Abstract
The T-DNA transfer apparatus of Agrobacterium tumefaciens mediates the delivery of the T-DNA into plant cells, the transfer of the IncQ plasmid RSF1010 into plant cells, and the conjugal transfer of RSF1010 between Agrobacteria. We show in this report that the Agrobacterium-to-Agrobacterium conjugal transfer efficiencies of RSF1010 increase dramatically if the recipient strain, as well as the donor strain, carries a wild-type Ti plasmid and is capable of vir gene expression. Investigation of possible mechanisms that could account for this increased efficiency revealed that the VirB proteins encoded by the Ti plasmid were required. Although, with the exception of VirB1, all of the proteins that form the putative T-DNA transfer apparatus (VirB1–11, VirD4) are required for an Agrobacterium strain to serve as an RSF1010 donor, expression of only a subset of these proteins is required for the increase in conjugal transfer mediated by the recipient. Specifically, VirB5, 6, 11, and VirD4 are essential donor components but are dispensable for the increased recipient capacity. Defined point mutations in virB9 affected donor and recipient capacities to the same relative extent, suggesting that similar functions of VirB9 are important in both of these contexts.
The capacity of Agrobacterium tumefaciens to transfer DNA—and proteins—into the cytoplasm, and ultimately the nucleus, of plant cells is an intriguing and unique example of horizontal genetic flow. In the case of wild-type A. tumefaciens the transfer of a portion of the Ti (tumor-inducing) plasmid into plant cells and the subsequent expression of this “T-DNA” results in uncontrolled proliferation of cells and the formation of crown gall tumors. The DNA transfer process is initiated when virulent bacteria are exposed to wound sites on the plant, which, during the wound repair process, release a variety of phenolics and sugars. These molecules induce the expression of a series of genes on the Ti plasmid of virulent Agrobacteria, and the resultant virulence (Vir) proteins are responsible for the formation of the transferred DNA intermediate and its delivery into plant cells (for reviews see refs. 1–3).
Detailed investigation of the T-DNA transfer process suggests that it evolved from a DNA transfer system homologous to conjugative plasmid transfer between bacteria (4). In both conjugal transfer of the IncP plasmid RP4 and T-DNA transfer, a site-specific endonuclease (TraI and VirD2, respectively) creates a single-strand nick and attaches to the 5′ end of the nicked DNA (5). These single-stranded DNA–protein complexes ultimately interact with single-stranded DNA-binding proteins (SSB and VirE2) at some time during the transfer process (6–8) and are transported into the recipient cells. Additionally, a series of membrane-associated proteins from the tra2 and virB operons (of RP4 and Ti, respectively) show strong homology to each other and to tra-encoded proteins of other conjugal plasmids (IncN, IncW) and to the pertussis toxin secretion apparatus in Bordetella pertussis (9, 10). In each case, these proteins are postulated to form a membrane-bound complex responsible for DNA or protein translocation (11).
The proposed VirB complex is under intensive investigation. Several studies have begun to characterize the localization of the 11 VirB proteins and the interactions of VirB proteins that are thought to form a multimeric protein complex that spans the inner and outer membranes (for review see ref. 11) and is involved in pilus formation (12). Further, it has been shown that the VirB proteins are responsible for the transfer of a variety of different substrates into plant cells. These include not only the T-DNA intermediate, but also the VirE2 (8, 13) and VirF proteins (14, 15). One intriguing result has been that for each of the processes so far studied each of the VirB proteins, except VirB1, is required (6, 16–19). This indicates that each of the VirB proteins, with the possible exception of VirB1, is necessary to construct a functional transport complex.
The proposal that T-DNA transfer is homologous to conjugal DNA transfer is supported further by the findings that the Ti plasmid virulence gene system supports the transfer of the IncQ plasmid RSF1010 into either plants (20) or other Agrobacteria (21) in a fashion that depends on the mob and oriT sequences of this plasmid. Moreover, a functional virB operon is required for either type of transfer (18, 22). In this study, we have examined interbacterial transfer of an RSF1010 derivative and demonstrated that its very low efficiency of conjugation can be increased dramatically by the presence of a wild-type Ti plasmid in the recipient Agrobacterium. Investigation of the molecular and genetic mechanisms underlying this increased recipient capacity showed that expression of the virB operon in the recipient Agrobacterium strain is crucial. Moreover, we show that only a subset of the VirB proteins is required for this phenomenon, thereby distinguishing between the VirB proteins that are required solely for DNA export and those required for both export and import of DNA.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
Agrobacterium tumefaciens strain A348 has the wild-type chromosomal background of strain C58 and contains pTiA6 (23) whereas strain UIA 143 has the C58 chromosomal background carrying recA∷Ery140, rendering it erythromycin-resistant, and does not contain a Ti plasmid (24). The PC10XX Agrobacterium strains are derived from A348 and carry deletions of individual virB ORFs (19). Strain At12506 also is derived from strain A348 and contains an insertion mutation in virD4 (29).
