Abstract
Familial multiple system tauopathy with presenile dementia (MSTD) is a neurodegenerative disease with an abundant filamentous tau protein pathology. It belongs to the group of familial frontotemporal dementias with Parkinsonism linked to chromosome 17 (FTDP-17), a major class of inherited dementing disorders whose genetic basis is unknown. We now report a G to A transition in the intron following exon 10 of the gene for microtubule-associated protein tau in familial MSTD. The mutation is located at the 3′ neighboring nucleotide of the GT splice-donor site and disrupts a predicted stem-loop structure. We also report an abnormal preponderance of soluble tau protein isoforms with four microtubule-binding repeats over isoforms with three repeats in familial MSTD. This most likely accounts for our previous finding that sarkosyl-insoluble tau protein extracted from the filamentous deposits in familial MSTD consists only of tau isoforms with four repeats. These findings reveal that a departure from the normal ratio of four-repeat to three-repeat tau isoforms leads to the formation of abnormal tau filaments. The results show that dysregulation of tau protein production can cause neurodegeneration and imply that the FTDP-17 gene is the tau gene. This work has major implications for Alzheimer’s disease and other tauopathies.
Keywords: alternative mRNA splicing, Four-repeat tau isoforms, Frontotemporal dementia, microtubule binding, Tau filaments
Multiple system tauopathy with presenile dementia (MSTD) is an autosomal-dominantly inherited neurodegenerative disease characterized by dementia, disinhibition, generalized bradykinesia, rigidity, and superior gaze palsy (1, 2). It belongs to the group of familial frontotemporal dementias with Parkinsonism linked to chromosome 17 (FTDP-17) (3–5). Many (if not all) cases of FTDP-17 show abundant filamentous deposits consisting of hyperphosphorylated microtubule-associated protein tau and the tau gene maps within the interval on chromosome 17q21–22, making it a strong candidate gene (5).
Six tau isoforms are expressed in normal adult human brain (6). They range from 352 to 441 amino acids and are produced from a single gene by alternative mRNA splicing. They contain three or four tandem repeats located in the carboxyl-terminal half, which constitute microtubule-binding domains. They also differ by the presence or absence of 29- or 58-amino acid inserts of unknown function that are located near the amino terminus.
In familial MSTD, abundant filamentous tau protein deposits form in the neocortex, some subcortical nuclei, brainstem, and spinal cord, where they are found in both nerve cells and glial cells, chiefly oligodendrocytes (1). Ultrastructurally, the tau filaments are twisted, with an irregular periodicity of 90–130 nm (1). Biochemically, they consist of three tau isoforms, each with four microtubule-binding repeats, while lacking tau isoforms with three repeats (1). A similar pattern of pathological tau bands is also found in progressive supranuclear palsy and corticobasal degeneration, two largely sporadic neurodegenerative diseases with abundant filamentous tau deposits (7, 8).
This pattern of pathological tau bands contrasts with the paired helical and straight filaments of Alzheimer’s disease and some other tauopathies in which six tau isoforms are found (9, 10). However, all of these diseases are similar in that the tau protein extracted from the filaments is hyperphosphorylated and unable to bind to microtubules (11, 12). Hyperphosphorylation of tau is believed to precede filament assembly (13); however, hyperphosphorylation alone is probably insufficient for assembly and other factors, such as sulfated glycosaminoglycans or nucleic acids, may be necessary for nucleating the assembly of tau into filaments (14–18). Incubation of recombinant three- and four-repeat tau isoforms with sulfated glycosaminoglycans gives rise to filaments with similar morphologies to the tau filaments of Alzheimer’s disease (14, 16).
The difference between three- and four-repeat tau isoforms derives from the alternative mRNA splicing of exon 10 of the tau gene (19, 20). This exon encodes the 31-amino acid repeat that is added after the first repeat of the three-repeat tau isoforms to give isoforms with four repeats (19). We have previously found the sequence of the tau exons themselves to be normal in familial MSTD (2). In view of the absence of three-repeat tau isoforms from tau filaments in familial MSTD, we have examined the sequences of the introns flanking exon 10.
