Abstract
TVA, the cellular receptor for subgroup A avian leukosis viruses (ALV-A) can mediate viral entry when expressed as a transmembrane protein or as a glycosylphosphatidylinositol-linked protein on the surfaces of transfected mammalian cells. To determine whether mammalian cells can be rendered susceptible to ALV-A infection by attaching a soluble form of TVA to their plasma membranes, the TVA-epidermal growth factor (EGF) fusion protein was generated. TVA-EGF is comprised of the extracellular domain of TVA linked to the mature form of human EGF. Flow cytometric analysis confirmed that TVA-EGF is a bifunctional reagent capable of binding simultaneously to cell surface EGF receptors and to an ALV-A surface envelope-Ig fusion protein. TVA-EGF prebound to transfected mouse fibroblasts expressing either wild-type or kinase-deficient human EGF receptors, rendered these cells highly susceptible to infection by ALV-A vectors. Viral infection was blocked specifically in the presence of a recombinant human EGF protein, demonstrating that the binding of TVA-EGF to EGF receptors was essential for infectivity. These studies have demonstrated that a soluble TVA-ligand fusion protein can mediate viral infection when attached to specific cell surfaces, suggesting an approach for targeting retroviral infection to specific cell types.
Retroviral entry is initiated by the binding of viral surface envelope proteins (ENVs) to specific cell surface receptors. Retroviruses utilize a number of different cell surface proteins as receptors including several transporter proteins with multiple transmembrane regions, and proteins with single transmembrane regions that are derived from Ig, low density lipoprotein receptor, and tumor necrosis factor receptor protein superfamilies (1, 2). TVA, the cellular receptor for subgroup A avian leukosis viruses (ALV-A), is a type I membrane protein with an 83-amino acid extracellular region that contains a 40-amino acid motif related to the ligand binding repeat regions of the low density lipoprotein receptor (3). The major viral interaction determinants of TVA have been mapped to the low density lipoprotein receptor-related motif, and analysis of different transmembrane and glycosylphosphatidylinositol-linked forms of this receptor have indicated that these determinants can be placed at various distances from the cell surface membrane without loss of function (refs. 4–6, and H. Wang, K. Gendron, D. Chu, H. E. Varmus, and P. Bates, personal communication).
There is accumulating evidence to indicate that TVA may be the only cell surface protein required for ALV-A entry. (i) TVA can confer susceptibility to ALV-A infection when expressed in cells of human, mouse and monkey origin (refs. 3 and 7, and K. Zingler and J.A.T.Y., unpublished data), indicating that if other cellular factors are involved then they must be highly conserved between different species. (ii) This receptor can elicit conformational changes in ALV-A ENV like those that are thought to be responsible for promoting virus-cell membrane fusion (8).
Given that TVA may be the only cellular protein needed to promote ALV-A infection, and that this receptor is able to function when placed at variable distances from the cell membrane, we decided to determine whether a soluble protein containing the extracellular region of TVA could allow viral entry when attached to cell surfaces by a specific protein-protein interaction. To test this idea TVA-EGF, a soluble protein comprised of the extracellular region of TVA fused to the mature form of human EGF, was generated. TVA-EGF conferred susceptibility to ALV-A infection when attached to cell surface EGF receptors (EGFRs), demonstrating that retroviruses can be targeted to cells by a soluble retroviral receptor-ligand fusion protein.
MATERIALS AND METHODS
Construction of the TVA-EGF Gene and Production of the TVA-EGF Protein.
A DNA fragment encoding a proline-rich linker, PPPELLGGP, placed at the N-terminal end of the mature form of human EGF was obtained by PCR amplification by using a human EGF minigene template (9) and the DNA primers 5′-GCGCGGCCGGGTACCTTATCGCAGTTCCCATTTCAGGTCGCG-3′ and 5′-GCGCGGCCGCCCACCCCCTGAACTCCTGGGGGGACCGGAGGTTC- AGAACTCTGATTCCGAATGC-3′. The DNA fragment was then digested with the EagI restriction enzyme and subcloned into the EagI site of a synthetic Tva gene (5) contained within the pCI mammalian expression vector (Promega) to generate plasmid pSS1. The calcium phosphate method (10) was used to transfect 45 μg of plasmid pSS1 into human 293 cells plated at 20% confluency on 150-mm tissue culture plates. Extracellular supernatants from transfected and nontransfected cells were collected 72 hr after transfection and 45 μl aliquots of these supernatants were subjected to electrophoresis on a 10% polyacrylamide gel containing SDS under nonreducing conditions. These proteins were then transferred to a nitrocellulose membrane and TVA-EGF was detected by immunoblotting with ALV-A surface ENV (SU)-Ig fusion protein (SUA-rIgG) (11) followed by a horseradish peroxidase-conjugated antibody specific for rabbit Igs (Amersham).