Agrobacteria were routinely grown at 25°C in Luria–Bertani (LB) broth or in AB induction medium (25) under appropriate antibiotic selection. Antibiotic concentrations were the following (liquid/solid medium in μg/ml): gentamicin (100/100), erythromycin (150/150), spectinomycin (25/75), carbenicillin (30/100), and kanamycin (10/50). Plasmid DNA was introduced into Agrobacterium either by electroporation (Bio-Rad, Gene Pulser) or by triparental matings (introduction of pJB31) using the Escherichia coli strain S17–1 that carries the transfer functions of plasmid RP4 integrated into the chromosome (26).
Plasmids.
pJB20 is an IncW broad host range cloning vector and is ampicillin- and tetracycline-resistant (28). pJB20–0, 20–7, 20–290, and 20–294 are derivatives of pJB20 carrying wild-type (20–0) and mutant versions of virB9 expressed by the virB promoter (28). pJB31 is a derivative of the IncQ plasmid RSF1010 and is spectinomycin-resistant (28). In some cases pJB61, an IncP, gentamicin-resistant plasmid (28), was used as the counterselectable marker in conjugation experiments. pCF218 is an IncP, tetracycline-resistant plasmid that overexpresses traR of pTiA6 (27).
Conjugation of pTiA6 into Strain UIA143.
Although most Ti plasmids can be mobilized between Agrobacterium strains via the plasmid’s tra genes, pTiA6 carries a 12.65-kb deletion that includes part of the tra operon and renders it nonconjugable (27). However, A348—carrying pTiA6—becomes conjugation competent when traR is overexpressed from another plasmid (27). For Ti conjugation, LB-grown donor A348(pCF218) and recipient UIA143 cells were grown to an OD600 ≈2.0. Two hundred microliters of donor and 50 μl of recipient were centrifuged and washed twice in water, and each was resuspended in 10 μl water. These were mixed, spotted on minimal AB medium (1% glucose), and incubated overnight at 25°C. Transconjugants were selected by streaking the conjugal mix on minimal AB plates containing octopine as the sole carbon and nitrogen source (utilization encoded by pTiA6) in the presence of erythromycin (resistance encoded by UIA143). Transfer of the Ti plasmid was confirmed by testing transconjugant colonies for tumor initiation on Kalanchoe daigremontiana and for VirB10 expression, probing immunoblots with anti-VirB10 antisera (42).
VirB-Dependent Conjugation of RSF1010 Derivative pJB31.
Conjugation assays were performed as described previously (28). LB-grown cultures of bacteria were diluted into AB induction medium to an OD600 of 0.25 and grown for 6 h in the presence of the inducer acetosyringone (AS, Aldrich). Unless otherwise stated donor and recipient were mixed at a ratio of 5:1 for testing recipient capacities or at a ratio of 1:5 for testing donor abilities. Significantly higher ratios of donor to recipient (25:1) were used when testing strain UIA 143 as a recipient, because this strain grows more rapidly than the Ti plasmid-containing strains used as donors. Five microliters of the conjugation mix were spotted on 1.5 ml agar-solidified AB induction medium, containing various levels of AS, in 24-well plates. After 3 days of incubation at 25°C, cells were collected by washing the wells twice with 0.5 ml of 0.9% saline solution. Cell suspensions were diluted if necessary and plated on LB plates with appropriate antibiotic selection to recover donors, recipients, and transconjugants. Colonies were scored after 3 days of incubation at 25°C. Donor cells routinely contained the spectinomycin-resistant RSF1010 derivative pJB31 (28). If the recipient did not carry a chromosomally or Ti-encoded antibiotic resistance it was transformed with the gentamicin-resistant pSW213 derivative pJB61 (not capable of moving through the VirB pore) as counterselectable marker (28). To confirm AS dependency of conjugations, control matings were done in the absence of AS, and these resulted in transconjugant numbers <1. DNase in the medium (1 μg/ml) did not influence transfer efficiencies. Plasmid DNA (≈250 ng) of either pJB31 or pJB61 added to recipient or donor cells, respectively, before spotting on induction media did not result in any transformants.
