Abstract
A colonization mutant of the efficient root-colonizing biocontrol strain Pseudomonas fluorescens WCS365 is described that is impaired in competitive root-tip colonization of gnotobiotically grown potato, radish, wheat, and tomato, indicating a broad host range mutation. The colonization of the mutant is also impaired when studied in potting soil, suggesting that the defective gene also plays a role under more natural conditions. A DNA fragment that is able to complement the mutation for colonization revealed a multicistronic transcription unit composed of at least six ORFs with similarity to lppL, lysA, dapF, orf235/233, xerC/sss, and the largely incomplete orf238. The transposon insertion in PCL1233 appeared to be present in the orf235/233 homologue, designated orf240. Introduction of a mutation in the xerC/sss homologue revealed that the xerC/sss gene homologue rather than orf240 is crucial for colonization. xerC in Escherichia coli and sss in Pseudomonas aeruginosa encode proteins that belong to the λ integrase family of site-specific recombinases, which play a role in phase variation caused by DNA rearrangements. The function of the xerC/sss homologue in colonization is discussed in terms of genetic rearrangements involved in the generation of different phenotypes, thereby allowing a bacterial population to occupy various habitats. Mutant PCL1233 is assumed to be locked in a phenotype that is not well suited to compete for colonization in the rhizosphere. Thus we show the importance of phase variation in microbe–plant interactions.
The use of microorganisms, including fluorescent Pseudomonas spp., to protect plants against soil-borne diseases is an alternative for the use of chemical pesticides. The biocontrol activity of these strains usually results from the production of one or more antifungal factors. The application of fluorescent Pseudomonas spp. and other plant-growth-promoting rhizobacteria is hampered by inconsistency of performance in the field (1, 2). Although the mechanisms underlying biocontrol are complex and diverse, the need to bring the plant-growth-promoting rhizobacteria cells and their antifungal factors to the right sites at the right time is universal. The importance of this process, designated as root colonization, is underscored in two studies. Schippers et al. (1) showed that inadequate colonization leads to decreased biocontrol activity, and Bull et al. (3) reported an inverse relation between the number of bacteria present on the wheat root and the number of take-all lesions seen on the plant. For these and other reasons, root colonization is often considered the limiting factor for biocontrol in the rhizosphere (1, 2).
Two approaches were used in our laboratory to identify traits involved in root colonization. The first approach is to guess which traits are involved in colonization, isolate mutants in these traits, and then test these mutants for colonization in competition with the parental strain. With this approach, motility (4) and synthesis of the O-antigen of lipopolysaccharide (LPS) (5) were shown to be essential for colonization. Moreover, mutants auxotrophic for amino acids or vitamin B1 (6) and mutants with a slightly increased generation time (7) also appeared to be reduced in colonization. Our second approach involves random transposon mutagenesis, using Tn5lacZ (8), of the efficient root colonizer Pseudomonas fluorescens WCS365 (9). Individual mutants were tested for their colonization ability in competition with the parental strain in a gnotobiotic system as described by Simons et al. (7). This approach enables us to obtain knowledge of bacterial colonization traits. In this paper, we describe such a mutant strain, PCL1233, and the results indicate the involvement of a site-specific recombinase in root colonization.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
The characteristics of all bacterial strains and plasmids used in this work are described in Table 1. Wild-type P. fluorescens WCS365 cells and derivatives of this strain were grown overnight at 28°C on solidified King B medium (18) or in liquid King B or standard succinate medium (SSM) (19) with vigorous shaking.