Cell Lines and Viruses.
B82 mouse L cells that lack EGFRs, T23 mouse L cells that express wild-type human EGFRs, and M5 mouse L cells that express kinase-deficient EGFRs containing the K721M mutation (12, 13), were kindly provided by G. Gill (University of California, San Diego, La Jolla). T23TVA cells were generated by cotransfecting T23 cells with plasmid pKZ261 encoding an epitope-tagged transmembrane form of TVA (4) and with pPur plasmid DNA encoding puromycin-N-acetyl-transferase (CLONTECH). After selection in medium containing 2 μg/ml puromycin, independent clones of T23TVA cells were isolated and characterized for cell surface TVA expression by flow cytometric analysis by using SUA-rIgG (11) and fluorescein isothiocyanate-conjugated swine anti-rabbit antibodies (Dako). The subgroup A-specific viruses RCASA-Neo encoding neomycin phosphotransferase and RCASH-A encoding hygromycin B phosphotransferase were described (2, 7).
Flow Cytometry.
B82, M5, and T23 cells were detached from tissue culture plates with Ca2+/Mg2+-free PBS containing 1 mM EDTA and placed on ice. All of the following steps were performed at 4°C. Approximately 3.5 × 105 cells of each type were incubated for 1 hr with extracellular supernatants that either lacked or contained different amounts of TVA-EGF in 1 ml of DMEM. The cells were then washed with ice-cold PBS and incubated for 1 hr with 500 μl of PBS containing 3 μg of purified SUA-rIgG (11). After washing with ice-cold PBS, cells were incubated for 30 min. with 500 μl of PBS containing 5 μl of fluorescein isothiocyanate-conjugated swine anti-rabbit antibody (Dako). The cells were then washed again, resuspended in PBS containing 1% formaldehyde, and the bound proteins were detected by flow cytometry by using a Becton Dickinson FACScan. These experiments were performed in the presence of excess amounts of SUA-rIgG to ensure this reagent is saturating (data not shown).
Competition experiments were performed by adding different amounts of a recombinant human EGF protein (Upstate Biotechnology, Lake Placid, NY) to 200 μl aliquots of extracellular supernatants containing TVA-EGF and incubating with M5 cells at 4°C for 1 hr. The bound TVA-EGF proteins were detected as described above. To eliminate the possibility that EGF might interfere nonspecifically with TVA/SUA-rIgG interactions, 3.5 × 105 T23TVA cells were incubated with 1 ml DMEM that either lacked or contained 5 μg of EGF before flow cytometric analysis with SUA-rIgG and fluorescein isothiocyanate-conjugated swine anti-rabbit antibody (Dako).
To follow the uptake of TVA-EGF proteins from cell surfaces, ≈3.5 × 105 M5 and T23 cells were first incubated at 4°C for 1 hr with 200 μl of extracellular supernatants containing TVA-EGF. Cells were then washed with ice-cold PBS and either left on ice, or were incubated at 37°C for 30, 60, or 120 min with 500 μl of DMEM that had been prewarmed to 37°C. At each of these time points, cells were immediately transferred to ice to block any subsequent receptor-mediated endocytosis. TVA-EGF proteins remaining on the cell surfaces were then detected by flow cytometry as described above.
TVA-EGF-Dependent Viral Infection.