RESULTS
The Ti Plasmid Increases the Capacity of A. tumefaciens to Serve as a Recipient in Conjugal Transfer of RSF1010.
During studies to examine the role of the virB genes in the conjugal transfer of the RSF1010 derivative pJB31 between Agrobacterium we noted that the numbers of transconjugants obtained were higher than published data (12, 21) when a Ti-plasmid containing strain, A348, was used as a recipient. To demonstrate that this was, in fact, because of the presence of the Ti plasmid, we compared conjugal efficiency by using as recipients either the Ti plasmidless strain UIA143 (24) or UIA143 carrying Ti plasmid pTiA6. In these, as in all the following conjugation experiments, the donor strain, A348, carried the IncQ derivative pJB31 (28). Donor and recipient bacteria were grown for 6 h in AB induction medium plus the vir-gene inducer AS, mixed, and transferred to solidified AB induction medium containing AS at various concentrations. After 3 days the conjugal mix was resuspended and plated onto selective media to determine the numbers of donors, recipients, and transconjugants (see Materials and Methods for details). The data demonstrate that in the conjugation assays using UIA143(pTiA6) as recipient, transconjugants are 3–4 log orders more abundant than when Ti-plasmidless UIA143 is the recipient (Table 1). In both cases the conjugal transfer of pJB31 is not observed in the absence of AS but increases with increasing AS concentrations. The results with the Ti plasmid-containing recipient are consistent with those of Beaupré et al. (28), who showed a similar high frequency of virB-mediated pJB31 conjugal transfer by using a pTiA6 containing strain, A348(pJB61), as a recipient.
Table 1.
Influence of the presence of pTiA6 on the capacity of A. tumefaciens UIA143 to function as recipient in virB-mediated transfer of the RSF1010 derivative pJB31
| [AS]†, μM | Recipient Ti status | Total number of output*
|
|||
|---|---|---|---|---|---|
| Donor, 108 | Recipient, 108 | Transconjugants | Transconjugants/output recipient ± SD‡ | ||
| 0 | − | 24.5 | 7.6 | 0 | 0 |
| 0 | + | 26.5 | 8.1 | 0 | 0 |
| 200 | − | 5.1 | 12.4 | <2 | (0.8 ± 0.8) 10−9 |
| 200 | + | 5.5 | 6.8 | 49,866 | (7.3 ± 1.5) 10−5 |
| 400 | − | 3.8 | 13.6 | 12 | (8.4 ± 3.5) 10−9 |
| 400 | + | 3.9 | 5.7 | 112,622 | (2.0 ± 0.8) 10−4 |
| 800 | − | 1.9 | 13.2 | 20 | (1.5 ± 0.6) 10−8 |
| 800 | + | 1.4 | 5.0 | 361,422 | (7.2 ± 0.7) 10−4 |
Donor, recipient, and transconjugants in mating mix after 3-day incubation.
Concentration of AS (μM) in the conjugation medium.
Data represent means (±SD) of triplicates from a single experiment. Three independent experiments with similar results were performed. Ratios between input donor [A. tumefaciens A348 (pJB31)] and input recipients were 25:1 for strain UIA143 and 7:1 for UIA143 (pTiA6).
A Subset of the VirB Proteins Is Required for Increased Efficiency of A. tumefaciens as Recipient.