Table 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Characteristic(s) |
|---|---|
| P. fluorescens | |
| WCS365 | Biocontrol strain in a Fusarium oxysporum f. sp. radicis–lycopersici–tomato system (L.C.D., unpublished results), causes systemic acquired resistance in Arabidopsis thaliana ecotype Columbia (H. Gerrits, personal communication, efficient colonizer of the potato root (9) |
| PCL1500 | Tn5lacZ (8) derivative of WCS365 that behaves as wild type in colonization assays (10) |
| PCL1233 | A Tn5lacZ derivative of WCS365 that was isolated as a colonization-impaired mutant (11) |
| PCL1224 | PCL1233 harboring plasmid pWTT2081* |
| PCL1228 | PCL1233 harboring pMP5213, used for homogenotization* |
| PCL1231 | Wild-type WCS365 obtained by homogenotization* |
| PCL1234 | PCL1233 harboring pMP5215, used for complementation of the mutation* |
| PCL1239 | WCS365 harboring pWTT2081* |
| PCL1250 | PCL1233 harboring pMP5244* |
| P. aeruginosa | |
| 7NSK2 | Plant-growth-promoting P. aeruginosa strain (12) |
| SSS | Derivative of P. aeruginosa 7NSK2 mutated in the sss gene, a homologue of xerC (13, 14). |
| E. coli | |
| XL1-Blue | SupE44 hsdR17 recA1 endA1 hyrA46 thi relA1 lac-F′ [proAB + lac1q lacZ M15 Tn10(ter−)], used for transformation and propagation of pBluescript phagemids (Stratagene) |
| DH5α | EndA1 gyrSA96 hrdR17(rK− mK−) supE44 recA1, used for propagation of plasmids (15) |
| Plasmids | |
| pME3049 | Suicide plasmid, used for picking up wild-type genes (16) |
| pIC20H/R | Used for cloning and subcloning of fragments (17) |
| pMP2740 | Plasmid used for homogenotization (17) |
| pWTT2081 | Plasmid stably maintained in the rhizosphere; used for complementation of PCL1233 (10) |
| PBluescript | Vector used for single-stranded DNA sequencing (10) |
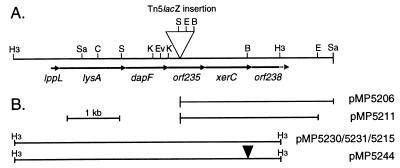
| pMP5206 | pIC20H containing a SalI fragment of the chromosomal region of PCL1233 flanked by Tn5lacZ (Fig. 1)* |
| pMP5211 | pME3049, which contains a 1.8-kb EcoRI fragment from pMP5206 used to isolate a larger DNA fragment from WCS365 by homologous recombination* (Fig. 1) |
| pMP5209 | pME3049 containing an 8.5-kb fragment of WCS365, obtained by homologous recombination and spanning the Tn5lacZ insertion* |
| pMP5230/31 | A 5-kb HindIII fragment derived from pMP5209 cloned into pBluescript used for single-stranded sequencing* (Fig. 1) |
| pMP5213 | The 8.5-kb EcoRI fragment of pMP5209 in pMP2740, used for homogenotization* |
| pMP5215 | Complementing 5-kb HindIII fragment, derived from pMP5209 and cloned into pWTT2081* |
| pMP5244 | pWTT2081 containing the 5-kb fragment of pMP5215 with a small insertion in sss* (Fig. 1) |
*This paper.
Various Escherichia coli strains were grown overnight at 37°C in liquid or on solidified Luria–Bertani medium (LBM) (20). When appropriate a final concentration of the following antibiotics was added to these media: nalidixic acid, 15 μg/ml; tetracyclin, 40 μg/ml; streptomycin, 500 μg/ml; spectinomycin, 200 μg/ml; kanamycin 50 μg/ml; and carbenicillic acid, 100 μg/ml. To distinguish between the wild-type P. fluorescens WCS365 cells and its Tn5lacZ derivatives, 5-bromo-4-chloro-3-indolyl β-d-galactoside was added to the medium to 40 μg/ml.
The ability to produce siderophores such as pyoverdin was tested on solidified LBM supplemented with 2.5 or 5 mM ZnSO4 (13). Addition of the salt was followed by adjustment of the pH of the medium.
Plasmid Constructions.
To isolate and study the DNA fragment from mutant PCL1233, which contains the Tn5lacZ (8) insertion, we isolated a flanking region of the transposon by using SalI to digest the chromosomal DNA of mutant PCL1233. Subsequently, ligation of this digested chromosomal DNA into pIC20H (17) and transformation to E. coli resulted in plasmid pMP5206 (Fig. 1 and Table 1). Plasmid pMP5206 was subsequently used to construct pMP5211 (Fig. 1 and Table 1). After introduction of pMP5211 into P. fluorescens WCS365 and selection for a single homologous recombination event, total DNA was isolated and digested with BamHI. Self-ligation and selection for kanamycin resistance (21) resulted in plasmid pMP5209, which contains an 8.5-kb EcoRI fragment of P. fluorescens WCS365 spanning the Tn5lacZ insertion (Table 1). Various subclones of pMP5209 were generated (Fig. 1 and Table 1). Plasmid pMP5215 was used for complementation studies. Plasmid pMP5244 was constructed after digestion of plasmid pMP5215 with BglII (Fig. 1 and Table 1). To create a blunt end, the BglII site present in the xerC/sss homologue was subsequently filled in by using the Klenow fragment of DNA polymerase I, and the blunt ends were religated.
Figure 1.
Restriction map of the DNA fragment from P. fluorescens WCS365, which includes the 5-kb HindIII fragment that complements the colonization defect. (A) The 5-kb HindIII fragment in which arrows indicate the direction and the size of the six ORFs. (B) The fragments used for complementation or single strand-DNA sequencing. B, BamHI; C, ClaI; E, EcoRI; Ev, EcoRV; H3 HindIII; K, KpnI; S, SmaI; Sa, SalI.