Triplicate samples of ≈1 × 105 B82, T23 or M5 cells were plated on 60-mm tissue culture plates for ≈24 hr. The plates were placed on ice and incubated at 4°C for 1 hr with 1 ml of ice-cold DMEM containing different amounts of TVA-EGF. Cells were then washed with ice-cold PBS, incubated with 1.5 ml of medium containing 400 μl of RCASA-Neo virus, and placed at 37°C for 28 hr. For control purposes, T23TVA cells were incubated at 4°C for 1 hr with 1.5 ml medium that either contained or lacked 50 μl of RCASA-Neo virus, and then incubated at 37°C for 28 hr. All of these cells were then selected in medium containing 200 μg/ml Geneticin (G418) (GIBCO/BRL). G418-resistant colonies each arising from an independent infection event were counted after 2 weeks. It is important to note that although the pKZ261 plasmid transfected into T23TVA cells encodes neomycin phosphotransferase, a cloned line of these cells was isolated that expressed cell surface TVA but was highly sensitive to G418-induced killing. This cell line was therefore a suitable control for infection experiments that employed the RCASA-Neo virus. In two independent experiments, each performed in triplicate, the number of G418 resistant T23TVA colonies obtained without virus was ≈10% of that number obtained after RCASA-Neo virus infection (data not shown).
The infection experiments employing the RCASH-A virus were performed essentially as described above except that 1 ml of TVA-EGF containing supernatants and 300 μl of RCASH-A virus were used. The cells were incubated with media that contained sufficient amounts of hygromycin B to select against the growth of any uninfected cells (900 μg/ml for B82 cells and 700 μg/ml for M5 cells). For control purposes, B82 cells expressing an epitope-tagged transmembrane form of TVA, generated by transfection with plasmid pKZ261 (4), were shown to be resistant to 900 μg/ml hygromycin B after infection by RCASH-A virus (data not shown).
To block TVA-EGF dependent viral entry, triplicate samples of ≈1 × 105 M5 cells plated on 60-mm plates were incubated at 4°C for 1 hr with 1 ml ice-cold DMEM that contained 200 μl of TVA-EGF supernatant and either 0, 0.5 μg or 5 μg of a recombinant human EGF protein. Also, T23TVA cells were incubated with 1 ml of DMEM that contained either 0 or 5 μg of EGF. The cells were then washed with ice-cold PBS, incubated with 1.5 ml of DMEM containing 150 μl of RCASA-Neo virus and transferred to 37°C for 28 hr. The number of infected G418-resistant colonies was determined as described above.
RESULTS
The TVA-EGF Protein.
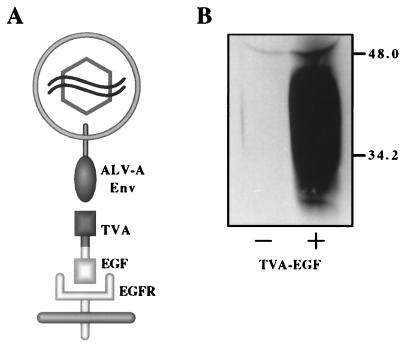
TVA-EGF was designed as a bifunctional reagent with two distinct domains: the N-terminal domain, comprised of the extracellular region of TVA containing the determinants necessary for binding the ALV-A SU and promoting viral entry (4–6, 14, 15), and the C-terminal domain, consisting of the mature form of human EGF (9) that binds to cell surface EGFRs (Fig. 1A). To increase the probability that these two domains of TVA-EGF would function independently, they were separated by a 9-amino acid proline-rich linker region derived from the hinge region of a rabbit Fc chain (see Materials and Methods).
Figure 1.
The TVA-EGF protein. (A) A model of TVA-EGF mediated ALV-A infection. TVA-EGF is comprised of the extracellular domain of TVA fused via a proline-rich linker region to the mature form of human EGF (see Materials and Methods). TVA-EGF was predicted to bind to ALV-A SU ENV and to cell surface EGFRs, promoting viral entry. (B) TVA-EGF was produced in the extracellular supernatants of transfected human 293 cells. Samples of extracellular supernatants taken from 293 cells that were either not transfected (−) or transfected with plasmid DNA encoding TVA-EGF (+) were subjected to SDS/PAGE. After transfer to a nitrocellulose membrane, the TVA-EGF protein was detected by immunoblotting with an ALV-A SU-Ig fusion protein (11) and a horseradish peroxidase-coupled secondary antibody followed by enhanced chemiluminescence (Amersham). Molecular mass markers are given in kDa.
