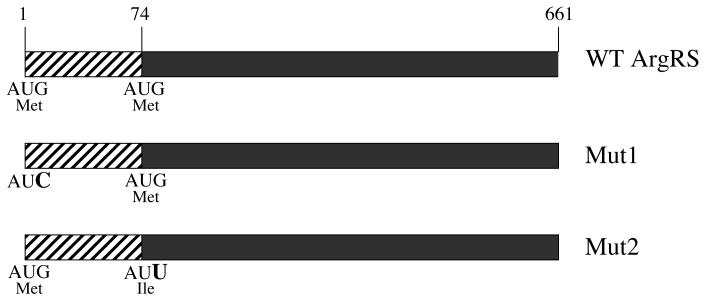SUMMARY
In mammalian cells, aminoacyl-tRNA synthetases (aaRSs) are organized into a high molecular weight multisynthetase complex whose cellular function has remained a mystery. In this study, we have taken advantage of the fact that mammalian cells contain two forms of ArgRS, both products of the same gene, to investigate the complex’s physiological role. The data indicate that the high molecular weight form of ArgRS, which is present exclusively as an integral component of the multisynthetase complex, is essential for normal protein synthesis and growth of CHO cells even when low molecular weight, free ArgRS is present and Arg-tRNA continues to be synthesized at close to wild type levels. Based on these observations, we conclude that Arg-tRNA generated by the synthetase complex is a more efficient precursor for protein synthesis than Arg-tRNA generated by free ArgRS, exactly as would be predicted by the channeling model for mammalian translation.
INTRODUCTION
In mammalian cells, aminoacyl-tRNA synthetases (aaRSs) are organized into a high molecular mass multisynthetase complex (Han et al., 2003; Lee et al., 2004). Although the number of aminoacyl-tRNA synthetases in the complex in vivo is unclear, a stable core of at least nine synthetases and three additional non-synthetase accessory proteins (p43, p38 and p18) can be isolated reproducibly (Mirande et al., 1985; Kerjan et al., 1994). Multisynthetase complexes with many fewer components than the mammalian one also exist in lower organisms such as Haloarcula marismortui (Goldgur and Safro, 1994), Methanococcus jannaschii (Lipman et al., 2000; Lipman et al., 2003), and Saccharomyces cerevisiae (Simos et al., 1996). Nevertheless, despite extensive study since its initial discovery over 35 years ago (Bandyopadhyay and Deutscher, 1971), the cellular function of the multisynthetase complex remains a mystery. It is known that removal of Arc1p, a yeast p43 homolog, leads to slowed growth and reduced MetRS activity (Simos el al., 1996; Deinert et al., 2001), and also that mutation of mammalian p38 results in loss of structural integrity of the multisynthetase complex and mouse neonatal death (Kim et al., 2002; Kim et al., 2003), suggesting that the role of the synthetase complex is important.
Among the mammalian aminoacyl-tRNA synthetases, ArgRS is unique in that two forms of this enzyme are present within a cell (Deutscher and Ni, 1982; Vellekamp et al., 1985; Cirakoglu and Waller, 1985). One form is ∼ 72-kDa, and is found exclusively as an integral component of the multisynthetase complex. The second form of ArgRS is ∼60-kDa, and exists free in cell extracts. It lacks the 12-kDa, N-terminal, hydrophobic extension characteristic of the larger form, known to be responsible for the latter’s association with the multisynthetase complex. Both forms of the enzyme display similar catalytic properties in vitro (Vellekamp et al., 1985; Lazard et al., 2000). Northern blot analysis indicates that the two forms of ArgRS are derived from a single transcript, but utilize alternate AUG initiation codons (Lazard and Mirande, 1993; Girjes et al., 1995; Zheng et al., 2006).
Our laboratory has proposed a model to explain the need for two forms of ArgRS in a single cell (Sivaram and Deutscher, 1990). In contrast to other aminoacyl-tRNAs, mammalian cells utilize Arg-tRNA for two discrete functions - protein synthesis and post-translational modification carried out by arginyl-tRNA protein transferase which arginylates the -NH2 terminus of certain proteins (Ferber and Ciechanover, 1987), a modification that serves as a signal for ubiquitin-dependent protein degradation. The model proposes that Arg-tRNA generated by the high molecular weight form of ArgRS, as part of the multisynthetase complex, is used for protein synthesis, whereas Arg-tRNA generated by the free form of ArgRS is used for the protein transferase reaction. One important corollary of this model is that the two pools of Arg-tRNA generated in this fashion remain separate, as that would appear to be the only apparent reason for needing distinct forms of ArgRS. This conclusion led to the concept of channeling of aminoacyl-tRNAs for protein synthesis (Sivaram and Deutscher, 1990). However, although channeling of aminoacyl-tRNA was subsequently confirmed by a variety of experimental approaches (Negrutskii and Deutscher, 1991; Negrutskii and Deutscher, 1992; Negrutskii et al., 1994; Stapulionis and Deutscher, 1995), the original suggestion that the multisynthetase complex is required to generate aminoacyl-tRNAs channeled for protein synthesis has not been verified.
In this paper, we take advantage of the existence of the two forms of ArgRS, and of stable clones deficient in one or the other form, to show that the high molecular mass ArgRS is essential for normal growth and protein synthesis in mammalian cells even when the necessary arginyl-tRNA can still be synthesized by the low molecular mass enzyme. This work demonstrates for the first time a direct role for the multisynthetase complex in mammalian translation, and further emphasizes the importance of aminoacyl-tRNA channeling in the translation process.
RESULTS
Properties of Arg-1 Mutant CHO Cells
In order to assess the growth potential and synthetase activity of cells containing only a single form of ArgRS, it was desirable to carry out experiments in a background that would minimally affect such measurements. For this purpose, we made use of the Arg-1 CHO mutant which is known to exhibit temperature-sensitive growth and diminished ArgRS activity relative to the parental Gat- strain (Thompson et al., 1977; Thompson et al., 1978) due to a Cys to Tyr substitution at position 599 in ArgRS (Lazard et al., 2000). We have confirmed that the Arg-1 mutant shows marked growth inhibition at 40°C, that it displays slightly slower growth at 34°C, and that its ArgRS activity is reduced ∼80% at 40°C (data not shown).