Plasmid pMP5213, harboring the total 8.5-kb EcoRI fragment isolated from WCS365 (Table 1), was introduced into mutant strain PCL1233. A selection was made for a double homologous recombination, which results in white kanamycin-sensitive colonies. One of these was designated PCL1231.
Triparental mating and electroporation were used to transfer the various plasmids to P. fluorescens WCS365. Electroporations were performed according to the manufacturer’s guidelines using a Bio-Rad Gene Pulser.
DNA Modifications.
Standard molecular techniques were performed as described (20). Single-strand sequencing of pMP5230/31, which contain a subclone of 5 kb of the originally isolated fragment, was performed by using the dideoxynucleotide chain-termination method (23). Computer analysis of the obtained sequence results was performed with the GCG Wisconsin software. The site of the Tn5lacZ insertion was determined by sequencing pMP5206 with a specific Tn5 primer constructed for the ends of the Tn5lacZ. To determine whether the integration host factor (IHF) was actually binding to the putative IHF binding site, an IHF binding study was performed (24).
Methods Used for Isolation and Characterization of Mutant PCL1233.
The method of transposon mutagenesis used to obtain colonization-impaired mutants has been described (7). Individual mutants were screened by inoculation of potato stem cuttings with a 1:1 (OD620 = 0.1) mixture of cells of the parental strain and one mutant in the compact gnotobiotic system (6, 7, 21). To determine the numbers of mutant and wild type cells on the root tip in a competitive root colonization, a time course study was performed by sampling and estimating the numbers of each strain on 10 plants approximately every other day starting at day 5. Competitive root tip colonization on tomato, radish, and wheat was determined in the gnotobiotic system described by Simons et al. (7), referred to herein as the standard system.
To determine colonization abilities in unsterile potting soil (Jongking Grond B.V., Aalsmeer, The Netherlands), germinated tomato seeds were bacterized with a 1:1 mixture (OD620 = 0.1) of mutant and parental cells and contained in plastic pots. The plants were grown for 10 days in a climate-controlled growth chamber. Root samples were taken, the adhering soil and bacteria were removed by shaking, and soil suspensions were plated on King B plates supplemented with nalidixic acid, cycloheximide (100 μg/ml), and 5-bromo-4-chloro-3-indolyl β-d-galactoside (80 μg/ml).
To determine whether mutant PCL1233 is actually a derivative of P. fluorescens WCS365 and whether differences exist in the ability of the mutant and the wild type to oxidize 95 different C-sources, the Biolog system (22) was used according to the manufacturer’s guidelines. Isolation of cell envelopes and visualization of cell-envelope protein patterns was performed according to Lugtenberg et al. (25). Analysis of LPS ladder patters was performed according to de Weger et al. (26). To measure motility, semisolid (0.35% agar) agar plates containing King B medium diluted 1:20 were used (4). Growth in competition in SSM was carried out as described by Dekkers et al. (27).
RESULTS AND DISCUSSION
Mutant Strain PCL1233 Is a Colonization-Impaired Mutant.
After screening on potato roots and preliminary identification as a competitive colonization mutant (11, 28), mutant strain PCL1233 was selected and tested for previously described colonization traits. The parent strain P. fluorescens WCS365 and the mutant strain PCL1233 were indistinguishable with respect to motility, the production of the O-antigen of LPS, amino acids, and vitamin B1. The parent and mutant were also indistinguishable with respect to growth rate in SSM in competition, in their cell envelope proteins, and in the oxidation of 95 different carbon sources in Biolog plates (22). We conclude that mutant PCL1233 does not differ from the wild-type strain P. fluorescens WCS365 in any of the established colonization traits. Mutant PCL1233 was therefore selected for further molecular analysis.
Root Colonization Characteristics of Mutant PCL1233.
Root colonization experiments in which cells of mutant PCL1233 and parental strain P. fluorescens WCS365 were coinoculated on sterile potato plantlets showed consistent and statistically significant, at least 50-fold, less colonized root tips for mutant PCL1233 compared with its wild-type parent (Tables 2 and 3). However, inoculation of mutant PCL1233 alone on potato, tomato, and wheat resulted in bacterial numbers at the root tips comparable to those measured for the wild type (Table 2), indicating that mutant strain PCL1233 is not colonization-defective because of supersensitivity to compounds exuded by the roots.
Table 2.