TVA-EGF was produced in the extracellular supernatants of transiently transfected human 293 cells. To confirm expression of the fusion protein, the extracellular supernatants of transfected and nontransfected 293 cells were assayed by immunoblotting with a ALV-A SU-Ig fusion protein (SUA-rIgG) (11). SUA-rIgG specifically detected TVA-EGF in extracellular supernatants of transfected, but not nontransfected, cells (Fig. 1B). The TVA-EGF proteins varied in size between ≈25–49 kDa, presumably because of heterogeneous posttranslational modifications of the TVA domain (3).
TVA-EGF Is a Bifunctional Reagent that Can Bind Simultaneously to the Ligand-Binding Region of Cell Surface EGFRs and to ALV-A SU.
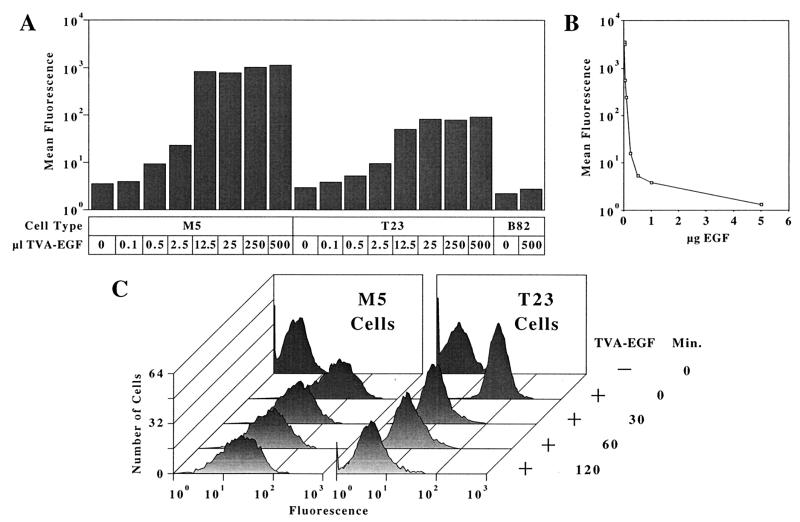
To determine whether TVA-EGF was capable of binding both to cell surface EGFRs and to ALV-A SU, flow cytometric analysis was performed. Mouse L cells that either did not express EGFRs (B82 cells), expressed wild-type human EGFRs (T23 cells), or expressed kinase-deficient human EGFRs containing the K721M mutation (M5 cells) (12, 13), were incubated with extracellular supernatants that contained increasing amounts of TVA-EGF. The TVA-EGF proteins that bound to the surfaces of these cells were then detected by flow cytometry by using SUA-rIgG (Fig. 2A). TVA-EGF bound to M5 and T23 cells in a saturable manner but did not bind to B82 cells (Fig. 2A). Thus, TVA-EGF was indeed a bifunctional reagent capable of binding both SUA-rIgG and the surfaces of cells expressing EGFRs.
Figure 2.
TVA-EGF binds specifically to the ligand-binding regions of cell surface EGFRs. (A) TVA-EGF binds in a saturable manner to M5 and to T23 cells. B82 mouse L cells lacking EGFRs, M5 cells expressing kinase-deficient human EGFRs, or T23 cells expressing wild-type human EGFRs (12, 13), were incubated with the indicated amounts of extracellular supernatants that contained TVA-EGF. The cells were then incubated with SUA-rIgG (11) and with fluorescein isothiocyanate-conjugated antibodies specific for rabbit Igs and analyzed by flow cytometry. Mean fluorescence values obtained with each cell population are shown. (B) EGF competes for TVA-EGF binding to M5 cells. M5 cells were incubated with extracellular supernatants containing TVA-EGF and increasing amounts of a recombinant human EGF protein. The level of cell surface-associated TVA-EGF in each cell population was measured by flow cytometry as described above. The data shown is representative of two independent experiments that were each performed in triplicate. (C) TVA-EGF disappeared more rapidly from the surfaces of T23 as opposed to M5 cells. M5 and T23 cells were preincubated at 4°C with extracellular supernatants that either lacked or contained TVA-EGF and then transferred to 37°C for the indicated periods of time. The TVA-EGF proteins that remained on the cell surface at each time point were detected by flow cytometry as described above. The data shown is representative of results obtained from three independent experiments.