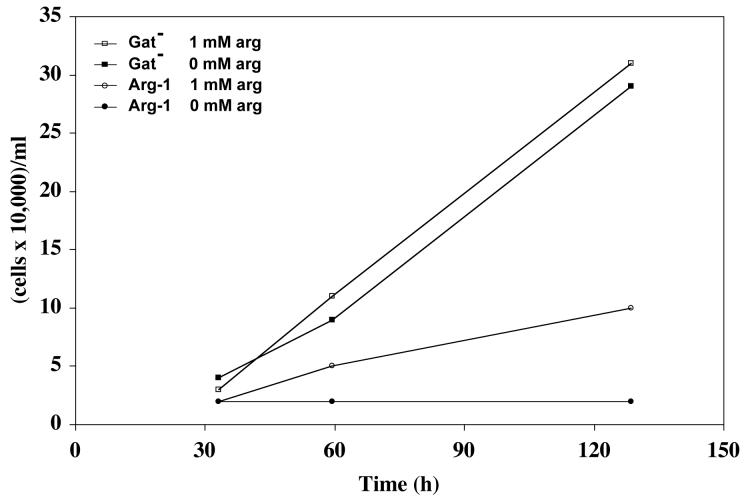
To reduce mutant cell growth even further, cells were cultured in a customized F-12 medium that contains no arginine and is supplemented with dialyzed, rather than normal, fetal bovine serum. As shown in Figure 1, the Gat- parental cells are able to grow at 39.5°C in both normal and arginine-free media. In contrast, the Arg-1 mutant grows at a reduced rate at 1 mM arginine and fails to grow at all in the absence of arginine. Interestingly, Arg-1 mutant cells also grow very poorly at 34°C in the absence of arginine, although they grow similarly to the parent at 1 mM arginine (data not shown). These data show that Arg-1 cells contain an ArgRS that is both thermosensitive and hyperauxotrophic for arginine, and they provide a condition (40°C, 0 arginine) in which Arg-1 cell growth can be eliminated entirely.
Figure 1. Growth of Gat- and Arg-1 cells in the presence and absence of arginine.
Cultures were seeded and grown in 24-well plates (1 ml each) at a cell density of 10,000 cells/ml/well at 39.5°C in customized arginine-free F-12 medium supplemented with 5% dialyzed FBS. Cells were grown either with addition of 1-mM arginine or without any arginine. At the points indicated, three wells were washed with PBS, trypsinized and counted.
Construction of Mutant Cells Expressing a Single Form of ArgRS
Earlier work showed that the two forms of ArgRS arise from a single mRNA through the utilization of alternative AUG initiation codons (Lazard and Mirande, 1993; Girjes et al., 1995) (Figure 2). ArgRS initiating from the first AUG codon would be expected to contain the N-terminal hydrophobic extension, enabling it to be assembled into the multisynthetase complex, whereas that initiating from the downstream AUG would lead to synthesis of the low molecular weight, free ArgRS. As also shown in Figure 2, plasmid constructs were generated in which both AUG codons in the argRS gene either remained intact (WT) or in which the upstream (Mut1) or downstream (Mut2) AUG codons were mutated. The Mut1 construct would be expected to lead to synthesis of only the free form of ArgRS, whereas Mut2 was expected to synthesize only high molecular weight ArgRS. The WT construct would be expected to give rise to both forms of ArgRS since both initiation codons are intact. Each of the plasmid constructs was stably transfected into Arg-1 cells, from which stable clones were selected using the antibiotic, G418. Individual clones (3 of each type) were randomly selected for further study.
Figure 2. Diagram of wild type and mutant ArgRS.
The two AUG initiation codons for high molecular weight ArgRS (AUG1) and for free ArgRS (AUG74) are indicated, together with the nucleotide change made to inactivate each codon. The cross-hatched region represents the N-terminal hydrophobic extension that distinguishes the two forms of ArgRS and is responsible for assembly of high molecular weight ArgRS into the multisynthetase complex.
ArgRS Activity in Mutant Cells
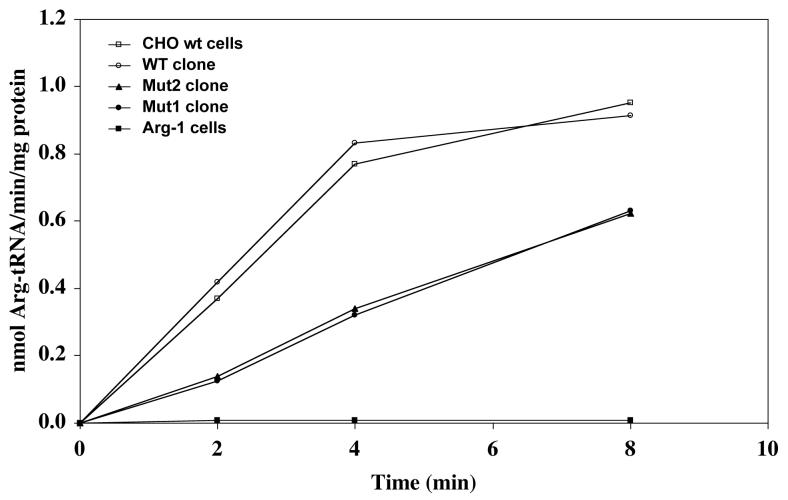
Assay of ArgRS activity in S10 extracts from the selected clones revealed that each of the clones of a particular type (WT, Mut1 or Mut2) had essentially identical activity (data not shown). Thus, only one clone of each type was chosen for detailed examination. An equal amount of S10 extract (30 μg) was assayed in each case following a 45 min preincubation at 40°C to first inactivate the thermosensitive ArgRS activity derived from the Arg-1 cell background. Consequently, any ArgRS activity observed would result from the transfected argRS genes. The data presented in Figure 3 confirm that the preincubation did, in fact, eliminate the background activity, allowing measurement of ArgRS activity due to transfection without interference from the endogenous enzyme. The data show that transfection with the WT argRS gene construct leads to the same rate of ArgRS activity as normally found in a wild type CHO cell control, whereas transfection with either of the mutant constructs leads to only about half the rate. These observations suggest that each form of ArgRS contributes about equally to the total rate of ArgRS activity present.
Figure 3. ArgRS activity of stable clones.
