Abstract
Production of structure-grade mammalian membrane proteins in substantial quantities has been hindered by a lack of methods for effectively profiling multiple construct expression in higher eukaryotic systems such as insect or mammalian cells. To address this problem, a specialized small-scale eukaryotic expression platform by Thomson Instrument Company (Vertiga-IM) was developed and coupled with a Guava EasyCyte microcapillary 96-well cytometer to monitor cell density and health and evaluate membrane protein expression. Two proof of concept experiments were conducted using the human β2-adrenergic receptor (β2AR) and gap junction protein connexin26 (Cx26) in a baculovirus expression system. First, cell surface expression was used to assess the expression levels of fourteen β2AR truncation variants expressed using the Vertiga-IM shaker. Three of these variants were then compared to wild type β2AR using three metrics: cell surface expression, saturation ligand binding and protein immunoblot analysis of dodecyl maltoside extracted material. Second, a series of systematic Cx26 truncation variants were evaluated for expression by protein immunoblot analysis. The cumulative results for these two systems show that the Vertiga-IM instrument can be used effectively in the parallel insect cell micro-expression of membrane protein variants, and that the expression of cell surface molecules as monitored with the Guava EasyCyte instrument can be used to rapidly assess the production of properly folded proteins in the baculovirus expression system. This approach expedites the in vitro evaluation of a large number of mammalian membrane protein variants.
Introduction
Technologies focused primarily on bacterial expression of prokaryotic proteins developed as part of pilot structural genomics projects are now being applied to determine the structures of more challenging eukaryotic and membrane protein targets. Although there are many cases where eukaryotic proteins can be expressed in bacteria, other protein targets of interest, particularly human membrane proteins, often require expression in eukaryotic systems such as Pichia pastoris, yeast, insect cells, and mammalian cells [1,2]. Consequently, it is essential to develop expression technologies to produce recombinant eukaryotic membrane proteins in an efficient and stable manner [3]. Perhaps one of the most useful eukaryotic expression systems for structural biology is the baculovirus-mediated gene delivery to insect cells, where the heterologous protein is expressed at a particular phase of the viral life cycle [4,5]. Unfortunately, the preparation and evaluation of recombinant protein in eukaryotic cells is time consuming and does not lend itself to high-throughput applications where multiple protein constructions must be examined. The ability to assay protein expression using early viral passages in a small scale format could save significant time and resources, and make the process more amenable to parallel screening efforts [6].
The goal of our current study was to develop and optimize an efficient small scale expression platform capable of screening multiple protein constructs using a baculovirus expression system to produce samples for structure determination studies. We were interested in developing a protocol to take advantage of the Thomson eukaryotic incubated shaker (Vertiga-IM; http://www.htslabs.com), a second generation system to that previously described for parallel bacterial expression [7,8]. We describe a process that utilizes small volumes of Spodoptera frugiperda (Sf9) insect cell suspension cultures that were incubated in 24-well blocks, rather than adherent cultures or as suspensions in large vessels [9–11]. With human β2-adrenergic receptor (β2AR), we were able to take advantage of the substantial effort that has already been devoted to the expression of G-protein coupled receptors (GPCRs) by including several previously characterized truncation variants [12,13]. In this report, we evaluate protein extraction, and plasma membrane ligand binding of truncation variants of β2AR (Figure 1A) with active detergent extractable material. In conjunction with the Vertiga-IM, we assayed for cell surface expression using a Guava EasyCyte microcapillary flow cytometer (http://www.guavatechnologies.com) thus providing a powerful and straightforward method for assessing expression levels of membrane protein in a small scale format. A second complimentary study was undertaken using the human gap junction protein connexin26 (Cx26). Gap junction channels allow direct cell-to-cell movement of ions and signalling molecules such as ATP to control the metabolic and electrical activities within tissues [14]. Connexin subunits (Figure 1B) contain four transmembrane domains (M1–M4), two extracellular loops (EL1 and EL2), a cytoplasmic loop (IL) between M2 and M3, and amino- and carboxy-tails. Six subunits assemble as a hemichannel, also called a connexon. The dodecameric channel is formed by the end-to-end docking of two connexions, and tight packing of the extracellular loops prevents exchange of molecules with the extracellular environment [15]. Unlike β2AR which has an extracellular N-terminus and an intracellular C-terminus, Cx26 has both the N- and C-terminus within the cell. Thus, microscale expression in the Vertiga-IM was assessed by traditional protein immunoblot analysis.
Fig. 1.