We now report the presence of a heterozygous G to A transition in the intron following exon 10 of the tau gene. It is located immediately adjacent to the splice-donor site. The mutation segregates with the disease and disrupts a predicted stem-loop structure which may lead to increased use of this splice site. We have investigated soluble tau from familial MSTD brain and have found an abnormal preponderance of four-repeat over three-repeat tau isoforms, consistent with increased use of the splice site. The present findings identify the genetic defect responsible for familial MSTD and indicate that a change in the ratio of four-repeat to three-repeat tau isoforms is sufficient to produce nerve cell and glial cell dysfunction, leading to tau filament formation, causing degeneration and resulting in a dementing disorder.
MATERIALS AND METHODS
Tau Gene Sequencing.
Genomic DNA from blood samples was isolated by proteinase K digestion, phenol-chloroform extraction, and isopropanol precipitation. Six DNA samples were extracted from histological sections, as described (21). Tau exons were amplified from genomic DNA using primers designed to flanking intronic sequence (M.G.S. and M.G., unpublished data). The primer sequences for exon 10 were: 5′-CGAGCTCGCTTGTTCACTCATCCTTTTT-3′ (sense) and 5′-CGAGCTCGCAGTGTCTCGCAAGGTGTA-3′ (antisense). PCR reactions contained 20 ng/μl DNA, 0.25 μM of each primer, and 1 unit of Pfu polymerase (Promega). Amplification was carried out over 30 cycles (denaturation, 95°C; annealing, 60°C, extension, 72°C), with a final 10-min extension at 72°C. Amplified products were run on a 2% low-melting agarose gel, the bands excised, diluted 1:3 in distilled water, and heated to 75°C. The DNA was purified using a Qiaquick PCR purification spin column (Qiagen, Chatsworth, CA) and used for double-stranded DNA sequencing. In some experiments the PCR products were digested with SacI, subcloned into M13 mp18, and used for single-stranded DNA sequencing.
Extraction of Soluble Brain Tau and Immunoblotting.
Two hundred micrograms of frontal cortex, temporal cortex, and hippocampus from three familial MSTD patients and three age-matched controls were Dounce homogenized in 0.5 ml of 2.5% perchloric acid. The homogenate was left to stand on ice for 20 min and spun at 13,000 rpm for 10 min. The supernatant was dialyzed against 50 mM Tris⋅HCl (pH 7.4), 0.1 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride overnight at 4°C. Tau protein was dephosphorylated by treating 100-μl aliquots of the supernatants with Escherichia coli alkaline phosphatase (13.5 units/ml, Sigma Fine Chemicals) for 3 h at 67°C (9). The six adult human brain tau isoforms were expressed in E. coli and purified as described (22). Tau proteins were analysed by 10% SDS-PAGE and blotted onto an Immobilon-P membrane (Millipore). Blots were incubated overnight at 4°C with anti-tau antibodies BR133 or BR134 (diluted 1:1,000) which recognise the amino- and carboxyl-termini of tau, respectively (9). Tau bands were visualised using the avidin-biotin Vectastain system (Vector Laboratories) and 3,3-diaminobenzidine as the substrate.
RESULTS
Sequencing of intronic sequences flanking exon 10 of the tau gene in familial MSTD identified a G to A transition in the nucleotide 3′ of the exon 10 splice-donor site. It was found in 11 affected family members and segregated with the disease haplotype in other family members (Figs. 1–3). The G to A change was not present in 50 Caucasian controls. No change was found in tau cDNA from familial MSTD brain, indicating that the tau exons are spliced correctly (data not shown).
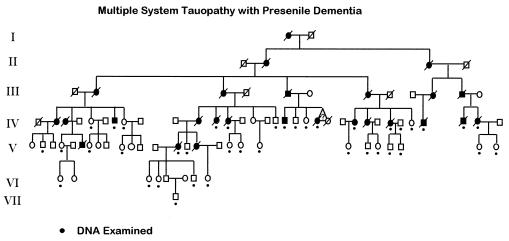
Figure 1.
Pedigree of the family with MSTD. Blackened symbols denote affected individuals. Black dots indicate individuals from whom DNA was available and tested by sequencing for the presence of the G to A mutation in the nucleotide adjacent to the exon 10 splice-donor site of the tau gene. The triangle identifies twins (it is not known whether they were mono- or dizygotic). Generation numbers are shown to the left.
