- T-complex
T-strand associated with VirD2 and VirE2 proteins
- GST
glutathione S-transferase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- PPIase
peptidyl-prolyl isomerase
References
- 1.Finlay B B, Cossart P. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 2.Winans S C. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward E R, Barnes W M. Science. 1988;242:927–930. [Google Scholar]
- 4.Yanofsky M F, Porter S G, Young C, Albright L M, Gordon M P, Nester E W. Cell. 1986;47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 5.Christie P J, Ward J E, Winans S C, Nester E W. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields S, Song O K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Golemis E A, Gyuris J, Brent R. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1996. pp. 20.1.1–20.1.28. [Google Scholar]
- 11.Baron C, Thorstenson Y R, Zambryski P. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Anderson L B, Xie Y-H. J Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballas N, Citovsky V. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 15.Winans S C, Allenza P, Stachel S E, McBride K E, Nester E W. Nucleic Acids Res. 1987;15:825–837. doi: 10.1093/nar/15.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Porter S G, Yanofsky M F, Nester E W. Nucleic Acids Res. 1987;15:7503–7517. doi: 10.1093/nar/15.18.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valvekens D, van Montagu M, van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hood E E, Helmer G L, Fraley R T, Chilton M D. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleif R F, Wensink P C. Practical Methods in Molecular Biology. New York: Springer; 1981. [Google Scholar]
- 21.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 22.Nagata T, Okada K, Takebe I, Matsui C. Mol Gen Genet. 1981;184:161–165. [Google Scholar]
- 23.Ooms G, Hooykaas J J, van Veen R J M, van Beelen P, Regensburg-Tuink T J G, Schilperoort R A. Plasmid. 1982;7:15–29. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 24.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piers K L, Heath J D, Liang X, Stephens K M, Nester E W. Proc Natl Acad Sci USA. 1996;93:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marks A R. Physiol Rev. 1996;76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- 27.Lippuner V, Chou I T, Scott S V, Ettinger W F, Theg S M, Gasser C S. J Biol Chem. 1994;269:7863–7868. [PubMed] [Google Scholar]
- 28.Hayman G T, Miernyk J A. Biochim Biophys Acta. 1994;1219:536–538. doi: 10.1016/0167-4781(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 29.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 30.Luban J. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 31.Shruvinton C E, Hodges L, Ream W. Proc Natl Acad Sci USA. 1992;89:11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito T, Ishiguro S, Ashida H, Kawamukai M, Matsuda H, Ochiai H, Nakagawa T. Plant Cell Physiol. 1995;36:377–382. doi: 10.1093/oxfordjournals.pcp.a078770. [DOI] [PubMed] [Google Scholar]
- 33.Bartling D, Heese A, Weiler E W. Plant Mol Biol. 1992;19:529–530. doi: 10.1007/BF00023405. [DOI] [PubMed] [Google Scholar]
- 34.Hunter T. Cell. 1998;92:141–143. doi: 10.1016/s0092-8674(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 35.Duina A A, Chang H-C J, Marsh J A, Linquist S, Gaber R F. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 36.Baker E K, Colley N J, Zuker C S. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handschumacher R E, Harding M W, Rice J, Drugge R J. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 38.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid F X. Nature (London) 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi N, Hayano T, Suzuki M. Nature (London) 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Farmer J J D, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 41.Hacker J, Fischer G. Mol Microbiol. 1993;10:445–456. doi: 10.1111/j.1365-2958.1993.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 42.Hoerauf A, Rascher C, Bang R, Pahl A, Solbach W, Brune K, Rollinghoff M, Bang H. Mol Microbiol. 1997;24:421–429. doi: 10.1046/j.1365-2958.1997.3401716.x. [DOI] [PubMed] [Google Scholar]
- 43.Freeman B C, Toft D O, Morimoto R I. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 44.Sykes K, Gething M-J, Sambrook J. Proc Natl Acad Sci USA. 1993;90:5853–5857. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis E S, Becker A, Heitman J, Hall M N, Brennan M B. Proc Natl Acad Sci USA. 1992;89:11169–11173. doi: 10.1073/pnas.89.23.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolinski K, Muir S, Cardenas M, Heitman J. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marivet J, Margis-Pinheiro M, Frendo P, Burkard G. Plant Mol Biol. 1994;26:1181–1189. doi: 10.1007/BF00040698. [DOI] [PubMed] [Google Scholar]
- 48.Ryffel B, Woerly G, Greiner B, Haendler B, Mihatsch M J, Foxwell B M J. Immunology. 1991;72:399–404. [PMC free article] [PubMed] [Google Scholar]
- 49.Montague J W, Hughes J, F M, Cidlowski J A. J Biol Chem. 1997;272:6677–6684. doi: 10.1074/jbc.272.10.6677. [DOI] [PubMed] [Google Scholar]
- 50.Krummrei U, Bang R, Schmidtchen R, Brune K, Bang H. FEBS Lett. 1995;371:47–51. doi: 10.1016/0014-5793(95)00815-q. [DOI] [PubMed] [Google Scholar]
- 51.Gheysen G, Villarroel R, van Montagu M. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- 52.Vogel A M, Yoon J, Das A. Nucleic Acids Res. 1995;23:4087–4091. doi: 10.1093/nar/23.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]