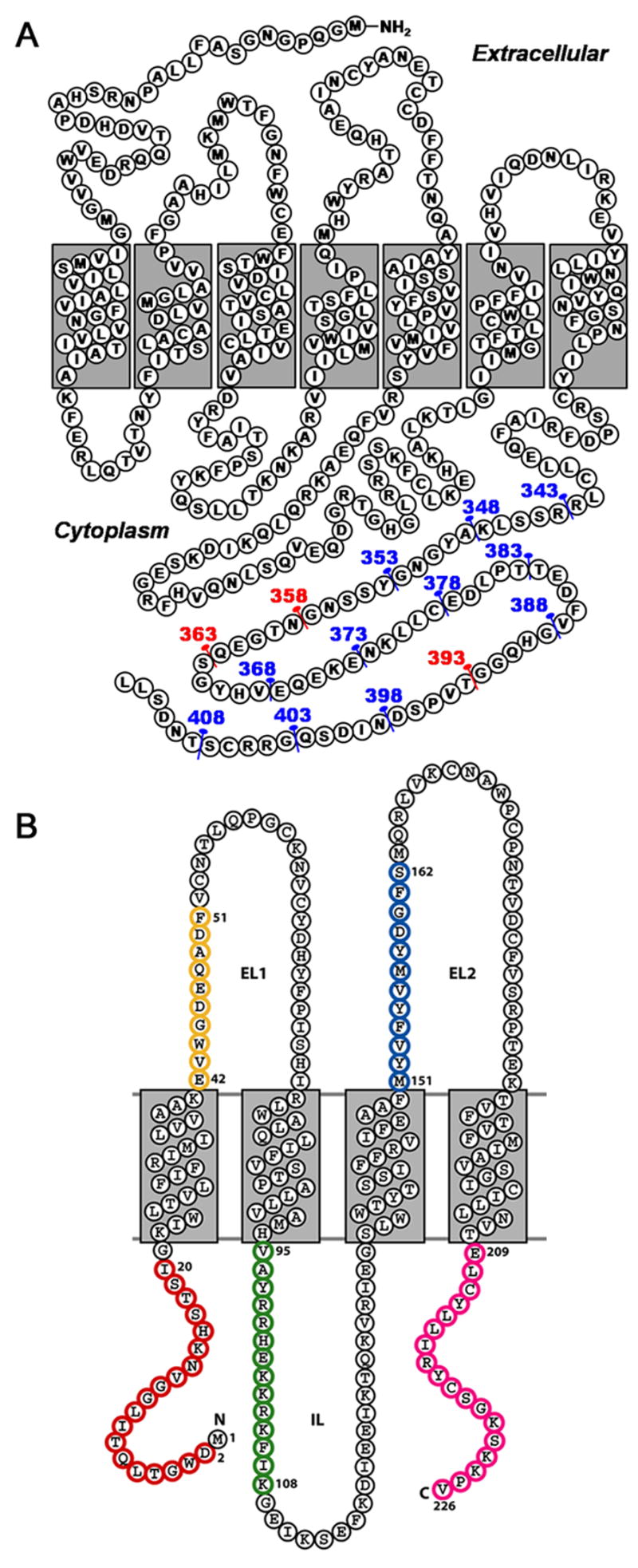
Membrane topology of receptor truncations and variants. (A) Membrane topology of the β2AR protein sequence with numbers and pins indicating the location of truncations for the 14 constructs used in this experimental series. Numbers and pins in red indicate the mutations focused on for further study as highlighted in Figures 3 and 4. (B) Membrane topology of a Cx26 subunit. The 4 transmembrane domains (M1 to M4) are denoted with gray boxes. Areas that were mutated are the N-terminus (red), extracellular loop 1 (yellow), the cytoplasmic M2–M3 loop (green), extracellular loop 2 (blue) and the C-terminus (magenta).
Materials and Methods
Construct design of human β2AR
The initial full length β2AR cDNA (a gift from B. Kobilka at Stanford University) contained the amino terminal hemagglutinin signal sequence and Flag epitope, as well as the 6-histidine tag [16]. The entire construct was amplified from its original vector using standard polymerase chain reaction (PCR) protocols with flanking primers designed to introduce 5′ BamHI and 3′ HindIII restriction sites immediately upstream from hemagglutinin signal sequence and downstream from the 6-histidine tag, respectively. The constructs were subsequently cloned into the pFastBacI (pFB) Bac-to-Bac transfer vector (Invitrogen) to generate the wild-type β2AR (pFB/wtβ2AR) construct under the control of the polyhedrin promotor. Fourteen C-terminal truncation variants in five amino acid steps starting with the 1–408 variant (Figure 1A) were generated from wtβ2AR using the QuikChange protocol (Stratagene). Receptor specific primers were designed to insert a 6-histidine tag followed by a stop codon immediately adjacent to the last amino acid of the receptor variant.