To demonstrate specific binding of TVA-EGF to EGFRs, a recombinant human EGF protein was tested for its capacity to block TVA-EGF binding to M5 cells. Extracellular supernatants containing TVA-EGF were mixed with various amounts of EGF before incubation with M5 cells, and the bound TVA-EGF was detected by flow cytometry (Fig. 2B). The level of cell-associated TVA-EGF (contained in a 200-μl aliquot of extracellular supernatant) was reduced by 50% in the presence of 200 ng of EGF and by 97% in the presence of 5 μg of EGF (Fig. 2B). In contrast, EGF did not compete with SUA-rIgG binding to T23TVA cells that stably express a transmembrane form of TVA (Materials and Methods), thus ruling out the possibility that the addition of EGF interfered with the SUA-rIgG/TVA interaction (data not shown).
Given that TVA-EGF appears to bind to cell surface EGFRs in the same way as EGF, it seemed likely that this retroviral receptor-ligand fusion protein would be more rapidly down-regulated from cell surfaces after binding to wild-type EGFRs as opposed to kinase-deficient EGFRs (12, 13, 16). Indeed, the majority of TVA-EGF prebound to wild-type EGFRs on T23 cells was cleared from cell surfaces within a 30 minute incubation period at 37°C, whereas a substantial amount of TVA-EGF still remained on the surface of M5 cells even after a 2-hr incubation period at this temperature (Fig. 2C). Taken together, these studies demonstrate that TVA-EGF binds in the same way as EGF to cell surface EGFRs and like EGF, this retroviral receptor-ligand fusion protein is capable of stimulating receptor-mediated endocytosis.
TVA-EGF Confers Susceptibility to ALV-A Infection upon Cells that Express EGFRs.
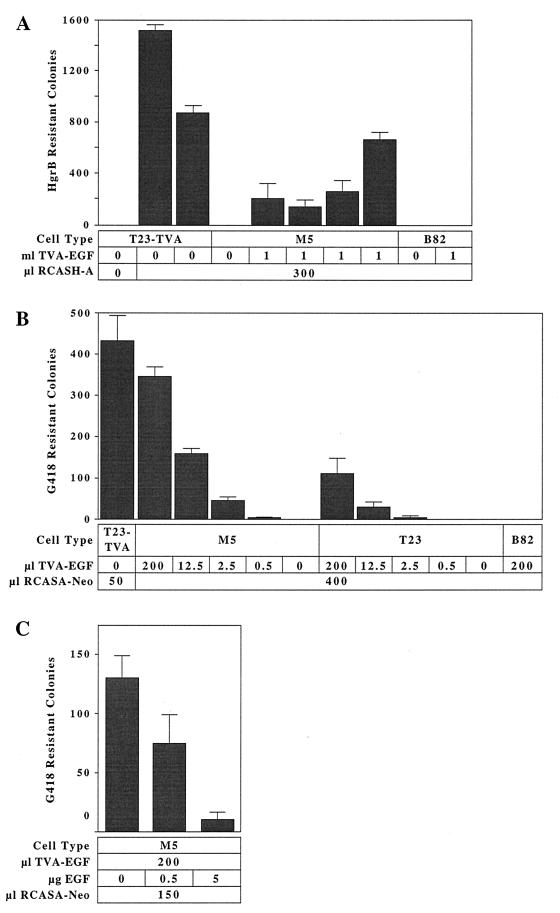
We next asked if TVA-EGF bound to cell surface EGFRs could mediate ALV-A entry. B82 and M5 cells were incubated with extracellular supernatants that either contained or lacked TVA-EGF and were then challenged with the ALV-A vector RCASH-A encoding hygromycin B phosphotransferase (7). M5 cells that had been incubated with supernatants containing, but not lacking, TVA-EGF, were infected by virus (Fig. 3A). In fact, TVA-EGF rendered M5 cells highly susceptible to ALV-A infection: the average number of infected M5 colonies obtained was ≈23% of that number observed after virus infection of T23TVA cells that express a fully functional epitope-tagged transmembrane TVA protein (Fig. 3A). In contrast, B82 cells that lack EGFRs were not infected by virus after TVA-EGF addition (Fig. 3A).
Figure 3.