Populations of mutant strain PCL1233 with the wild-type (WCS365) on root tips with different plant species and rooting media
| Colonization conditions | Mean log10(CFU + 1)/ cm of root tip
|
|
|---|---|---|
| WCS365 | PCL1233 | |
| Gnotobiotic compact sand column in competition on potato | ||
| Exp. 1 | 5.5a | 0.3b |
| Exp. 2 | 5.4a | 1.4b |
| Strains alone in the gnotobiotic compact sand column | ||
| Potato | — | 5.6a |
| 5.7a | — | |
| Tomato | — | 4.3a |
| 4.4a | — | |
| Wheat | — | 4.1a |
| 5.0a | — | |
| Standard gnotobiotic sand column in competition | ||
| Potato | 3.6a | 1.1b |
| Tomato | 4.2a | 1.6b |
| Wheat | 4.7a | 3.8b |
| Radish | 4.3a | 2.9b |
| Potting soil in competition | ||
| Tomato | 4.1a | 1.4b |
Inoculation was carried out on sterile stem cuttings of potato and germinated seeds of radish, tomato, and wheat (7). In every experiment, 10 plants were inoculated. When values from the same experiment are followed by a different letter, they are significantly different at P = 0.05 according to the Wilcoxon Mann–Whitney test (29).
Table 3.
Populations of wild-type (WCS365 or PCL1239), complemented mutant (PCL1234), and mutant complemented with a deleted xerC (PCL1250), on potato root tips 14 days after inoculation of potato stem cuttings with a 1:1 mixture
| Colonization test | Mean log10(CFU + 1)/ cm of root tip
|
|
|---|---|---|
| Wild type | Various xerC derivatives | |
| WCS365 vs. PCL1233 | 4.6a | 2.9b |
| 6.1a | 3.1b | |
| PCL1239 vs. PCL1234 | 4.6a | 4.6a |
| 5.1a | 5.5a | |
| PCL1239 vs. PCL1250 | 5.6a | 3.8b |
| 5.0a | 3.4b | |
| 7NSK2 vs. SSS | 5.3a | 1.6b |
| 5.6a | 4.5b | |
Statistical analysis was performed as described in Table 2.
The results in competition with the parent (Table 2) show that the mutation also exhibits an inability to colonize roots of tomato, radish, and wheat, indicating that the colonization trait impaired in mutant PCL1233 has a broad host range. Also in potting soil, mutant PCL1233 appeared to be defective in tomato root tip colonization when coinoculated with the parental strain (Table 2). Similar results in potting soil were described by Glandorf (11) for the colonization of the lower parts of potato roots. These results in soil indicate that the colonization trait impaired in mutant PCL1233 is likely to also play a role under more natural conditions. A time course experiment in which PCL1233 was coinoculated 1:1 with the parental strain on the potato root showed that the wild-type numbers isolated from the root tip remained constant or decreased slowly from 6.7 log10(CFU + 1)/cm of root (where CFU is colony-forming units) at day 5 to 6.4 at day 14. The numbers of mutant cells at the growing root tip decreased in time: values of 5.9, 5.4, 5.1, 5.1, and 3.8 log10(CFU + 1)/cm of root were found on days 5, 7, 10, 12, and 14, respectively.
Identification of the DNA Fragment with the Colonization Gene.
Two approaches were followed to test whether the Tn5lacZ insertion is responsible for the colonization defect. (i) Strain PCL1234 was tested in competition with PCL1239, to rule out the possibility of a genetic and/or physiological burden for the complemented mutant in the rhizosphere (10). Introduction of pMP5215 in strain PCL1233 caused complementation of the colonization defect in PCL1233 (Table 3). (ii) The wild-type fragment present in pMP5213 was exchanged for the mutated fragment present in mutant PCL1233. After selection, strain PCL1231, a white derivative of PCL1233, was isolated that behaved indistinguishably from P. fluorescens WCS365 in a competitive colonization assay (results not shown), indicating that the colonization defect of PCL1233 is linked to the Tn5lacZ insertion.
Homologues of ORF1–6.
Sequence analysis of 4,525 bp of pMP5230/31 revealed the presence of six ORFs, designated ORF1–6, of which ORF6 is incomplete (Fig. 1A). The average G+C content of the predicted ORFs is 63.7%, which is consistent with values found in other Pseudomonas species (30). In view of the close proximity of the six ORFs and the fact that no putative promoter sequences were found in the intergenic regions between the six ORFs, they probably form one multicistronic transcriptional unit. All ORFs, except ORF5, were preceded by a putative Shine–Dalgarno sequence that is correctly spaced from the various start codons. Lack of a Shine–Dalgarno sequence in front of ORF5 suggests that ORF4 and ORF5 are translationally coupled.
The first gene in the operon (Fig. 1A) shows an ORF of 171 bp; the deduced amino acid sequence indicates a 57-amino acid protein with a calculated molecular weight of 6,073. The predicted product shows 53% identity and 66.7% similarity at the amino acid level to LppL, a lipopeptide described for Pseudomonas aeruginosa (31).