Construct design of Cx26
A similar procedure to the above was used for the Cx26 series (Figure 1B; Supplementary Figure S1) which were designed to test a larger series of truncations in all of the extra-membrane regions of a protein. Wild-type human Cx26 (NCBI: AF479776) with a C-terminal 6-histidine tag in pVL1393 was used as a template for production of all the variants. Virus was then generated by recombination with linearized baculovirus DNA (BaculoGold; BD Bioscience). A total of 19 N-terminal, 20 C-terminal, 19 intracellular loop (IL), and 38 extracellular loop (EL; 19 each in EL1 and EL2, respectively) truncations were evaluated.
Baculovirus viral DNA was generated by site-specific recombination of the transfer vector into the viral genome in DH10Bac cells (Invitrogen) according to the manufacturer’s protocol. The resulting recombinant bacmid DNA was isolated and transfected into Sf9 insect cells using cellfectin (Invitrogen). A viral titer was determined for each construct after three rounds of viral amplification, which was normalized to 1 × 109 infectious particles (IP) per milliter [17].
Vertiga-IM Development and Expression of Constructs
The Vertiga-IM (Thomson) was developed as a second generation small scale protein expression system [7,8] to enable small scale screening of insect or mammalian (e.g. CHO & HEK293) cells in suspension. Three parameters were optimized using the Vertiga-I as a starting point: orbital path, throw distance and temperature control. The throw distance for the circular orbit was optimized to one-half inch for use with the Thomson 24-well plates. However, the throw is adjustable to accommodate different vessel formats. Finally, the temperature control was upgraded from the peltier system used on the Vertiga-I to a compression refrigeration system which is capable of maintaining a temperature from 5–40°C (+/− 0.1°).
Stocks of insect cell cultures were maintained in shake flasks and passaged every three days with ESF 921 insect cell media (Expression Systems). For each data point a 5 ml aliquot of mid-log phase insect cell culture at a density of 2 × 106 cells/ml was infected with a high-titer (1 × 109 IP/mL) stock of each construct (β2AR or Cx26) at a final multiplicity of infection (MOI) of 2. During the 72 hour induction the cells were maintained in suspension in a 24-well sterile block (Thomson) sealed with BreathEasy membranes (USA Scientific) by shaking at 300 rpm at 27 °C in the Vertiga-IM. Medium scale expression was performed in 50 ml shake flasks (Corning) maintained at 27 °C and 115 rpm in a standard incubator (New Brunswick).
Flow cytometric analysis of β2AR cell surface expression
Twenty micrograms of M2-anti-Flag monoclonal antibody (Sigma) were derivatized with Zenon Alexa-488 (Invitrogen) according to the manufacturer’s protocol. The reaction was diluted to 1 ml with Tris-buffered saline (TBS: 20 mM Tris pH 7.5, 130 mM NaCl) containing 4% bovine serum albumin to yield a stock solution of 20 μg/ml Alexa-488 labelled Flag (Alexa488-Flag) antibody. For the cell surface expression assay, 10 μl of culture were transferred from the 24-well Thomson block to a 96-well Costar round bottom plate and mixed with 15 μl of the Alexa488-Flag stock solution. The mixture was incubated at 4 °C for 30 min and then diluted five-fold with TBS to a final volume of 200 μl.
The cultures were then assayed for fluorescence using a Guava Easycyte microcapillary flow cytometer, utilizing laser excitation and emission wavelengths of 488 and 532 nm, respectively. The gain was adjusted so that negative expression and antibody isotype control cell populations fluoresced with mean fluorescence intensity (MFI) between 0 and 10. Cell populations expressing above 10 MFI were thus expressing above the noise level of the assay. For each assay point, 2000 cellular events were collected and the resulting histograms inspected manually to delineate the populations used to compute the MFI reported (Figure 2A).
Fig. 2.