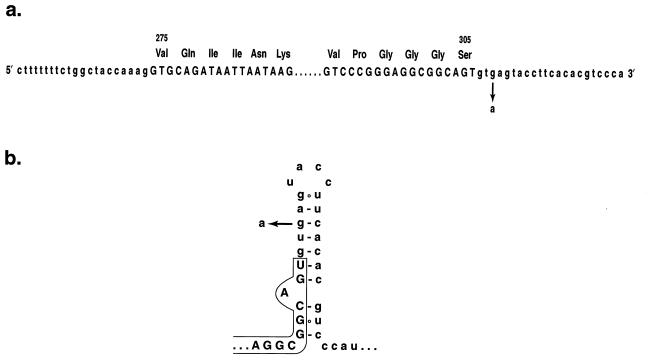
Examination of the nucleotide sequence of exon 10 and the 5′ intron junction identified a predicted stem-loop structure [ΔG = −3.2 to −4.3 kcal/mol (23)] that encompasses the last 6 nucleotides at the 3′ end of exon 10 and 19 nucleotides of the intron, including the GT splice-donor site (Fig. 3).
Figure 3.
Nucleotide sequence of the exon 10–intron junctions of the tau gene (a) and structure of the predicted stem-loop in the pre-mRNA (b). The exon sequences are shown in capital and the intron sequences in small letters. Amino acid numbering corresponds to the 441-amino acid isoform of human brain tau. The G to A transition responsible for familial MSTD is shown.
The G to A transition destabilises this stem-loop structure (ΔG= −0.6 to −1.7 kcal/mol). This may result in the more frequent use of the splice site and could lead to increased production of tau isoforms with four repeats over isoforms with three repeats.
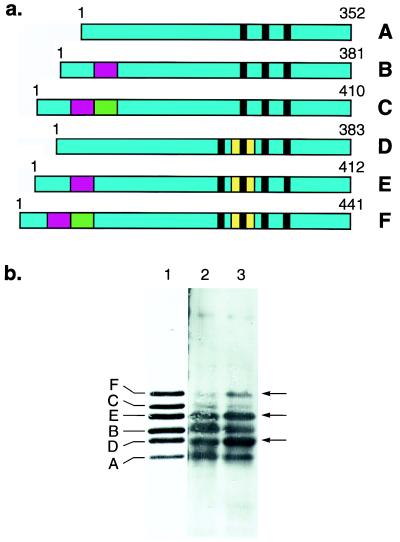
This question was examined directly using soluble tau extracted from cerebral cortex of control brain and of familial MSTD brain (Fig. 4). Following alkaline phosphatase treatment to dephosphorylate the protein, tau from control brain showed the characteristic pattern of four strong and two weak bands (Fig. 4, lane 2) which aligned with the six recombinant human brain tau isoforms (Fig. 4, lane 1). Similar levels of three-repeat and four-repeat tau isoforms were found, with a slight preponderance of isoforms with three repeats, in agreement with previous results (22). Soluble tau from familial MSTD brain also ran as four strong and two weak bands (Fig. 4, lane 3). However, unlike tau from control brain, a clear preponderance of tau isoforms with four repeats over isoforms with three repeats was observed, as reflected in a striking pattern of alternating stronger and weaker tau bands. Relative to soluble tau from control brain, the levels of four-repeat isoforms (shown arrowed in Fig. 4) were increased in familial MSTD brain, whereas the levels of three-repeat isoforms were reduced. The total amount of soluble tau did not appear to differ significantly between control brain and familial MSTD brain. Similar results were obtained with soluble tau extracted from three different brain regions of three cases with familial MSTD.
Figure 4.
(a) Schematic representation of the six human brain tau isoforms, with the alternatively spliced exons shown in red (exon 2), green (exon 3), and yellow (exon 10). The microtubule-binding repeats are indicated by black bars. (b) Immunoblots of dephosphorylated soluble tau protein from the frontal cortex of a control subject (lane 2) and a patient with familial MSTD (lane 3) using anti-tau serum BR133. Similar results were obtained with anti-tau serum BR134. Six tau isoforms are present in lanes 2 and 3. They align with the six recombinant human brain tau isoforms (lane 1). In the frontal cortex from the familial MSTD patient, tau isoforms with four repeats (isoforms D, E, and F) are more abundant and tau isoforms with three repeats (isoforms A, B, and C) are less abundant than in frontal cortex from the control. Arrows indicate the positions of tau isoforms with four repeats.