ORF2 (Fig. 1A; 1,248 bp) could encode a protein of 416 amino acids with a calculated molecular weight of 45,222. The putative amino acid sequence shows homology with LysA from various bacteria (32–35). The best identity is found with LysA of P. aeruginosa (32), of which 84% of the amino acids are identical and 92.5% are similar. lysA encodes meso-diaminopimelate decarboxylase, the last enzyme in l-lysine biosynthesis.
ORF3 (Fig. 1A; 828 bp) could encode a protein of 276 amino acids with a calculated molecular weight of 29,920 and substantial amino acid sequence similarity (71.5%) and identity (54%) with diaminopimelate epimerase or DapF of E. coli K12 (36). This enzyme is involved in the formation of meso-diaminopimelate, an essential component of peptidoglycan in rod-shaped bacteria and the direct precursor of l-lysine. Part of this dapF gene was also found in the biocontrol strain P. aeruginosa 7NSK2 (13). Sequence homology of ORF3 with the dapF is even more striking, with 80% identity and 90% similarity over 108 amino acids.
The site of the Tn5lacZ insertion was shown to be in ORF4 (Fig. 1A; 720 bp), which encodes a putative protein of 240 amino acids, designated Orf240, with a molecular weight of 26,820. The Tn5lacZ insertion is located 150 bp downstream of the orf240 start codon. The function of this ORF, designated orf235 in E. coli (14) (29% identity and 53% similarity at the amino acid level) and orf233 in P. aeruginosa 7NSK2 (13) (67% identity and 81.5% similarity at the amino acid level), is unknown.
ORF5 contains 897 bp and encodes a putative protein of 299 amino acids with a calculated molecular weight of 33,772. The putative protein sequence of this ORF, designated Sss, shows a high similarity (Fig. 2) to the Sss protein of P. aeruginosa (71% identity and 83% similarity) (13) and the XerC protein of E. coli (48% identity and 68% similarity) (14). Both XerC and Sss are members of the λ integrase family of site-specific recombinases. Other well known but slightly less homologous members of this family are XerD, formerly known as XrpB (38), FimE, and FimB (37, 39) (see Fig. 2), and λ integrase (40).
Figure 2.
Comparison of the C-terminal part of the deduced amino acid sequence of the Sss protein of P. fluorescens WCS365 (boldface type) with that of other site-specific recombinases. Amino acids found in more than four sequences are marked by an asterix. The conserved tyrosine residue is boxed. The protein is compared with FimB and FimE (37, 39) of E. coli, Sss of P. aeruginosa 7NSK2 (SssPa) (13), and XerC (14) and XerD (38) of E. coli.
Because ORF6 is not necessary for complementation, it was only partially sequenced. The deduced amino acid sequence of this ORF shows homology with Orf238 of E. coli, which, as in P. fluorescens WCS365, is also located downstream of xerC (14). The deduced amino acid sequence of the first 66 amino acids of ORF6 shows significant similarity (44%) and identity (34%) to Orf238, the function of which in E. coli is not known (14). A striking resemblance is observed for E. coli, P. aeruginosa 7NSK2, and P. fluorescens WCS365 in that the genetic organization of the dapF, orF240/235/233 genes, and genes located further downstream is identical for all three strains, although the genetic information preceding these four ORFs is different.
Regulation of the Promoter.
A putative σ70 promoter sequence (−35 region TAGGCA 17 bp −10 region TATACT) was found in front of ORF1. A putative IHF binding site (41) was present between the σ70 binding site and the ATG start codon of the first ORF. However, when a DNA fragment containing this IHF binding site was incubated with purified E. coli IHF protein, no IHF was bound to the DNA (results not shown).
A 300-bp region upstream of the ATG start codon of the lppL gene of P. aeruginosa is 71% homologous at the DNA level to the region upstream of the lppL homologue in strain WCS365. Even the putative −10 and −35 sequences, including the spacing of −10 and −35, are completely homologous, indicating strong conservation of this particular promoter region. This latter observation suggests an important role for this upstream region, perhaps in binding of certain regulatory elements. In P. aeruginosa, lppL is also located upstream of the lysA gene. A location in the outer membrane is suggested for LppL but its function is still unknown (31). A role in the regulation of lysA mediated by an attenuation-type system of regulation was proposed by Jann et al. (31); this could explain the conservation of the promoter region upstream of the lppL/lysA genes in both Pseudomonas species.