The TVA-EGF protein mediates ALV-A infection when attached to cell surface EGFRs. (A) B82 and M5 cells incubated with medium that contained or lacked TVA-EGF, and T23TVA cells expressing a transmembrane form of TVA, were challenged with the subgroup A specific RCASH-A virus encoding hygromycin B phosphotransferase (7). The numbers of hygromycin B-resistant colonies, representing individual infection events, were counted and the results of independent experiments, each performed in triplicate, are shown. (B) B82, T23 and M5 cells were incubated with medium containing the indicated amounts of TVA-EGF and then challenged with the subgroup A-specific RCASA-Neo virus encoding neomycin phosphotransferase (2). The number of infected G418-resistant colonies was determined. T23TVA cells were also challenged with this virus, and the average number of infected G418-resistant colonies was determined after subtracting the small number of drug-resistant colonies that arose without virus addition (see Materials and Methods). These data are representative of three independent infection experiments that were each performed in triplicate. (C) EGF blocks TVA-EGF-dependent viral entry. M5 cells were incubated with extracellular supernatants containing TVA-EGF in the presence of different amounts of recombinant EGF and then challenged with the RCASA-Neo virus. The resulting G418-resistant infected colonies were counted, and the average numbers from two independent experiments that were each performed in triplicate are shown.
To assess the effect of increasing amounts of TVA-EGF on the efficiency of subgroup A virus infection, M5, T23, and B82 cells were incubated with extracellular supernatants containing different amounts of TVA-EGF and challenged with the ALV-A virus RCASA-Neo encoding neomycin phosphotransferase (2). TVA-EGF conferred susceptibility to viral infection upon M5 and T23 cells in a dose-dependent manner (Fig. 3B). The average number of infected cells observed when M5 cells were incubated with an excess amount of TVA-EGF (200 μl; Fig. 2A) was ≈10% of that obtained with T23TVA cells (Fig. 3B). However, the level of infection obtained with T23 cells was lower than that seen with M5 cells (Fig. 3B). As expected, B82 cells that lack EGFRs were not infected by the RCASA-Neo virus after exposure to TVA-EGF (Fig. 3B).
To confirm that TVA-EGF-mediated viral entry requires that this fusion protein binds to the ligand-binding region of EGFRs, a recombinant human EGF protein was tested for its ability to block viral infection. EGF blocked TVA-EGF dependent viral entry into M5 cells in a dose-dependent manner (Fig. 3C). To exclude any nonspecific effect of EGF addition on RCASA-Neo virus infection, T23TVA cells were also treated with EGF before viral challenge. The number of infected T23TVA cells was not affected by pretreatment with EGF (data not shown). Taken together, these studies demonstrate that TVA-EGF can efficiently mediate ALV-A entry when bound to the ligand-binding domain of cell surface EGFRs.
DISCUSSION
In this report we have shown that the TVA-EGF fusion protein is a bifunctional reagent that binds both to cell surface EGFRs and to ALV-A SU, promoting viral entry. Thus these studies have demonstrated that cells can be rendered susceptible to viral infection when a soluble retroviral receptor protein is attached to their surfaces by a protein-protein interaction.
Several independent lines of evidence have demonstrated that TVA-EGF binds to the ligand-binding region of cell surface EGFRs and that this binding is necessary for mediating ALV-A entry. (i) TVA-EGF binds to T23 cells expressing wild-type EGFRs and to M5 cells expressing kinase-deficient EGFRs, but not to B82 cells lacking EGFRs. (ii) A recombinant human EGF protein specifically blocked TVA-EGF binding to M5 cells and TVA-EGF mediated viral entry. (iii) Like EGF (12, 13, 16), TVA-EGF is taken up more rapidly from cell surfaces (presumably after receptor-mediated endocytosis) after binding to wild-type EGFRs as opposed to kinase-deficient EGFRs.
The highest level of TVA-EGF dependent viral entry was observed by using M5 cells incubated with excess amounts of the soluble retroviral receptor-ligand fusion protein (Fig. 3 A and B). These levels reached ≈10–23% of those obtained with the T23TVA cell line, stably expressing TVA. However, in three independent experiments, T23 cells incubated with excess amounts of TVA-EGF were on average 2.5-fold less susceptible to ALV-A infection than were M5 cells (Fig. 3B). This difference between the two cell types may reflect the fact that there may be as many as 12.4-fold more EGFRs on the surfaces of M5 cells than on the surfaces of T23 cells (Fig. 2 A and C). However it is worth noting that although this data is representative of most of the flow cytometry experiments performed, some experiments indicated that there are equivalent amounts of EGFR on the surfaces of M5 and T23 cells (data not shown). The reason for this difference between experiments is not known and is currently under investigation.