DISCUSSION
The present findings suggest a role for pre-mRNA structure in the regulation of the alternative splicing of tau. The presence of a stem-loop structure at the 5′ end of the exon 10 intron is predicted to be required for the normal splicing of exon 10. Disruption of this stem-loop by the G to A mutation of familial MSTD may lead to increased splicing of exon 10. A role for pre-mRNA structure in the regulation of alternative splicing is well documented in other systems (24, 25). Moreover, experimental studies have shown that the formation of stem-loop structures at the 5′ splice sites leads to inefficient splicing (26, 27). In addition, the G to A transition probably results in increased binding of the U1 snRNA to the 5′ splice site of exon 10 (28). This may also result in increased splicing of exon 10.
The predicted biological effect of the G to A transition in the tau gene in familial MSTD is an increased production of four-repeat tau isoforms and a reduced production of three-repeat tau isoforms, with no significant change in the total level of tau protein. This is what we observed when we analysed soluble tau in brain tissue from three cases with familial MSTD. The DNA from patients with familial MSTD is heterozygous for the G to A transition in the tau gene. Tau protein isoform ratios produced from the wild-type allele are therefore most likely normal, accounting for the presence of three-repeat tau isoforms in the soluble tau from familial MSTD brain. It is at present unclear whether only four-repeat tau isoforms are produced from the mutant allele or whether there is merely an increase in the levels of tau isoforms with four repeats.
In normal adult human brain six tau isoforms are expressed, with a slight preponderance of tau isoforms with three repeats over isoforms with four repeats (6, 22). Thus, a changed ratio in the levels of tau isoforms appears to be sufficient to lead to assembly into filaments. We have shown previously that filaments from familial MSTD brain contain only tau isoforms with four microtubule-binding repeats (1), implying that an abnormal preponderance of tau isoforms with four repeats over isoforms with three repeats leads to filamentous assembly of four-repeat isoforms.
The most unexpected implication of these findings is that a precise regulation of tau isoform ratios is essential for preventing assembly of tau into filaments. The mechanisms underlying assembly of only four-repeat tau isoforms into filaments are at present unknown. Tau protein is known to bind to microtubules and to promote microtubule assembly, with tau isoforms with four repeats being better at binding to microtubules and at promoting microtubule assembly than isoforms with three repeats (22, 29–32). Tau is a natively unfolded protein that is believed to become structured upon binding to microtubules (33, 34). It is unlikely that tau would assemble into filaments while bound in structured form to microtubules.
We are therefore led to conclude that when there is a preponderance of tau isoforms with four repeats over isoforms with three repeats, at least a proportion of the four-repeat tau does not bind to microtubules. It is possible that tau isoforms with three repeats and isoforms with four repeats bind to distinct sites on microtubules (29). An increase in four-repeat tau isoforms may lead to an excess in protein over available binding sites, thus increasing the time four-repeat tau spends in its natively unfolded state in the cytoplasm. Over time, this may lead to the hyperphosphorylation of four-repeat tau isoforms, rendering them completely unable to bind to microtubules. We have previously shown that in familial MSTD filamentous tau is hyperphosphorylated at the same sites as in Alzheimer’s disease (1). Interaction with other factors, such as sulfated glycosaminoglycans, may then result in nucleation and filament formation. We have previously shown that in familial MSTD brain cells with tau deposits are immunoreactive for heparan sulfate (1).
Pathological tau protein bands very similar to those in familial MSTD are found in progressive supranuclear palsy and corticobasal degeneration (7, 8). It appears likely that defects leading to an increase in the alternative splicing of exon 10 of the tau gene or changes in exon 10 itself also underlie these neurodegenerative diseases. Interestingly, a genetic polymorphism in the intron preceding exon 10 has been described in progressive supranuclear palsy (35, 36).