Three global regulators have been described to affect switching induced by fimB and fimE, namely IHF, the histone-like protein H-NS, and the leucine-responsive element (Lrp) (37, 42–46). Lrp differentially regulates switching in various media and is involved in the stimulation of switching upon addition of aliphatic amino acids (47). Furthermore, the isolation of fimbriate E. coli is favored under certain growth conditions (ref. 46 and references therein). Gally et al. (47) showed further that the fim switch is differently regulated by environmental conditions such as temperature and medium (47). For XerC, the presence of l-arginine functioning as a corepressor of ArgR positively influences site-specific recombination at cer (48). As a result of these findings and the presence of the lysA and dapF genes upstream of the sss homologue in P. fluorescens WCS365, we tested a possible regulatory role for the amino acid l-lysine in the regulation of switching induced by Sss. However, l-lysine added at 1 mM to the gnotobiotic system had no effect on the competitive colonization behavior of wild type and mutant (results not shown). If l-lysine suppressed transcription of the operon, one should expect the parent to behave as the mutant.
The ORF5 Site-Specific Recombinase Functions in Competitive Colonization.
Because the mutation is located in orf240 and pMP5215 complements for colonization, orf240 and the xerC/sss homologue are the only candidates that can be responsible for the colonization defect of mutant PCL1233. A small insertion was introduced in the xerC/sss gene present on the complementing plasmid (pMP5244). This mutation is located downstream of the actual transposon insertion in mutant PCL1233. Transfer of plasmid pMP5244 to PCL1233 resulted in strain PCL1250. In contrast with PCL1234, PCL1250 is not complemented for root tip colonization (Table 3). Therefore, xerC/sss is crucial for colonization, whereas a role of orf240 cannot be excluded.
The sss mutant of P. aeruginosa 7NSK2 was reported to have a prolonged lag phase (13) and inoculation of this mutant on maize led to a 4-fold reduction in colonization in a system quite different from ours (13, 49). In contrast, we observed a normal logarithmic-growth phase for mutant PCL1233. When the colonizing ability of the sss mutant of P. aeruginosa 7NSK2 was tested in our gnotobiotic system, it appeared to be at least 10- to 1,000-fold impaired in its ability to colonize potato root tips after inoculation with a 1:1 mixture of mutant and parental cells (Table 3). These results confirm independently the role of Sss in colonization. Furthermore, sss mutants were described not to be able to produce the siderophore pyoverdin when grown in LBM supplemented with Zn2+ (13). Incubation of P. fluorescens WCS365 and mutant PCL1233 on LBM supplemented with Zn2+ showed that neither P. fluorescens WCS365 nor mutant PCL1233 produces a siderophore under these circumstances, whereas 7NSK2 does. Höfte et al. (49) attributed the observed slightly decreased root colonization capacity for the sss mutant of 7NSK2 to an altered ability of the mutant to induce pyoverdin and related outer membrane proteins (49). In contrast to their results, (i) the effect of an sss mutation on colonization is much larger in our more sensitive root tip assay and (ii) we did not find the siderophore effect described by Höfte et al. (12).
Members of the λ integrase family of site-specific recombinases such as XerC and XerD have been described to have various functions in E. coli. XerC and XerD act cooperatively in the process of monomerization of ColE1 plasmids and of the circular chromosome at, respectively, cer and dif sites (38). xerC mutants of E. coli are reported to have a slow recovery from stationary phase and to show a variable growth rate in the exponential phase when compared with a xerC+ strain (50). This is clearly not the case for mutant PCL1233, which also does not show cell division defects (results not shown).
The observed homologies indicate that the xerC/sss homologue of P. fluorescens WCS365 is a member of the λ integrase family of site-specific recombinases. These enzymes promote conservative reciprocal recombination (which does not require DNA synthesis) between two small (approximately 15 bp) homologous DNA sequences. The presence and orientation of two of these sequences will lead to inversion or excision of the DNA fragment situated between these small recognition sites (51). Comparison of all site-specific recombinases shows that homology is particularly striking in the C-terminal part (Fig. 2), in which a perfectly conserved tyrosine residue is present and thought to form a linkage with the DNA (40).