Preliminary experiments indicated that the virA and virB operons each were required for A. tumefaciens to exhibit maximal efficiency as a recipient (data not shown). Because VirA is a sensor kinase involved in the AS-mediated activation of vir gene transcription, we focused our studies on the various VirB proteins as well as VirD4, which also is hypothesized to be part of the DNA transfer complex (11). A series of mutants derived from A348, each with a different virB gene deleted (the PC10XX series, ref. 19), as well as a virD4 insertional mutant (29), were used as recipients in conjugation assays. These experiments demonstrate that VirB3, 4, 7, 8, 9, and 10 are absolutely required for a strain to serve as a high efficiency recipient (Table 2); deletion of any one of these genes caused a dramatic loss in transconjugant numbers. Deletions of the virB1, 2, or 6 genes resulted in strains that were 2 log order better recipients than the Ti-plasmidless strains, but only 2–10% of wild type. In contrast, we observed that the loss of expression of either VirB5, 11, or VirD4 in the recipient caused little or no decrease in conjugation frequencies with the transconjugant numbers remaining at a highly elevated level (>30% of wild type) if the respective mutants served as recipients. To confirm the results obtained with the virB deletion strains, a series of strains with nonpolar transposon insertions into various virB genes (6, 16) also were tested as recipients in conjugation assays. Strains with such insertions (virB4-, virB5-, virB6-, virB8-, virB9-, and virB10-) showed a similar capacity to serve as recipients as was observed in the case of the virB deletions (data not shown).
Table 2.
Influence of various virB gene deletions on the capacity of A. tumefaciens strain A348 to function as recipient in virB-mediated transfer of the RSF1010 derivative pJB31
| Recipient strain† | Gene disrupted | Absolute number of output*
|
||||
|---|---|---|---|---|---|---|
| Donor, 108 | Recipient, 108 | Tcs | Tcs/output recipient ± SD‡ | % of A348 value | ||
| PC1001 | virB1 | 4.0 | 1.8 | 375 | (2.12 ± 0.24) 10−6 | 3.58 |
| PC1002 | virB2 | 3.9 | 2.7 | 425 | (1.52 ± 0.39) 10−6 | 2.48 |
| PC1003 | virB3 | 5.3 | 1.3 | 5 | (3.71 ± 2.84) 10−8 | 0.06 |
| PC1004 | virB4 | 6.0 | 1.4 | 24 | (1.71 ± 0.73) 10−7 | 0.28 |
| PC1005 | virB5 | 5.0 | 1.2 | 4,083 | (3.54 ± 0.49) 10−5 | 57.65 |
| PC1006 | virB6 | 5.0 | 1.6 | 1,080 | (6.69 ± 169) 10−6 | 10.90 |
| PC1007 | virB7 | 5.1 | 1.6 | 0 | 0 | 0 |
| PC1008 | virB8 | 5.3 | 1.4 | 20 | (1.41 ± 0.91) 10−7 | 0.23 |
| PC1009 | virB9 | 5.7 | 1.2 | <2 | (1.01 ± 0.95) 10−8 | 0.02 |
| PC1010 | virB10 | 5.7 | 1.1 | <1 | (4.1 ± 7.1) 10−9 | 0.01 |
| PC1011 | virB11 | 5.5 | 1.3 | 3,023 | (2.39 ± 0.77) 10−5 | 38.02 |
| A348 | 4.8 | 1.2 | 6,910 | (6.14 ± 2.08) 10−5 | 100.00 | |
| At12506 | virD4 | 3.1 | 0.2 | 697 | (3.48 ± 1.93) 10−5 | 110.00 |
| A348 | 2.9 | 0.3 | 950 | (3.16 ± 1.50) 10−5 | 100.00 | |
Tcs, transconjugants.
Output donor, recipient, and transconjugants were recovered after 3 days growth on induction media containing 250 μM AS at 25°C.
Recipients contained the gentamicin resistant vector pJB61 as counterselectable marker.
Data represent the means of triplicates (±SD) from a single experiment. A total of four independent experiments showing similar results were performed. Although absolute numbers varied between individual experiments, the relative frequencies between the mutant strains and the wild type showed the same tendencies in every experiment. Input recipient and donor (A. tumefaciens pJB31) were added in a ratio of 1:5 (≈5 × 105 and 2.5 × 106, respectively).
A Different Subset of virB Genes Is Required for A. tumefaciens to Serve as Donor of the IncQ Plasmid.
In the experiments described above the donor strain, A. tumefaciens A348(pJB31), carries an intact virB operon whereas the recipient carries a deletion in individual virB genes. This experimental setup could, theoretically, allow intercellular complementation to take place: the virB gene products made in the donor could be exported to the mutant recipient and function there. If such intercellular complementation could take place, then it should also be observed if the recipient strain carries an intact virB operon while the donor strain carries specific virB deletions. Therefore, pJB31 was mated into each virB deletion mutant, and these were then tested for their capacity to serve as a donor to a wild-type Ti plasmid-containing recipient. These data show that virB genes 2–11 each are required for A. tumefaciens to serve as a donor (Table 3) demonstrating that VirB proteins are not exchangeable between donor and recipient during the conjugation process. In the case of the virB1 deletion mutant a reduced but still significant number of transconjugants was observed in comparison to wild type. These results are consistent with the results of plant tumorigenesis studies reported by Berger and Christie (19), demonstrating that strains without VirB1 were virulent, though attenuated in comparison to wild type.