Activity was determined as described in “Experimental Procedures” using 30 μg of S10 extract per 100 μl of reaction mixture. [3H]-arginine (∼60 cpm/pmol) incorporation into rabbit liver tRNA was measured at 40°C. Prior to the assay, extracts were preincubated at 40°C for 45 min to inactivate the thermosensitive, endogenous ArgRS in the Arg-1 mutant strain. Data presented are the average of 3 experiments.
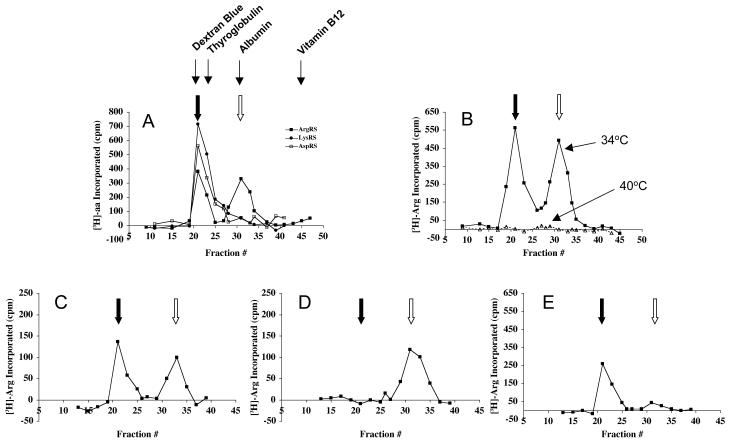
To confirm this conclusion, and to directly determine the amount of each form of ArgRS present in each of the strains, S10 cell extracts were fractionated on a Sephacryl S-300 gel filtration column, and the fractions assayed for ArgRS activity. As is well known, cells normally contain two forms of ArgRS, one eluting in the void volume of a gel filtration column as part of the multisynthetase complex, and one corresponding to the free form at a molecular mass of ∼60 kDa (e.g. Deutscher and Ni, 1982; Mirande et al., 1985; Sivaram and Deutscher, 1990). Western blot analysis indicates that the complex contains exclusively ArgRS of ∼72 kDa, whereas the late-eluting activity is exclusively low molecular weight ArgRS (Nathanson and Deutscher, 2000).
As shown in Figure 4A, extracts from wild type CHO cells contain the same two forms of ArgRS, present in about equal amounts. The high molecular weight ArgRS is associated with LysRS and AspRS, two other synthetases known to be associated with the complex. When assayed at 34°C, Arg-1 cells display the same two forms of ArgRS in the same relative amounts (Figure 4B); however, when column fractions were preincubated at 40°C for 45 min prior to assay, ArgRS activity in both peaks was completely inactivated, as expected for the endogenous thermosensitive enzyme (Figure 3). Thus, any activity peaks observed upon fractionation of extracts from the stable clones following preincubation of fractions at 40°C are due to the transfected genes. As shown in Figure 4C, transfection with the WT clone led to two peaks of ArgRS activity in the same positions and relative amounts as in wild type cells. In marked contrast, the mutant clones each express primarily a single form of ArgRS (Figures 4D-E), corresponding to that initiated from the AUG initiation codon remaining in each mutant (Figure 2). Thus, Mut1 cells express only the free form of ArgRS and no complexed ArgRS is detectable. Mut2 cells express largely the high molecular weight form of ArgRS. A small amount of free enzyme is reproducibly observed in these cells which we suspect represents some high molecular weight ArgRS that did not assemble into the complex. Note also that the absolute amount of ArgRS activity varies in the different experiments due to loading different amounts of extract on the column (see legend to Figure 4 for amounts loaded).
Figure 4. ArgRS activities determined by gel filtration.
Extracts from (A) CHO wt (1.0 mg total protein), (B) Arg-1 (1.2 mg), (C) WT (0.29 mg), (D) Mut1 (0.30 mg), and (E) Mut2 (0.50 mg) cells were applied to a column (0.7 × 50 cm) of Sephacryl S-300 equilibrated with 50 mM Tris-HCl, pH 7.5, 10% glycerol (v/v), 0.2 mM DTT, 0.2 mM EDTA, 100 mM NaCl. Fractions of 350 μl were collected. Portions of these fractions (60 μl) were assayed for 10 min at 34°C (A and B) or 40°C (B, C, D, and E). For part of B, C, D and E, fractions were preincubated at 40°C for 45 min prior to the aminoacylation assay. The filled arrow indicates the elution position of the high MW form of ArgRS present in the multisynthetase complex; the empty arrow indicates where free ArgRS is eluted. The elution positions of four protein size markers used to calibrate the column are shown. These are Dextran Blue 2000 (MW 2000 kDa), thyroglobulin (MW 650 kDa), serum albumin (MW 67 kDa), and vitamin B12 (MW 1.3 kDa). Note the different scales for ArgRS activity due to different amounts of extract loaded in the various experiments. Panel A also shows the elution profiles of LysRS and AspRS.
These data demonstrate that each form of ArgRS is dependent on a distinct AUG codon for translation initiation, and that the ArgRS proteins expressed as a result of transfection distribute either in the multisynthetase complex or as the free form exactly as predicted from their structure. Most importantly, the availability of a stable, mutant clone expressing only the free form of ArgRS enabled us to assess the role of the multisynthetase complex in vivo.