Most DNA rearrangements involved in phenotypic switching regulate expression of phase-variable cell surface antigens (52) such as fimbriae, flagella, LPS, and lipoprotein. FimE and FimB play a role in phase variation of type 1 fimbriae in E. coli. Both proteins are thought to be site-specific recombinases (37) acting on a specific 314-bp DNA element located in the promoter region of fimA, the main fimbrial subunit gene (39, 53). From the homology found with FimB and FimE (Fig. 2), we postulate that the Sss protein is involved in DNA rearrangements that lead to phenotypes with different colonization abilities. DNA rearrangements can also regulate the production of two forms of LPS related to pathogenicity in the intracellular pathogen Francisella tularensis (54) and antigenic variation of surface lipoprotein antigens in Mycoplasma bovis (55). Also the regulation of the alternate expression of two flagellin genes in Salmonella typhymurium is regulated by an invertible DNA element modulated by recombination (56). The fact that most DNA rearrangements involved in phenotypic switching regulate expression of phase-variable cell surface antigens and our observation that mutant PCL1233 is only impaired in competitive colonization but not when inoculated alone lead us to speculate that PCL1233 is impaired in competition for nutrients or for sites on the root surface. Flagella, crucial for colonization (4), presumably play a role in competition for nutrients through chemotaxis. Moreover, the composition of the LPS may affect motility because O-antigen-negative mutants of P. fluorescens WCS365 are often less motile than their parental strain (L.C.D., unpublished results). Fimbriae, LPS, and cell-surface proteins could be involved in competition for sites on the root surface. We favor the hypothesis that a cell-surface-related molecule or trait is subject to phase variation by Sss and that, as a result, competition for nutrients or sites on the root surface is affected. This hypothesis is also consistent with the observation that the mutant is impaired only in competition. The discovery that Sss plays a role in root colonization shows the importance of phase variation in this plant–microbe interaction.
Colony sector formation is often associated with DNA rearrangements. Interestingly, we observed that old colonies of WCS365 contain morphologically distinct sectors. Sectoring has been observed less frequently in mutant strain PCL1233 than in WCS365 (L.C.D., unpublished results). Dybvig (52) postulated that bacteria are able to generate subpopulations in certain environments, by means of DNA rearrangements, that result in different abilities to adjust to sudden environmental changes. By this mechanism, such a bacterial population is at all times able to respond adequately to environmental changes even if only a few cells of a subpopulation are surviving. According to this notion, mutant PCL1233 is locked in a genetic configuration that is less rhizosphere competent when compared with cells of the parental strain.
Acknowledgments
We thank N. Goosen for supplying us with purified E. coli IHF and technical assistance with binding studies, H. Spaink and L. de Weger for useful discussions, and M. Höfte for P. aeruginosa 7NSK2 and its mutant. We thank I. Brand for isolation of mutant PCL1233. The investigations were partly funded by the European Union Biotech Program BIO2-CT93.0196. C.C.P. is funded by the Netherlands Organization for Scientific Research-Ministry of Agriculture, Nature Management and Fisheries Priority Program Crop Protection, Project 805.45.008.
ABBREVIATIONS
- LPS
lipopolysaccharide
- IHF
integration host factor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y12268).
References
- 1.Schippers B, Bakker A W, Bakker P A H M. Annu Rev Phytopathol. 1987;25:339–358. [Google Scholar]
- 2.Weller D M. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 3.Bull C T, Weller D M, Thomashow L S. Phytopathology. 1991;81:954–959. [Google Scholar]
- 4.de Weger L A, van der Vlugt C I M, Wijfjes A H M, Bakker P A H M, Schippers B, Lugtenberg B J J. J Bacteriol. 1987b;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Weger L A, Bakker P A H M, Schippers B, van Loosdrecht M C M, Lugtenberg B J J. In: Signal Molecules in Plants and Plant–Microbe Interactions. Lugtenberg B J J, editor. Berlin: Springer; 1989. pp. 197–202. [Google Scholar]
- 6.Simons M, Permentier H P, de Weger L A, Wijffelman C A, Lugtenberg B J J. Mol Plant-Microbe Interact. 1997;10:102–106. [Google Scholar]
- 7.Simons M, van der Bij A J, Brand J, de Weger L A, Wijffelman C A, Lugtenberg B J J. Mol Plant-Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 8.Lam S T, Ellis D M, Ligon J M. Plant and Soil. 1990;129:11–18. [Google Scholar]
- 9.Geels F P, Schippers B. Phytopathol Z. 1983;108:193–206. [Google Scholar]
- 10.van der Bij A J, de Weger L A, Tucker W T, Lugtenberg B J J. Appl Environ Microbiol. 1996;62:1076–1080. doi: 10.1128/aem.62.3.1076-1080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glandorf D C M. Ph.D. thesis. Utrecht, The Netherlands: University of Utrecht; 1992. [Google Scholar]