Table 3.
Frequencies of pJB31 transfer from various virB mutant strains serving as donors in conjugations in which the recipient carries an intact virB operon
| Donor strain (pJB31) | Gene function disrupted | Transconjugants/output donor* | % of A348 value |
|---|---|---|---|
| A348 | None | (8.6 ± 0.22) 10−6 | 100 |
| PC1000 | virB1 | (4.43 ± 2.80) 10−7 | 5.15 |
| Other mutant strains tested: | |||
| PC1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, 1010, 1011, At12506 | virB2, virB3, virB4, virB5, virB6, virB7, virB8, virB9, virB10, virB11, virD4 | No transconjugants recovered† | 0 |
Donor strains and the gentamicin-resistant recipient strain, A348(pJB61), were mixed at a ratio of 1:5; conjugation took place on induction medium (250 μM AS) for 3 days at 25°C.
Data represent means of triplicates (±SD) from a single experiment. A total of three independent experiments were performed, each showing the same basic result.
Even at concentrations of 800 μM AS, no transconjugants were recovered.
Effect of Point Mutations in virB9 on Capacity of A. tumefaciens to Serve as a Recipient.
An important question is whether the same activities of the virB proteins that serve to allow DNA export also are required to increase the capacity of a strain to serve as a conjugal recipient. As a first step to examine this, we tested the capacity of a series of previously characterized virB9 point mutants (28) to serve as recipients. These mutant strains showed the same relative decrease in their capacity to incite tumors on plants and to transfer the RSF1010 derivative, pJB31, between Agrobacterium strains. In those assays mutant #7 showed the least activity (<0.1% of wild type) whereas mutants #290 and #294 had significantly higher activities (≈8 and 2% of wild type, respectively). Interestingly, these mutants also exhibited reduced amounts of high molecular weight forms of VirB10 after chemical cross-linking, suggesting that the VirB core complex was not assembling properly. Moreover, the relative drop in the quantity of the VirB10 high molecular weight species correlated with the loss of virulence described above (28).
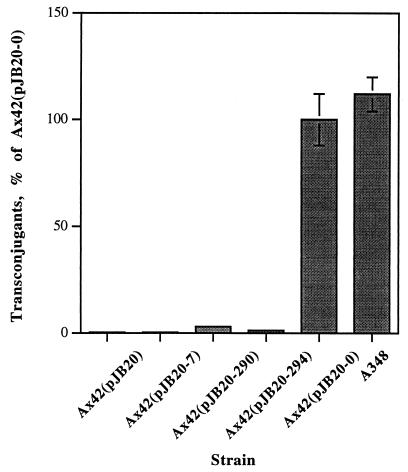
To determine their effects on the activity of Agrobacterium as a recipient, plasmids containing either wild-type or mutant virB9, under the control of the virB promoter (the pJB20 series, ref. 28), were electroporated into a strain containing a nonpolar transposon insertion in virB9 (Ax42) (16). Conjugation assays using these strains as recipients showed that the wild-type virB9 restored the capacity of Ax42 to function as a high efficiency recipient during conjugation, nearly equaling the levels seen when using the wild-type strain A348 as recipient. The different virB9 point mutations supported substantially lower levels of conjugation compared with wild-type recipients (Fig. 1). Moreover, the relative order of the mutants’ effectiveness as a recipient was the same as the relative order for both T-DNA transfer to plants and IncQ plasmid conjugal transfer to recipient Agrobacteria (28).
Figure 1.
Comparison of the relative capacities of different virB9 point mutants to function as donors or recipients in VirB-mediated transfer of the IncQ derivative pJB31. Donor capacities of these mutants were published previously (28). Recipient data were determined by using Ax42 pJB61 strains that expressed the various virB9 alleles (-7, -290, -294) from the vector pJB20. The wild-type strain A348 (pJB31) served as donor in these experiments. The percentage of transfer efficiency of strain Ax42 complemented with wild-type virB9 (pJB20–0) is set to 100%.