Growth of Mutant Cells
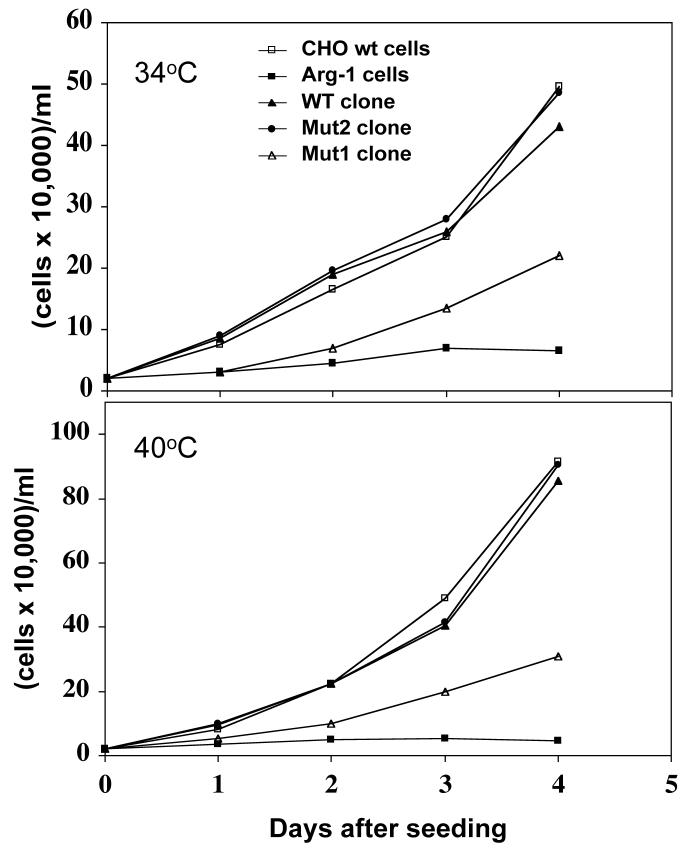
As shown above (Figure 1), at the non-permissive temperature in the absence of arginine, Arg-1 cells do not grow due to the presence of a thermosensitive and hyperauxotrophic ArgRS instead of the normal enzyme. Transfection of these cells with a WT argRS construct restored growth in arginine-free media at both 34°C and 40°C to that observed with wild type CHO cells (Figure 5), supporting the conclusions from the activity measurements that the transfected ArgRS proteins are fully active in vivo. Likewise, transfection with the mutant argRS construct (Mut2) that expresses only the high molecular weight complexed form of ArgRS also grows normally at both temperatures (Figure 5). On the other hand, transfection with the Mut1 argRS construct that expresses only the free form of ArgRS leads to poor growth under the same conditions (Figure 5). These observations indicate that the high molecular weight form of ArgRS is required for normal cell growth even though the free form of ArgRS is still present at normal levels.
Figure 5. Growth of stable clones.
The growth of the stable clones was compared to that of CHO wt and Arg-1 untransfected cells. Clones with the high MW form of ArgRS (Mut2), the low MW form (Mut1) or with both forms (WT) were grown at either 34°C or 40°C in the presence of arginine-free F-12 medium supplemented with 5% dialyzed fetal bovine serum. Cultures were seeded and grown in 6-well plates (2 ml each) at a cell density of 40,000 cells per well. At the time points indicated, one well from each cell line was washed with PBS, trypsinized and counted.
Aminoacyl-tRNA Synthetases and Aminoacyl-tRNAs in Mutant Cells
To examine in more detail the basis of the poor growth phenotype, aminoacyl-tRNA synthetases and aminoacyl-tRNAs in Mut1 cells were analyzed. Since these mutant cells do not have an active, complexed form of ArgRS, we first examined whether the multisynthetase complex itself remained intact and active. S-10 supernatant fractions from WT, Mut1 and Mut2 cells were each subjected to gel filtration on Sephacryl S-300 columns and analyzed for LeuRS, LysRS and AspRS activity, three enzymes known to be part of the complex. These experiments showed that despite the absence of an active ArgRS, the multisynthetase complex remained intact and that each of the three synthetases retained full activity in Mut1 cells (data not shown).
A portion of cellular tRNA is found associated with the multisynthetase complex following gel filtration (Bandyopadhyay and Deutscher, 1971). This amounted to 46% in wild type CHO cells (data not shown). In Arg-1 cells transfected with the WT argRS construct, this portion amounted to 43% of total tRNAArg (average of 3 experiments). A similar value of 37% was found in cells transfected with the Mut2 gene. However, only 20% of tRNAArg was associated with the complex in Mut1 cells. These data indicate that only about half as much tRNAArg is stably bound to the multisynthetase complex in the absence of an active ArgRS.
Protein synthesis is dependent on charged tRNAs. Therefore, it was of considerable interest to determine the level of tRNA arginylation in vivo in the various cells under study (Table 1). This was done using the periodate oxidation technique (Rizzino et al., 1974; Andrulis et al., 1979). The total amount of tRNAArg in each of the four cell lines examined was essentially identical (Table 1). Based on periodate oxidation, both wild type CHO cells and the WT clone were found to be aminoacylated with arginine to ∼90%, in agreement with other measurements in mammalian cells. In contrast, the level of arginylation was significantly reduced in the mutant clones, amounting to 78% in cells expressing only the complexed, high molecular weight ArgRS, and 68% in cells expressing only the free form. Thus, elimination of either of the two forms of ArgRS does lead to a decrease in the amount of arginyl-tRNA in vivo, although the extent of reduction is relatively modest. Most importantly, these data show that even though arginyl-tRNA continues to be made in the Mut1 cell at approximately 75% of normal levels (68÷90%) by the free ArgRS, growth is dramatically reduced, supporting the conclusion that for efficient protein synthesis, arginyl-tRNA must be generated by the ArgRS present in the multisynthetase complex.
Table 1. Levels of aminoacylation of tRNAArg in vivo.
Percent aminoacylation was obtained by dividing the amino acid acceptor activity (expressed as pmol of amino acid accepted per μg of tRNA) of the periodate-treated tRNA by the activity of the untreated tRNA. Based on statistical data analysis using the Paired Student’s t-Test, the differences between the % aminoacylation values were highly significant (P < 0.05) except between CHO wt and WT clone (P > 0.05)
| tRNA source | Untreateda | After periodate treatmenta | % aminoacylation | σb | # experiments |
|---|---|---|---|---|---|
| CHO wt | 7.6 | 6.8 | 89 | +/- 4.82 | 7 |
| WT clone | 7.4 | 7.0 | 95 | +/- 4.30 | 11 |
| Mut2 clone | 7.4 | 5.8 | 78 | +/- 6.76 | 10 |
| Mut1 clone | 7.8 | 5.3 | 68 | +/- 5.52 | 9 |
Acceptor activity is expressed as picomoles of 3H-Arg accepted per μg of tRNA used
Standard deviation
Channeling of Aminoacyl-tRNA for Protein Synthesis in Mutant Cells
That arginyl-tRNA must be generated by complexed ArgRS for efficient translation is exactly what would be predicted by the channeling model of mammalian translation. In this model, which has been verified experimentally, aminoacyl-tRNA generated within the translation apparatus is preferentially utilized for protein synthesis compared to exogenous aminoacyl-tRNA added to cells by electroporation (Negrutskii and Deutscher, 1991) or through use of permeabilized cells (Negrutskii and Deutscher, 1992; Negrutskii et al., 1994; Stapulionis and Deutscher, 1995). In the current situation, we propose that arginyl-tRNA synthesized by the free form of ArgRS, although generated within the cell, also is exogenous to the translation apparatus leading to less efficient utilization for protein synthesis and to slower growth. To test this idea directly, the various transfected cells were permeabilized with saponin, labeled with [3H]-amino acids, and aminoacyl-tRNA levels and protein synthesis measured, as described previously (Negrutskii and Deutscher, 1992; Negrutskii et al., 1994; Stapulionis and Deutscher, 1995). As a control for a true exogenous substrate, the corresponding [14C]-aminoacyl-tRNAs were added and protein synthesis measured. The amount of [14C]-exogenous aminoacyl-tRNA added was adjusted to be equal to that of the endogenous aminoacyl-tRNA.