Our findings also suggest an explanation for Pick’s disease, a frontotemporal dementia that is characterized neuropathologically by the presence of Pick bodies which consist of abundant filamentous deposits made of hyperphosphorylated tau protein (37, 38). Biochemically, these filaments only contain tau isoforms with three microtubule-binding repeats (39, 40). By analogy with familial MSTD, it appears likely that defects leading to reduced alternative splicing of exon 10 of the tau gene underlie Pick’s disease.
In contrast to familial MSTD, Alzheimer’s disease and several other dementias with tau pathology are characterized by the presence of tau filaments in which all six brain tau isoforms are found (9, 10), indicating that a change in tau isoform ratios is not the only mechanism that can lead to assembly into filaments. Recently, a Val to Met change at residue 337 in tau (in the numbering of the 441-amino acid isoform of human brain tau) had originally been described in Seattle family A, which also belongs to the group of FTDP-17 dementias (41–43). Although this change has been interpreted as a probable benign polymorphism (43), it is possible that it is pathogenic, especially since it is located in the microtubule-binding region of tau, where valine is found at this position in all known tau sequences, from Caenorhabditis elegans to humans (6, 44). The Seattle family A is characterized by tau filaments that contain all six tau isoforms, with morphologies and staining characteristics that are indistinguishable from those of the paired helical filaments and straight filaments of Alzheimer’s disease (9, 10). As in the case of familial MSTD and other FTDP-17 dementias (1, 5), these tau filaments occur in the absence of extracellular Aβ deposits (41).
The presence of all six tau isoforms in the filamentous tau deposits in Seattle family A (10) is consistent with the Met to Val mutation at residue 337 being present in all six tau isoforms produced from the mutant allele (43). It appears likely that Met337 tau binds less well to microtubules than wild-type tau. This may in turn lead to its hyperphosphorylation, followed by assembly into paired helical filaments and straight filaments. Thus, the inability to bind to microtubules appears to be the shared primary abnormality in tau protein resulting from the different mutations in the tau gene in familial MSTD and in Seattle family A.
In Alzheimer’s disease, it is well established that filamentous tau protein deposits form within nerve cells that degenerate and that a good correlation exists between the number of tau deposits and the presence of dementia (45–47). The findings reported here in familial MSTD go beyond mere correlations in that they establish that nerve cell death and dementia result from an abnormal preponderance of tau isoforms with four repeats over isoforms with three repeats. Although several possible mechanisms can be envisaged, it appears likely that it is the presence of deposits consisting of tau filaments in nerve cells and glial cells that causes cell death in familial MSTD. The same may be true of Alzheimer’s disease and the other tauopathies. Compounds that inhibit the formation of tau filaments may therefore prevent nerve cell degeneration in all tauopathies.
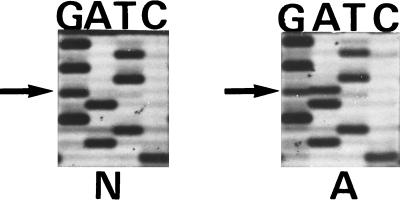
Figure 2.
Double-stranded DNA sequence of the exon 10-intron junction of the tau gene from an unaffected (labeled N) and an affected (labeled A) member of the family with MSTD. The position of the heterozygous G to A mutation is marked by the arrow.
Acknowledgments
We thank F. Epperson for help in collecting DNA samples. We are grateful to K. Nagai and G. Varani for helpful discussions. This work was supported by the U.K. Medical Research Council (to M.G.S., M.G., A.K.), The Royal Society of London (to M.G.S.), Public Health Service Grants AG10133 and NS14426 (to B.G.), and the Metropolitan Life Foundation (to M.G.).
ABBREVIATIONS
- MSTD
multiple system tauopathy with presenile dementia
- FTDP-17
frontotemporal dementias with Parkinsonism linked to chromosome 17
Note Added in Proof
Poorkaj et al. (48) have now reported two separate exonic mutations in the tau gene in two FTDP-17 families. Hutton et al. (49) have reported six different mutations in the tau gene in ten FTDP-17 families. Three of these mutations are located in the intron following exon 10, where they disrupt a predicted stem-loop. The other three mutations are found in exons.