DISCUSSION
In this report we demonstrate that the presence of the Ti plasmid in a recipient strain of Agrobacterium dramatically increases its capacity to serve as a recipient in VirB-mediated RSF1010 conjugal transfer. The molecular and genetic mechanism that is responsible for this increase was experimentally identified as the capacity of the recipient strain to express the virB operon and, in particular, a subset of the VirB proteins. The significance of these proteins is demonstrated further by the observation that point mutations in virB9 that interfere with donor functions (28) also interfere with recipient functions. Interestingly, analysis of these virB9 mutants indicated that recipient capacity was reduced quantitatively to the same extent as in the loss of donor function in strains carrying these mutations. This suggests that at least some of the basic structural requirements of the transfer apparatus are the same in donor and recipient.
Several controls were carried out to ensure that the observed increase in plasmid transfer was, in fact, conjugal in nature. Such transfer remained strictly dependent on AS and was not inhibited by the presence of DNase in the medium (data not shown). Externally added plasmid DNA could not be taken up by the Ti-containing recipient (data not shown), indicating that the transfer is a true conjugative event and not a result of an enhanced transformation capacity. Moreover the presence of the Ti plasmid in the recipient did not overcome virB requirements in the donor cell, which indicates that no intercellular complementation between donor and recipient occurs. In our hands a virB1 mutant donor strain conjugated at frequencies of about 5% compared with the wild type if the recipient carried the Ti plasmid. The extracellular localization of VirB1 and its proposed activity as a transglycosylase (30) makes this protein a likely candidate for intercellular complementation: the secreted form of the VirB1 could move from recipient to donor (or vice versa) and thereby contribute to the structure and function of the VirB complex. However, even if donor and recipient strains both carried deletions in virB1, conjugation still took place at low rates (data not shown), consistent with the observations of Berger and Christie (19) that VirB1 is the only nonessential VirB protein.
Although VirB2–11 and VirD4 were required for Agrobacterium to serve as a conjugal donor, only a subset of these proteins was required for the increase in recipient function compared with Ti plasmid-free strains. Conjugation rates remained at a considerably increased level even if the recipient could not express VirB5, VirB6, VirB11, or VirD4. This clearly distinguishes recipient cell requirements from donor cell requirements, for which expression of VirB2–11 and VirD4 is absolutely essential. Loss of the expression of either VirB7, 9, or 10 caused the most significant drop in conjugation frequencies. The VirB7-VirB9 heterodimer (30, 37, 38) is hypothesized to form the nucleation center around which the other VirB proteins assemble to build the transfer apparatus (11, 39). There is evidence that VirB9 is required for stabilization of VirB10 and expression of VirB7 through VirB10 is the minimal requirement to obtain crosslinked high molecular weight complexes of VirB10 (28, 40). The extreme drop in conjugation frequencies suggests that these proteins might form the core of the transfer apparatus and that without these proteins the whole complex cannot assemble. Although the absence of either VirB1 or VirB2 in the recipient decreased conjugation rates compared with recipients carrying the wild-type locus, these rates were still approximately 2 log orders higher than in Ti plasmid-free recipients or recipients carrying mutations in VirB3, 4, 7, 8, 9, or 10. Taken together, the results of this report suggest that the increased capacity to function as recipient may reflect enhancement in the uptake of the transferred DNA intermediate in the presence of some part(s) of the putative transfer apparatus. A recent report by Daugelavicius et al. (48) showed that expression of the Tra proteins involved in mating pair formation in the RP4 system altered the cell envelope permeability, suggesting that these proteins form some sort of channel. It would be interesting to determine whether VirB3, 4, 7, 8, 9, and 10 are sufficient to increase permeability in Agrobacteria.
Four proteins—VirB5, 6, 11, and VirD4—all of which are absolutely required for donor functions, are only moderately involved in the increase in recipient function. This suggests that these proteins are likely to be specific for the export processes of the putative VirB complex. Of these, VirD4 (41, 42) and VirB11 (43–45) are associated with the inner membrane, and VirB11 shows ATPase activity that is required for virulence whereas VirD4 appears to contain an ATP-binding domain suggestive of an ATPase (11). Our results suggest that these proteins may be involved in substrate recognition and/or energization of DNA export. In contrast, loss of the third ATPase of the VirB-D4 system, VirB4 (29, 46, 47), caused a significant drop in conjugation frequencies. This suggests that VirB4 may provide energy for the assembly of the pore complex. The absence of VirB5 and VirB6 only moderately affected the capacity of Agrobacteria carrying an otherwise wild-type Ti plasmid to serve as a recipient in the VirB-mediated RSF1010 transfer process. Little is known about the roles these genes play in the construction and activity of the VirB pore (11). Our results indicate that these proteins may be involved in processes specific to export.