- 12.Höfte M, Seong K Y, Jurkevitch E, Verstraete H. Plant and Soil. 1991;130:249–257. [Google Scholar]
- 13.Höfte M, Dong Q, Kourambas S, Krishnapillai V, Sherratt D, Mergeay M. Mol Microbiol. 1994;14:1011–1020. doi: 10.1111/j.1365-2958.1994.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 14.Colloms S D, Sykora P, Szatmari G, Sherratt D J. J Bacteriol. 1990;172:6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer H W, Roulland-Dussoix D. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 16.Voisard C, Bull C T, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. In: Molecular Ecology of Rhizosphere Microorganisms. O’Gara F, Dowling D N, Boesten B, editors. New York: VCH; 1994. pp. 67–89. [Google Scholar]
- 17.Marsh J L, Erfle M, Wykes E J. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 18.King E O, Ward M K, Raney D E. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 19.Meyer J M, Abdallah M A. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.de Weger L A, Dekkers L C, van der Bij A J, Lugtenberg B J J. Mol Plant-Microbe Interact. 1994;7:32–38. [Google Scholar]
- 22.Bochner B R. Nature (London) 1989;339:157–158. doi: 10.1038/339157a0. [DOI] [PubMed] [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goosen N, van Ulsen P, Zulianello L, van de Putte P. Methods Enzymol. 1996;274:232–242. doi: 10.1016/s0076-6879(96)74006-5. [DOI] [PubMed] [Google Scholar]
- 25.Lugtenberg B J J, Meyers J, Peters R, van der Hoek P, van Alphen L. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 26.de Weger L A, Jann B, Jann K, Lugtenberg B. J Bacteriol. 1987;169:1441–1446. doi: 10.1128/jb.169.4.1441-1446.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekkers L C, Bloemendaal C P, de Weger L A, Wijffelman C A, Spaink H P, Lugtenberg B J J. Mol Plant-Microbe Interact. 1998;11:45–56. doi: 10.1094/MPMI.1998.11.1.45. [DOI] [PubMed] [Google Scholar]
- 28.Lugtenberg B J J, de Weger L A. In: Pseudomonas: Molecular Biology and Biotechnology. Galli E, Silver S, Witholt B, editors. Washington, DC: Am. Soc. Microbiol.; 1992. pp. 13–19. [Google Scholar]
- 29.Sokal R R, Rolhf F J. Biometry. San Francisco: Freeman; 1981. [Google Scholar]
- 30.Palleroni N J. In: General Properties and Taxonomy of the Genus Pseudomonas. Clarke P H, Richmond M A, editors. London: Wiley; 1975. pp. 1–36. [Google Scholar]
- 31.Jann A, Cavard D, Martin C, Cami B, Patte J-C. Mol Microbiol. 1990;4:667–682. doi: 10.1111/j.1365-2958.1990.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 32.Martin C, Cami B, Yeh P, Stragier P, Parsot C, Patte J. Mol Biol Evol. 1988;5:549–559. doi: 10.1093/oxfordjournals.molbev.a040515. [DOI] [PubMed] [Google Scholar]
- 33.Marcel T, Archer J A C, Mengin-Lecreulx D, Sinskey A J. Mol Microbiol. 1990;4:1819–1830. doi: 10.1111/j.1365-2958.1990.tb02030.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto J, Shimizu M, Yamane K. Agric Biol Chem. 1991;55:1615–1626. [PubMed] [Google Scholar]
- 35.Mills D A, Flickinger M C. Appl Environ Microbiol. 1993;59:2927–2937. doi: 10.1128/aem.59.9.2927-2937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richaud C, Printz C. Nucleic Acids Res. 1988;16:10367. doi: 10.1093/nar/16.21.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorman C J, Higgins C F. J Bacteriol. 1987;169:3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blakely G, May G, McCulloch R, Arciszewska L K, Burke M, Lovett S T, Sherratt D. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 39.Klemm P. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Pierson L S, III, et al. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goosen N, van de Putte P. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 42.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 43.Eisenstein B I, Sweet D S, Vaughn V, Friedman D I. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawula T H, Orndorff P E. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spears P A, Schauer D, Orndorff P E. J Bacteriol. 1986;168:179–185. doi: 10.1128/jb.168.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stirling C J, Szatmari G, Stewart G, Smith M C M, Sheratt D J. EMBO J. 1988;7:4389–4395. doi: 10.1002/j.1460-2075.1988.tb03338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Höfte M, Boelens J, Verstraete W. J Plant Nutr. 1992;15:2253–2262. [Google Scholar]
- 50.Blakely G, Colloms S, May G, Burke M, Sheratt D. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 51.Sadowski P. J Bacteriol. 1986;165:341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dybvig K. Mol Microbiol. 1993;10:465–471. doi: 10.1111/j.1365-2958.1993.tb00919.x. [DOI] [PubMed] [Google Scholar]
- 53.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cowley S C, Myltseva S V, Nano F E. Mol Microbiol. 1996;20:867–874. doi: 10.1111/j.1365-2958.1996.tb02524.x. [DOI] [PubMed] [Google Scholar]
- 55.Lysnyansky I, Rosengarten R, Yogev D. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeig J, Silverman H, Hilmen H, Simon M. Science. 1977;196:170–175. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]