References
- 1.Spillantini M G, Goedert M, Crowther R A, Murrell J R, Farlow M R, Ghetti B. Proc Natl Acad Sci USA. 1997;94:4113–4118. doi: 10.1073/pnas.94.8.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murrell J R, Koller D, Foroud T, Goedert M, Spillantini M G, Edenberg H J, Farlow M R, Ghetti B. Am J Hum Genet. 1997;61:1131–1138. doi: 10.1086/301594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelmsen K C, Lynch T, Pavlov E, Higgins M, Nygaard T G. Am J Hum Genet. 1994;55:1159–1165. [PMC free article] [PubMed] [Google Scholar]
- 4.Foster N L, Wilhelmsen K, Sima A A F, Jones M Z, D’Amato C, Gilman S, Spillantini M G, Lynch T, Mayeux R P, Gaskell P C, et al. Ann Neurol. 1997;41:706–715. doi: 10.1002/ana.410410606. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini M G, Bird T D, Ghetti B. Brain Pathol. 1998;8:387–402. doi: 10.1111/j.1750-3639.1998.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goedert M, Spillantini M G, Jakes R, Rutherford D, Crowther R A. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 7.Flament S, Delacourte A, Verny P, Hauw J-J, Javoy-Agid F. Acta Neuropathol. 1991;81:591–596. doi: 10.1007/BF00296367. [DOI] [PubMed] [Google Scholar]
- 8.Ksiezak-Reding H, Morgan K, Weidenheim K, Mattiace L A, Liu W K, Yen S H, Davies P, Dickson D W. Am J Pathol. 1994;145:1496–1508. [PMC free article] [PubMed] [Google Scholar]
- 9.Goedert M, Spillantini M G, Cairns N J, Crowther R A. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 10.Spillantini M G, Crowther R A, Goedert M. Acta Neuropathol. 1996;92:42–48. doi: 10.1007/s004010050487. [DOI] [PubMed] [Google Scholar]
- 11.Bramblett G T, Goedert M, Jakes R, Merrick SE, Trojanowski J Q, Lee V M-Y. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H, Ihara Y. J Neurochem. 1993;61:1183–1185. doi: 10.1111/j.1471-4159.1993.tb03642.x. [DOI] [PubMed] [Google Scholar]
- 13.Braak E, Braak H, Mandelkow E M. Acta Neuropathol. 1994;87:554–567. doi: 10.1007/BF00293315. [DOI] [PubMed] [Google Scholar]
- 14.Goedert M, Jakes R, Spillantini M G, Hasegawa M, Smith M J, Crowther R A. Nature (London) 1996;383:550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 15.Pérez M, Valpuesta J M, Medina M, Montejo de Garcini E, Avila J. J Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa M, Crowther R A, Jakes R, Goedert M. J Biol Chem. 1997;272:33118–33124. doi: 10.1074/jbc.272.52.33118. [DOI] [PubMed] [Google Scholar]
- 17.Kampers T, Friedhoff P, Biernat J, Mandelkow E M, Mandelkow E. FEBS Lett. 1996;399:344–349. doi: 10.1016/s0014-5793(96)01386-5. [DOI] [PubMed] [Google Scholar]
- 18.Ginsberg S D, Crino P B, Lee V M-Y, Eberwine J H, Trojanowski J Q. Ann Neurol. 1997;41:200–209. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- 19.Goedert M, Spillantini M G, Potier M C, Ulrich J, Crowther R A. EMBO J. 1989;8:393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreadis A, Brown M W, Kosik K S. Biochemistry. 1992;31:10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- 21.Nichols W G, Gregg R E, Brewer H B, Benson M D. Genomics. 1990;8:318–323. doi: 10.1016/0888-7543(90)90288-6. [DOI] [PubMed] [Google Scholar]