The difference in susceptibility to ALV-A infection observed between M5 and T23 cells increased to an average of 12-fold when nonsaturating amounts of TVA-EGF were used (Figs. 2A and 3B). This difference in infectivity may be due at least in part to differing numbers of EGFRs present on these cell surfaces and/or to the fact that TVA-EGF is cleared from the surfaces of T23 cells more rapidly than from those of M5 cells (Fig. 2C). Another factor that might contribute to this effect is that ALV-A/TVA-EGF complexes bound to wild-type EGFRs may be targeted after endocytosis to cellular compartments that do not permit efficient viral entry, including lysosomes where these complexes would be degraded (16). Experiments are now in progress to distinguish between these possibilities. If the decrease in efficiency of TVA-EGF mediated ALV-A infection of T23 cells is due to rapid down-regulation of wild-type EGFRs, then this might indicate that receptors that are not rapidly down-regulated after ligand binding might be the preferred cell surface targets for this method of viral delivery. For example, the erbB2, erbB3, and erbB4 receptors that bind to EGF-like ligands are apparently down-regulated from cell surfaces with the same slow kinetics as kinase-deficient EGFRs (17).
Before these studies, there was no indication that a soluble viral receptor could support viral entry when added exogenously to cells. In fact, one study showed that cells expressing a soluble form of the mouse hepatitis virus receptor were susceptible to viral infection, whereas cells that had been incubated with the soluble mouse hepatitis virus receptor were not (18). The use of a soluble viral receptor-ligand fusion protein as a bifunctional reagent to facilitate viral entry offers a strategy for delivering viral vectors to specific cell types. For example, replacement of the EGF portion of TVA-EGF with other factors that bind to specific cell surface markers, such as additional cell type-specific ligands or single-chain antibodies, may allow targeted infection of other specific cell types by ALV-A vectors. This method may also allow for the delivery of other types of retroviral vectors into cells, because murine leukemia virus and HIVs (HIV-1), pseudotyped with ALV-A ENV, can utilize TVA as a receptor for viral entry (ref. 19, and K. Zingler and J.A.T.Y., unpublished data).
Several other methods for retrovirus targeting were described previously (20–36). In one case, ecotropic murine leukemia virus virions were chemically modified with lactose, a procedure that resulted in the specific infection of human cells that expressed the asialoglycoprotein receptor (20). In another, a low level of viral infection was achieved by forming an antibody bridge between the ecotropic murine leukemia virus SU protein and specific host cell surface receptors (21, 22). A more common approach has been to design retroviral SU proteins that contain cell type-specific ligands or single chain antibodies to direct them to specific cell surface proteins (23–35). This latter approach requires that the inserted amino acid sequences do not impair critical ENV functions such as oligomerization and cell surface transport during biosynthesis, uptake into viral particles, and stimulating virus-cell membrane fusion (36). Indeed, this method generally gives rise to viruses that infect the desired cell types with a low efficiency (36). The approach that we have described in this report does not involve modifying the viral ENVs and thus by preserving native ENV-receptor interactions should avoid these types of complications.
Acknowledgments
We thank Gordon Gill for providing the mouse L cell lines and John Daley at the Dana–Farber Cancer Institute for assistance with flow cytometry. We also thank Sara Klucking for providing RCASA-Neo virus, Vince Solomon for help with producing TVA-EGF protein, John Naughton for help with the figures, and members of the Young lab and David Knipe, John Collier, and Caroline Alexander for critically reading the manuscript. This work was supported by National Institutes of Health Grant CA 62000 and by a Harvard Medical School Funds for Discovery Exploratory Award.