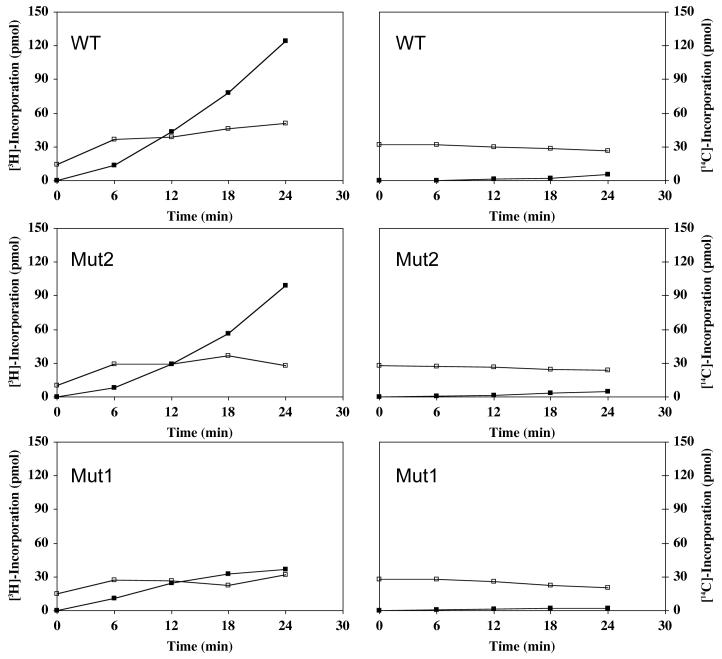
The data for this experiment are presented in Figure 6. In cells labeled with [3H]-amino acids (left panels), there is a very rapid increase in the level of [3H]-aminoacyl-tRNA after which it remains constant throughout the course of the experiment, as expected, since aminoacyl-tRNA continues to be made as it is used. After a short lag, protein synthesis takes off. While the rates of translation in WT and Mut2 cells are similar, it is reduced 3- to 4-fold in Mut1, even though the amount of aminoacyl-tRNA is essentially the same in all three strains. It should be noted that since there is no increase in free ArgRS (Figure 4) or in arginyl-tRNA protein transferase (data not shown) in the Mut1 cells, it is unlikely that enhanced protein degradation plays any role in the reduction of translation. Thus, these data demonstrate that protein synthesis is much less efficient in cells lacking the high molecular weight ArgRS, thereby accounting for its slower growth. As reported previously (Negrutskii and Deutscher, 1991; Negrutskii and Deutscher, 1992; Negrutskii et al., 1994; Stapulionis and Deutscher, 1995), exogenously-supplied [14C]-aminoacyl-tRNA is utilized for protein synthesis extremely poorly - rates are 3% or less of those in WT or Mut2 cells despite the presence of the same amount of aminoacyl-tRNA (right panels). Taken together, these observations confirm channeling of aminoacyl-tRNA and support the conclusion that efficient translation requires a fully active multisynthetase complex.
Figure 6. Channeling of aminoacyl-tRNA in permeabilized CHO cells.
WT, Mut2 and Mut1 cells (grown at 40°C to inactivate any endogenous ArgRS activity) were harvested, permeabilized, and incubated for protein synthesis as described in “Experimental Procedures”. At each time point, two 100 μl portions were taken, cooled on ice and precipitated with TCA. One portion, left on ice, measured aminoacyl-tRNA plus protein synthesized. The other portion was heated for 10 min at 95°C to solubilize labeled aminoacyl-tRNA. This portion was used to measure protein synthesis (■). The difference between the two samples represents aminoacyl-tRNA (□). Each data point represents ∼ 2 × 106 cells. Panels on the left were incubated with [3H] amino acids, those on the right with the corresponding [14C] aminoacyl-tRNAs.
DISCUSSION
The existence of two forms of ArgRS in a single cell, each of which could be eliminated by mutation, as well as the availability of the Arg-1 CHO cell mutant, provided the ideal system to examine the biosynthesis of the two forms of ArgRS, to ascertain their contribution to overall ArgRS activity, and to elucidate their functional roles. Moreover, the presence of a thermosensitive and hyperauxotrophic ArgRS in the Arg-1 cell background enabled us to completely eliminate any involvement by the endogenous ArgRS. Thus, using this system, we were able to clearly show that both forms of ArgRS are independently expressed in cells, that their synthesis is dependent on translation from two separate initiation codons, and that in CHO cell extracts each form of ArgRS contributes about half of the total ArgRS activity. Most importantly, these studies revealed that the two forms of ArgRS are functionally distinct.