- 22.Goedert M, Jakes R. EMBO J. 1990;9:4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serra M J, Turner D H. Methods Enzymol. 1995;250:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 24.Clouet d’Orval B, D’Auberton-Carafa Y, Sirant-Pugnet D, Gallego M, Brody E, Marie J. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 25.Libri D, Piseri A, Fiszman M. Science. 1991;252:1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- 26.Eperon L P, Graham I R, Griffiths A D, Eperon I C. Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 27.Goguel V, Wang Y, Rosbash M. Mol Cell Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 29.Goode B L, Feinstein S C. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butner K A, Kirschner M W. J Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gustke N, Steiner B, Mandelkow E M, Biernat J, Meyer H E, Goedert M, Mandelkow E. FEBS Lett. 1992;307:199–205. doi: 10.1016/0014-5793(92)80767-b. [DOI] [PubMed] [Google Scholar]
- 32.Lee G, Rook S L. J Cell Sci. 1992;102:227–237. doi: 10.1242/jcs.102.2.227. [DOI] [PubMed] [Google Scholar]
- 33.Schweers O, Schönbrunn-Hanebeck E, Marx A, Mandelkow E. J Biol Chem. 1994;269:24290–24297. [PubMed] [Google Scholar]
- 34.Goode B L, Denis P E, Panda D, Radeke M J, Miller H P, Wilson L, Feinstein S C. Mol Biol Cell. 1997;8:353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrad C, Andreadis A, Trojanowski J Q, Dickson D W, Kang D, Chen X, Wiederholt W, Hansen K, Masliah E, Thal L J, et al. Ann Neurol. 1997;41:277–281. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 36.Higgins J J, Livan I, Pho L T, Li W, Nee L E. Neurology. 1998;50:270–273. doi: 10.1212/wnl.50.1.270. [DOI] [PubMed] [Google Scholar]
- 37.Delacourte A, Robitaille Y, Sergeant N, Buée L, Hof P R, Wattez A, Laroche-Cholette A, Mathieu J, Chagnon P, Gauvreau D. J Neuropathol Exp Neurol. 1996;55:159–168. doi: 10.1097/00005072-199602000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Probst A, Tolnay M, Langui D, Goedert M, Spillantini M G. Acta Neuropathol. 1996;92:588–596. doi: 10.1007/s004010050565. [DOI] [PubMed] [Google Scholar]
- 39.Sergeant N, David J P, Lefranc D, Vermersch P, Wattez A, Delacourte A. FEBS Lett. 1997;412:578–582. doi: 10.1016/s0014-5793(97)00859-4. [DOI] [PubMed] [Google Scholar]
- 40.Delacourte A, Sergeant N, Wattez A, Gauvreau D, Robitaille Y. Ann Neurol. 1998;43:193–204. doi: 10.1002/ana.410430209. [DOI] [PubMed] [Google Scholar]
- 41.Sumi S M, Bird T D, Nochlin D, Raskind M A. Neurology. 1992;42:120–127. doi: 10.1212/wnl.42.1.120. [DOI] [PubMed] [Google Scholar]
- 42.Bird T D, Wijsman E M, Nochlin D, Leehey M, Sumi S M, Payami H, Poorkaj P, Nemens E, Raskind M A, Schellenberg G D. Neurology. 1997;48:949–954. doi: 10.1212/wnl.48.4.949. [DOI] [PubMed] [Google Scholar]
- 43.Poorkaj, P., Bird, T. D., Wijsman, E., Nemens, E., Garruto, R. M., Anderson, L., Andreadis, A., Wiederholt, W. C., Raskind, M. A. & Schellenberg, G. D. (1998) Ann. Neurol., in press. [DOI] [PubMed]
- 44.Goedert M, Baur C P, Ahringer J, Jakes R, Hasegawa M, Spillantini M G, Smith M J, Hill F. J Cell Sci. 1996;109:2661–2672. doi: 10.1242/jcs.109.11.2661. [DOI] [PubMed] [Google Scholar]
- 45.Goedert M, Trojanowski J Q, Lee V M-Y. In: The Molecular and Genetic Basis of Neurological Disease. Rosenberg R N, Prusiner S B, DiMauro S, Barchi R L, editors. Boston: Butterworth–Heinemann; 1997. pp. 613–627. [Google Scholar]
- 46.Braak H, Braak E. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 47.Arriagada P V, Growdon J H, Hedley-White E T, Hyman B T. Neurology. 1992;42:631–639. doi: 10.1212/wnl.42.3.631. [DOI] [PubMed] [Google Scholar]
- 48.Poorkaj P, Bird T D, Wijsman E, Nemens E, Garruto R M, Anderson L, Andreadis A, Wiederholt W C, Raskind M, Schellenberg G D. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 49.Hutton M, Lendon C L, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Nature (London) 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]