There are at least two alternative, and not mutually exclusive, biochemical mechanisms that could be responsible for the recipient-mediated increase in conjugation efficiency reported here: enhanced contact between donor and recipient or facilitated uptake of the transferred DNA by the recipient. Close contact seems to be a prerequisite for successful DNA transfer in either bacterial conjugation or T-DNA transfer. Attachment of Agrobacterium to plant cells is necessary for transformation and requires chromosomal genes (31) that are not necessary for conjugative transfer between bacteria (K. J. Fullner and E. W. Nester, personal communication). During bacterial conjugation of F-like plasmids, pili on the donor site initiate the first contact with the recipient, followed by closer apposition of donor and recipient cell outer membranes and the formation of a conjugal junction (32). Earlier results indicate that alterations in recipient capacities in conjugative systems are correlated with the stability of the formed mating pairs, a process strongly influenced by alterations in lipopolysaccharides or mutations in the outer membrane protein, OmpA (33, 34). Although a recent report described the VirB- and VirD4-dependent formation of pili in Agrobacterium, their actual function remains elusive (18). It is highly unlikely that pili-like structures on the recipient are responsible for the increase in conjugation rates as the formation of pili is disrupted by deletion of any of the virB genes or in a virD4 mutant strain (18). However, the processed form of VirB1 and presumably the pilin homologue VirB2 are of extracellular location (35, 36) and may facilitate or stabilize contact if expressed in the recipient, and loss of these could reduce mating efficiency as reflected in the results reported above (see Table 2).
The observed increase in RSF1010 conjugative transfer to recipients expressing the virB genes poses the more general question of whether the presence, in the recipient, of genes involved in conjugative mating pair formation can increase transfer efficiencies. This could be particularly important in understanding the horizontal gene flow among prokaryotes in general. The frequencies reported here are considerably lower than those reported, for example, in the RP4-mediated transfer of RSF1010 (49). It is possible that improvement in transfer frequencies of the type mediated by the VirB proteins is detectable only when baseline conjugal frequencies are low. This possibility is under investigation. Alternatively, it is possible that the lack of an entry exclusion function in the virB operon allows the donor strain the capacity to access the VirB proteins on the recipient cell surface. The results of studies to define entry exclusion (eex) determinants that prevent IncN plasmid transfer to recipients already containing an IncN plasmid suggest this may not be the case. An IncN plasmid in which eex functions were disrupted by a Tn5 insertion did not render the recipient more effective than plasmidless strains at receiving genetically distinct IncN plasmids (50).
Given that all the components of the VirB complex interact closely and are likely to depend on each other for stabilization and maintenance of their position relative to one another (11), we are aware that more specific conclusions are speculative. Nevertheless, these results indicate that, in contrast to all other VirB-mediated activities studied to date, only a subset of VirB proteins are required for the process of increasing the capacity of an Agrobacterium cell to serve as conjugal recipient. This observation, plus the fact that mutations in virB9 genes cause quantitatively similar losses in donor and recipient function, suggest it will be feasible to study a simplified version of a transfer complex. This is an important goal considering the complexity of the full complex and the almost complete lack of information about the biochemical mechanisms underlying DNA transfer through such complexes. Additionally, it will be interesting to determine whether expression of the VirB proteins in alternate recipients (bacterial, yeast, or even plants) can improve the Agrobacterium-mediated transfer of DNA to them as well.
Acknowledgments
We thank Dr. Peter Christie, Dr. Stephen Winans, Dr. Eugene Nester, and Dr. Stephen Farrand for providing various bacterial strains and plasmids. We extend special thanks to Dr. Lisa Stahl, Dr. Lois Banta, and Dr. Fevzi Daldal for their advice and support during this project and for reading earlier versions of this manuscript. This work was supported by the National Science Foundation through grants to A.N.B. (MCB95-13662) and to J. Bohne (by a grant from the Deutsche Forschungsgemeinschaft).