In earlier work (Sivaram and Deutscher, 1990), our laboratory predicted that the two forms of ArgRS would be separate translation products and that the high molecular weight form would be used to generate Arg-tRNA for translation, whereas the low molecular weight ArgRS would generate Arg-tRNA for N-terminal arginylation of certain proteins. In addition, based on these predictions, it was suggested that the Arg-tRNA generated by the two forms of ArgRS would not mix, leading to the concept of aminoacyl-tRNA channeling during translation (Sivaram and Deutscher, 1990), an idea that was subsequently confirmed by experiment (Negrutskii and Deutscher, 1991; Negrutskii and Deutscher, 1992; Negrutskii et al., 1994; Stapulionis and Deutscher, 1995). The role of the high molecular weight ArgRS has been confirmed by this work. However, based on these studies, there may be more overlap between the two Arg-tRNA pools than was originally envisioned. Thus, we find that cells can grow, albeit slowly, in the absence of high molecular weight ArgRS, implying that Arg-tRNA generated by the low molecular weight ArgRS is able to be used for translation, but much less efficiently (see Figure 6). It is possible that this “leaky” channeling only occurs when the complexed ArgRS is missing, and that under normal conditions, when both enzymes are present, only the pool of Arg-tRNA generated by complexed ArgRS is used for translation. This latter possibility is supported by the channeling experiments shown in Figure 6 which clearly demonstrate that exogenous aminoacyl-tRNA is a very poor substrate for translation compared to endogenous molecules. In a similar manner, Arg-tRNA synthesized by free ArgRS could be considered to be exogenous to the translation system. If so, this would explain why it is used less efficiently.
This study does not deal explicitly with the function of the low molecular weight form of ArgRS. However, since cells lacking this enzyme grow normally, either the function(s) carried out by free ArgRS are not essential for normal growth or Arg-tRNA generated by the high molecular weight enzyme can be used instead. Studies are now in progress to examine these various possibilities.
Over the years, a number of possible functions have been proposed for the multisynthetase complex (Han et al., 2003; Lee et al., 2004). These include: 1) channeling of aminoacyl-tRNAs for protein biosynthesis; 2) acting as a molecular reservoir to regulate the non-translational activities of aaRSs; 3) serving as a means to stabilize translation components; and 4) facilitating transport of tRNAs from the nucleus to the cytoplasm since the complex has also been found in the nucleus (Nathanson and Deutscher, 2000). The studies presented here provide strong support for the first role listed. Clearly, additional work will be necessary to determine whether the multisynthetase complex serves the other proposed functions as well, and to understand how it enhances the efficiency of translation, as reported here.
EXPERIMENTAL PROCEDURES
Materials
3H-labeled arginine, leucine, lysine, tyrosine, valine, and 14C-labeled arginine, leucine, lysine, and tyrosine were purchased from Amersham Biosciences. Unlabeled amino acids, phosphocreatine, creatine phosphokinase, ATP, GTP, saponin, DTT, and trypan blue were obtained from Sigma. Halt Protease Inhibitor Cocktail (EDTA-Free) was purchased from Pierce. Complete Mini EDTA-free Protease Inhibitor Cocktail Tablets were from Roche. Cell culture reagents were from Gibco. Chinese hamster ovary cells were obtained from the American Type Culture Collection (ATCC). The CHO mutant strain, Arg-1, containing a temperature sensitive ArgRS (Thompson et al., 1977; Thompson et al., 1978), was generously provided by Dr. L.H. Thompson (Livermore, California). The Arg-1 parental strain (Gat-), having a triple auxotrophic requirement for glycine, adenosine, and thymidine due to a defect in folate metabolism, was also provided by Dr. L.H. Thompson. Rabbit liver tRNA was prepared as described previously (Negrutskii et al., 1994).
Cell Culture
For experiments using normal growth medium (containing arginine), CHO cells were grown as monolayers in Ham’s F-12 medium (Gibco) containing 2 mM L-glutamine supplemented with 5% (v/v) fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (HyClone). For experiments in arginine-free medium, CHO cells were maintained as monolayers in customized arginine-free F-12 medium formulated in the lab and supplemented with dialyzed 5% (v/v) fetal bovine serum. Preparation of arginine-free F-12 medium was according to Gibco’s recipe by adding all components except arginine. The pH of the customized nutrient mixture was adjusted to pH 7.2-7.4 and filtered through a 0.1 μm Acrodisc syringe filter membrane (Pall). Cells were cultured in Nunc flasks at 34°C or 40°C in air containing 5% CO2 and were transferred every 2-3 days. Cells were harvested at 80-90% of confluence by washing with phosphate-buffered saline (PBS), followed by treatment with 2 ml pre-warmed trypsin containing EDTA (Invitrogen). Approximate cell numbers were determined in a hemacytometer, and their intactness was determined by trypan blue exclusion.
Growth Measurements
Exponentially-growing cells were harvested, resuspended in the appropriate medium and seeded into either 24-well plates (1 ml each at a cell density of 10,000 cells per well) or 6 well plates (2 ml each at a cell density of 40,000 cells per well). Cells were trypsinized and counted daily in a hemacytometer following staining with trypan blue. The generation of dead cells could also be readily monitored. In experiments with medium containing no arginine or reduced levels of arginine, the customized arginine-free F-12 medium was used, and the appropriate amount of arginine from a sterile arginine stock solution was added just prior to the experiment. Dialyzed fetal bovine serum (filter sterilized) was also used in these experiments. In all other cases, normal fetal bovine serum was used.
Preparation of S10 and S100 Supernatant Fractions
After harvesting and washing in PBS, the cell pellet was quickly frozen in an ethanol-dry ice bath and thawed in Buffer A (20 mM HEPES-KOH buffer, pH 7.4, containing 0.5 mM spermine, 130 mM KCl, 1% thiodiglycol, 0.5 mM EDTA, 2 mM CaCl2, and protease inhibitors (Roche; 1 tablet per 10 ml)). The cell suspension was incubated on ice for 15 min with occasional stirring. The S10 and S100 fractions were prepared by centrifuging at 10,000xg for 10 min or at 100,000xg for 2.5 h, respectively. The supernatant fraction was collected in each case, aliquoted, quick-frozen in liquid nitrogen and stored at -80°C. The total protein concentration was determined using the Bradford assay (Bradford, 1976).
Gel Filtration Chromatography
S10 or S100 supernatant fractions (1-1.5 mg total protein) were loaded into a column (0.7 × 50 cm) of Sephacryl S-300 equilibrated with 50 mM Tris-HCl, pH 7.5, 10% glycerol (v/v), 0.2 mM dithiothreitol, 0.2 mM EDTA, and 100 mM NaCl and eluted at a flow rate of 2.5 ml/h. Fractions of 350 μl were collected. Portions (60 μl) were assayed for aminoacyl-tRNA synthetase activity.