ABBREVIATIONS
- ALV-A
subgroup A avian leukosis virus
- EGF
epidermal growth factor
- EGFR
EGF receptor
- ENV
envelope protein
- SU
surface ENV
- SUA-rIG
ALV-A SU-Ig fusion protein
- TVA
the cellular receptor for ALV-A
References
- 1.Weiss R A, Tailor C S. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 2.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A T. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 3.Bates P, Young J A T, Varmus H E. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 4.Zingler K, Bélanger C, Peters R, Agard D, Young J A T. J Virol. 1995;69:4261–4266. doi: 10.1128/jvi.69.7.4261-4266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bélanger C, Zingler K, Young J A T. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rong L, Bates P. J Virol. 1995;69:4847–4853. doi: 10.1128/jvi.69.8.4847-4853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young J A T, Bates P, Varmus H E. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern D F, Hare D L, Cecchini M A, Weinberg R A. Science. 1987;235:321–324. doi: 10.1126/science.3492043. [DOI] [PubMed] [Google Scholar]
- 10.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheng Y C, Axel R. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 11.Zingler K, Young J A T. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W S, Lazar C S, Poenie M, Tsien R Y, Gill G N, Rosenfeld M G. Nature. 1987;328:820–823. doi: 10.1038/328820a0. [DOI] [PubMed] [Google Scholar]
- 13.Glenney J R, Chen W S, Lazar C S, Walton G M, Zokas L M, Rosenfeld M G, Gill G N. Cell. 1988;52:675–684. doi: 10.1016/0092-8674(88)90405-9. [DOI] [PubMed] [Google Scholar]
- 14.Connolly L, Zingler K, Young J A T. J Virol. 1994;68:2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert J M, Bates P, Varmus H E, White J M. J M J Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honegger A M, Dull T J, Felder S, Obberghen E V, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J. Cell. 1987;51:199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- 17.Baulida J, Kraus M H, Alimandi M, Di Fiore P P, Carpenter G. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 18.Dveksler G S, Gagneten S E, Scanga C A, Cardellichio C B, Holmes K V. J Virol. 1996;70:4142–4145. doi: 10.1128/jvi.70.6.4142-4145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landau N R, Littman D R. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neda H, Wu C H, Wu G Y. J Biol Chem. 1991;266:14143–14146. [PubMed] [Google Scholar]
- 21.Roux P, Jeanteur P, Piechaczyk M. Proc Natl Acad Sci USA. 1989;86:9079–9083. doi: 10.1073/pnas.86.23.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etienne-Julan M, Roux P, Carillo S, Jeanteur P, Piechaczyk M. J Gen Virol. 1992;73:3251–3255. doi: 10.1099/0022-1317-73-12-3251. [DOI] [PubMed] [Google Scholar]
- 23.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J M, Danos O, Verdier G, Cosset F L. J Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasahara N, Dozy A M, Kan Y W. Science. 1994;266:1373–1375. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Kasahara N, Kan Y W. Proc Natl Acad Sci USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matano T, Odawara T, Iwamoto A, Yoshikura H. J Gen Virol. 1995;76:3165–3169. doi: 10.1099/0022-1317-76-12-3165. [DOI] [PubMed] [Google Scholar]
- 27.Valsesia-Wittmann S, Morling F J, Nilson B H, Takeuchi Y, Russell S J, Cosset F L. J Virol. 1996;70:2059–2064. doi: 10.1128/jvi.70.3.2059-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilson B H, Morling F J, Cosset F L, Russell S J. Gene Ther. 1996;3:280–286. [PubMed] [Google Scholar]
- 30.Chu T H, Dornburg R. J Virol. 1995;69:2659–2663. doi: 10.1128/jvi.69.4.2659-2663.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu T H, Martinez I, Sheay W C, Dornburg R. Gene Ther. 1994;1:292–299. [PubMed] [Google Scholar]
- 32.Somia N V, Zoppe M, Verma I M. Proc Natl Acad Sci USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell S J, Hawkins R E, Winter G. Nucleic Acids Res. 1993;21:1081–1085. doi: 10.1093/nar/21.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S J, Cosset F L, Piechaczyk M. J Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ager S, Nilson B H, Morling F J, Peng K W, Cosset F L, Russell S J. Hum Gene Ther. 1996;7:2157–2164. doi: 10.1089/hum.1996.7.17-2157. [DOI] [PubMed] [Google Scholar]
- 36.Cosset F L, Russell S J. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]