ABBREVIATION
- AS
acetosyringone
References
- 1.Binns A N, Howitz V R. Curr Top Microbiol Immunol. 1994;192:119–138. doi: 10.1007/978-3-642-78624-2_6. [DOI] [PubMed] [Google Scholar]
- 2.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zupan J R, Zambryski P. Plant Physiol. 1995;107:1041–1047. doi: 10.1104/pp.107.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stachel S E, Zambryski P C. Cell. 1986;47:155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- 5.Lessl M, Lanka E. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 6.Binns A N, Beaupré C F, Dale E M. J Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie P J, Ward J E, Winans S C, Nester E W. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 9.Kado C I. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 10.Winans S C, Burns D L, Christie P J. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fullner K J, Nester E W. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 14.Melchers L S, Maroney M J, den Dulk-Ras A, Thompson D V, van Vuuren H A J, Schilperoort R A, Hooykaas P J J. Plant Mol Biol. 1990;14:249–259. doi: 10.1007/BF00018565. [DOI] [PubMed] [Google Scholar]
- 15.Regensburg-Tuïnk A J G, Hooykaas P J J. Nature (London) 1993;363:69–71. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- 16.Dale E M, Binns A N, Ward J E., Jr J Bacteriol. 1993;175:887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward J E, Jr, Dale E M, Christie P J, Nester E W, Binns A N. J Bacteriol. 1990;172:5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullner K J, Lara J C, Nester E W. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 19.Berger B R, Christie P J. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchanan-Wolloston V, Passiatore J E, Cannon F. Nature (London) 1987;328:172–175. [Google Scholar]
- 21.Beijersbergen A, Dulk-Ras A D, Schilperoort R A, Hooykaas P J J. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- 22.Ward J E, Jr, Dale E M, Binns A N. Proc Natl Acad Sci USA. 1991;88:9350–9354. doi: 10.1073/pnas.88.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciaky D, Montaya A L, Chilton M-D. Plasmid. 1978;1:238–253. doi: 10.1016/0147-619x(78)90042-2. [DOI] [PubMed] [Google Scholar]
- 24.Farrand S K, O’Morchoe S P, McCutchan J. J Bacteriol. 1989;171:5314–5321. doi: 10.1128/jb.171.10.5314-5321.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winans S C, Kerstetter R A, Ward J E, Nester E W. J Bacteriol. 1989;171:1616–1622. doi: 10.1128/jb.171.3.1616-1622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Pühler A. Biotechnology. 1983;1:784–791. [Google Scholar]
- 27.Alt-Mörbe J, Stryker J L, Fuqua C, Li P-L, Farrand S K, Winans S C. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beaupré C F, Bohne J, Dale E M, Binns A N. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullner K J, Stephens K M, Nester E W. Mol Gen Genet. 1994;245:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- 30.Baron C, Thorstenson Y R, Zambryski P C. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winans S C. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dürrenberger M, Villiger W, Bächi T. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 33.Anthony K G, Sherburne C, Sherburne R, Frost L S. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 34.Sherburne C, Taylor D E. J Bacteriol. 1997;179:952–955. doi: 10.1128/jb.179.3.952-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirasu K, Kado C I. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 36.Baron C, Llosa M, Zhou S, Zambryski P C. J Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson L B, Hertzel A V, Das A. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spudich G M, Fernandez D, Zhou X-R, Christie P J. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandnez D, Spudich G M, Zhou X-R, Christie P J. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward J E, Jr, Dale E M, Nester E W, Binns A N. J Bacteriol. 1990;172:5200–5210. doi: 10.1128/jb.172.9.5200-5210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto S, Toyoda-Yamamoto A, Ito K, Takebe I, Machida Y. Mol Gen Genet. 1991;228:24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- 42.Lin T-S, Kado C I. Mol Microbiol. 1993;9:803–812. doi: 10.1111/j.1365-2958.1993.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 43.Christie P J, Ward J E, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashkova S, Spudich G M, Christie P J. J Bacteriol. 1997;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens K M, Rousch C, Nester E W. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang T A T, Christie P J. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger B R, Christie P J. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daugelavicius R, Bamford J K H, Grahn A M, Lanka E, Bamford D H. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winans S C, Walker G. J Bacteriol. 1985;161:411–416. doi: 10.1128/jb.161.1.411-416.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]