Aminoacylation Assays
Aminoacyl-tRNA synthetase activity was determined at either 34°C or 40°C in reaction mixtures containing: 250 mM Tris-HCl, pH 7.5, 10 mM ATP, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mg/ml bovine serum albumin (BSA), 1.5 mg/ml rabbit liver tRNA, and 0.1 mM 3H-labeled amino acid (20-100 cpm/pmol). Each reaction was initiated by addition of cell lysate to the complete reaction mixture. A 100 μl portion was removed for the zero time point and the remaining mixture was placed in a 34°C or 40°C water bath. Additional 100 μl portions were removed at the indicated time points. Reactions were terminated on ice by the addition of ice-cold 10% trichloroacetic acid (TCA) containing 0.5% Casamino acids. Aminoacyl-tRNA precipitates were collected onto Glass Microfibre 696 (2.4 cm) filters (VWR) presoaked with 10% TCA containing 0.5% Casamino acids. The filters were washed nine times with 2.5% cold TCA and twice with ethanol:ether (1:1). After drying the filters, radioactivity was determined in a scintillation counter. The zero time point was used for background subtraction.
Construction of Plasmids
cDNA encoding the full-length, wild-type ArgRS sequence was generated from total RNA (isolated from exponentially growing CHO wt cells) via RT-PCR using primers complementary to the ArgRS cDNA sequence from CHO wt cells, and ligated into the EcoRI site of a pCMV-Script vector. For construction of a plasmid expressing only the high molecular weight form of ArgRS (Mut2), the second AUG codon (corresponding to Met74) was changed via site-directed mutagenesis into an AUU codon. For construction of a plasmid expressing only the low molecular weight form of ArgRS (Mut1), the first AUG codon (corresponding to Met1) was mutated into an AUC codon. The final constructs were verified by DNA sequencing.
DNA Transfection
CHO mutant Arg-1 cells were grown at 34°C in F-12 medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin and 100 μg/ml streptomycin. Arg-1 cells were stably transfected using FuGENE 6 (Roche) transfection reagent according to the manufacturer’s protocol with different pCMV-Script multicopy plasmid constructs: one encoding only the high molecular weight form of ArgRS (Mut2), one encoding only the low molecular weight form of ArgRS (Mut1), and one encoding both forms of ArgRS (WT). Antibiotic G418 (at a final concentration of 0.4 μg/μl) was used to select and maintain the stable Arg-1 cell lines. Stable clones were isolated from these lines using limited dilution and seeding with 0.5 cells/well.
Permeabilization of Cells
One day after reaching confluency, cells (grown at 40°C) were washed once with 25 ml of Dulbecco’s phosphate-buffered saline (PBS) and harvested from the flasks by incubation with 2 ml of TrypLE Express (Gibco) for 10 min at room temperature. The released cells were washed with ice-cold permeabilization Buffer S (130 mM sucrose, 50 mM potassium acetate, 50 mM KCl, 20 mM HEPES pH 7.4), pelleted by centrifugation, and resuspended in ice-cold Buffer S using ∼ 60 μl of buffer per 107 cells. The total volume of cell suspension was measured by pipetting, and an equal volume of saponin solution (150 μg/ml) in Buffer S was added to give ∼1.2 μg of saponin per 106 cells. The suspension was mixed and incubated at 37°C for 6 min, cooled on ice and centrifuged for 1 min at 400-500xg. The permeabilized cells were washed once with ice-cold PSW buffer (130 mM sucrose, 50 mM potassium acetate, 50 mM KCl, 20 mM HEPES pH 7.4, 5 mM ATP, 13 mM phosphocreatine, 6.1 mM MgCl2, 2.6 mM CaCl2, 5.3 mM EGTA, 5 mM glucose), suspended in 4-5 volumes of PSW buffer and immediately used for assays. A small portion was used to measure cell permeability by trypan blue exclusion and for cell counting.
Protein Synthesis in Permeabilized Cells
Protein synthesis experiments using permeabilized cells were carried out in PSW buffer supplemented with the following components: 0.1 mM GTP, 250 μM each of 20 amino acids, [3H]-labeled arginine, tyrosine, leucine, lysine, and 30 μg/ml creatine phosphokinase. Permeabilized cells were incubated at 28°C (at this temperature they are capable of carrying out high rates of protein synthesis), and at various times, duplicate portions of 100 μl were taken, precipitated with ice-cold 2.5% TCA and 0.5% Casamino acids, and cooled on ice. One portion was boiled for 10 min at 95°C to destroy labeled aminoacyl-tRNA and then cooled on ice. Precipitates were collected on microfibre filters. Hot TCA-insoluble radioactivity was considered to represent protein synthesis, whereas that from the non-boiled samples represents both protein synthesis and aminoacyl-tRNA.
Isolation of Charged tRNA and Periodate Oxidation
The extent of aminoacylation in vivo was measured by the periodate oxidation method (Rizzino, et al., 1974; Andrulis, et al., 1979). tRNA was isolated under acidic conditions by resuspending the cell pellet in buffer containing 0.3 M sodium acetate (pH 5.0), 10 mM Na2EDTA, and 0.15 M NaCl. Cells were lysed using TRI-Reagent (Sigma), chloroform was added and the mixture was vortexed for 15 sec, followed by incubation at room temperature for 15 min. After centrifugation at 10,000xg at 4°C for 15 min, the aqueous phase was collected, mixed with 2.5 vol of ice-cold 95% ethanol and placed at - 20°C. The precipitate was collected by centrifugation at 10,000xg at 4°C. Each pellet was resuspended in 0.1 M sodium acetate, pH 5.0, followed by two isopropanol precipitations to remove high molecular weight RNA species. The isolated tRNA was then dissolved in 0.1 M sodium acetate, pH 5.0, and divided into two equal portions. To one portion was added 40 mM sodium periodate (J.T.Baker) to a final concentration of 3 mM, while the control received 0.1 vol of distilled water. The tubes were incubated in the dark at 37°C for 30 min. After incubation, periodate was destroyed by addition of excess glucose and samples were incubated in the dark for 5 min at 37°C. Samples were mixed with 2.5 vol of ice-cold 95% ethanol and precipitated for 90 min at -20°C. Precipitates were collected by centrifugation at 10,000xg at 4°C for 10 min. tRNA was collected and deacylated in 0.1 M Tris-HCl, pH 9.0, by incubation at 37°C for 1 h. Samples were mixed with 2.5 vol of ice-cold 95% ethanol and precipitated at -20°C. tRNA was resuspended in 0.1 M sodium acetate, pH 5.0. The percentage of tRNA that originally existed in a charged form in vivo was then measured using an aminoacylation assay at 37°C, as described above.
ACKNOWLEDGMENTS
We thank David Fisher and Dr. Alice Hudder for construction of plasmids and for their efforts in the initial stages of this project, Dr. Oleksii Dubrovskyi for helpful discussion, and Dr. Larry H. Thompson for CHO cell strains. This work was supported by Grant GM16317 and by funds provided by the Lucille P. Markey Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrulis IL, Hatfield GW, Arfin SM. Asparaginyl-tRNA aminoacylation levels and asparagine synthetase expression in cultured Chinese hamster ovary cells. J. Biol. Chem. 1979;254:10629–10633. [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay AK, Deutscher MP. Complex of aminoacyl-transfer RNA synthetases. J. Mol. Biol. 1971;60:13–122. doi: 10.1016/0022-2836(71)90451-7. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cirakoglu B, Waller J-P. Multiple forms of arginyl- and lysyl-tRNA synthetases in rat liver: a re-evaluation. Biochim. Biophys. Acta. 1985;829:173–179. doi: 10.1016/0167-4838(85)90186-4. [DOI] [PubMed] [Google Scholar]
- Deinert K, Fasiolo F, Hurt EC, Simos G. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- Deutscher MP, Ni RC. Purification of a low molecular weight form of rat liver arginyl-tRNA synthetase. J. Biol. Chem. 1982;257:6003–6006. [PubMed] [Google Scholar]
- Ferber S, Ciechanover A. Role of arginine-tRNA in protein degradation by the ubiquitin pathway. Nature (London) 1987;326:808–811. doi: 10.1038/326808a0. [DOI] [PubMed] [Google Scholar]
- Girjes AA, Hobson K, Chen P, Lavin MF. Cloning and characterization of cDNA encoding a human arginyl-tRNA synthetase. Gene. 1995;164:347–350. doi: 10.1016/0378-1119(95)00502-w. [DOI] [PubMed] [Google Scholar]
- Goldgur Y, Safro M. Aminoacyl-tRNA synthetases from Haloarcula marismortui: an evidence for a multienzyme complex in a procaryotic system. Biochem. Mol. Biol. Int. 1994;32:1075–1083. [PubMed] [Google Scholar]
- Han JM, Kim JY, Kim S. Molecular network and functional implications of macromolecular tRNA synthetase complex. Biochem. Biophys. Res. Commun. 2003;303(4):985–993. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- Kerjan P, Cerini C, Semeriva M, Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim. Biophys. Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kang YS, Lee JW, Kim HJ, Ahn YH, Park H, Ko YG, Kim S. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. PNAS USA. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW, Lee SW, Han JM, Lee HW, Kim S. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- Lazard M, Agou F, Kerjan P, Mirande M. The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication. J. Mol. Biol. 2000;302:991–1004. doi: 10.1006/jmbi.2000.4102. [DOI] [PubMed] [Google Scholar]
- Lazard M, Mirande M. Cloning and analysis of a cDNA encoding mammalian arginyl-tRNA synthetase, a component of the multisynthetase complex with a hydrophobic N-terminal extension. Gene. 1993;132:237–245. doi: 10.1016/0378-1119(93)90201-d. [DOI] [PubMed] [Google Scholar]
- Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- Lipman RS, Sowers KR, Hou YM. Synthesis of cysteinyl-tRNA (Cys) by a genome that lacks the normal cysteine-tRNA synthetase. Biochemistry. 2000;39:7792–7798. doi: 10.1021/bi0004955. [DOI] [PubMed] [Google Scholar]
- Lipman RS, Chen J, Evilia C, Vitseva O, Hou YM. Association of an aminoacyl-tRNA synthetase with a putative metabolic protein in archaea. Biochemistry. 2003;42:7487–7496. doi: 10.1021/bi0344533. [DOI] [PubMed] [Google Scholar]
- Mirande M, Le Corre D, Waller J-P. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur. J. Biochem. 1985;147:281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Nathanson L, Deutscher MP. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multisynthetase complex. J. Biol. Chem. 2000;275:31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- Negrutskii BS, Deutscher MP. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii BS, Deustcher MP. A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3601–3604. doi: 10.1073/pnas.89.8.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii BS, Stapulionis R, Deutscher MP. Supramolecular organization of the mammalian translation system. Proc. Natl. Acad. Sci. U.S.A. 1994;91:964–968. doi: 10.1073/pnas.91.3.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino AA, Bresalier RS, Freundlich M. Derepressed levels of the isoleucine-valine and leucine enzymes in hisT1504, a strain of Salmonella typhimurium with altered leucine transfer ribonucleic acid. J. Bact. 1974;117:449–455. doi: 10.1128/jb.117.2.449-455.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Sivaram P, Deutscher MP. Existence of two forms of rat liver arginyl-tRNA synthetase suggests channeling of aminoacyl-tRNA for protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3665–3669. doi: 10.1073/pnas.87.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapulionis R, Deutscher MP. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Lofgren DJ, Adair GM. CHO cell mutants for arginyl-, asparaginyl-, glutaminyl-, histidyl- and methionyl-tRNA synthetases: identification and initial characterization. Cell. 1977;11:157–168. doi: 10.1016/0092-8674(77)90326-9. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Lofgren DJ, Adair GM. Evidence for structural gene alterations affecting aminaocyl-tRNA synthetases in CHO cell mutants and revertants. Somat. Cell Genet. 1978;4:423–435. doi: 10.1007/BF01538864. [DOI] [PubMed] [Google Scholar]
- Vellekamp G, Sihag RK, Deutscher MP. Comparison of the complexed and free forms of rat liver arginyl-tRNA synthetase and origin of the free form. J. Biol. Chem. 1985;260:9843–9847. [PubMed] [Google Scholar]
- Zheng YG, Wei H, Ling C, Xu MG, Wang ED. Two forms of human cytoplasmic arginyl-tRNA synthetase produced from two translation initiations by a single mRNA. Biochemistry. 2006;45:1338–1344. doi: 10.1021/bi051675n. [DOI] [PubMed] [Google Scholar]