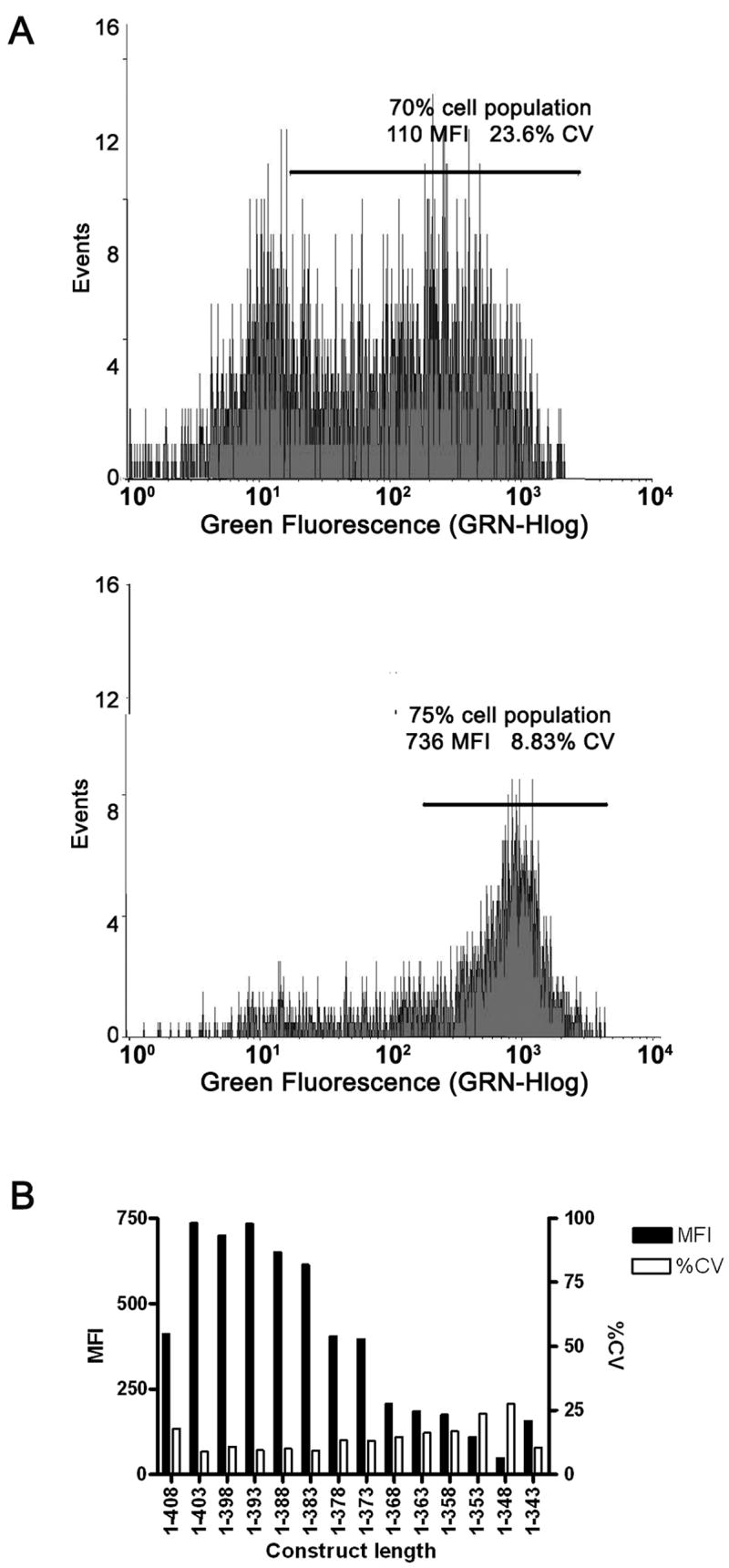
Example of β2AR cell surface expression. (A) Histogram of cell count (events) vs. mean fluorescence intensity (MFI). For cases where the expressing cells are not clearly delineated close to the limit of detection of 10 MFI the final reported MFI is derived from the mean fluorescence of the highest 70% of the total cell population (top panel, expressing cell population framed by solid line). For cases where a clearly expressing population of cells is detectable (bottom panel, expressing cell population framed by solid line) the reported MFI is derived from all cells in the expressing population which is frequently much greater than 70%. (B) Cell surface expression of β2AR arranged from the longest construct (1–408) to the shortest (1–343). Both MFI and %CV are calculated from two protocols described in A the latter protocol was preferred and utilized when possible. In general as the MFI decreases the %CV increases indicating a progressive loss of a clearly expressing cell population.
B2AR plasma membrane ligand binding assays
Cell pellets were suspended in ice-cold binding buffer (TME: 50 mM Tris-HCl, 10 mM MgCl2, 0.5 mM EDTA, pH 7.4), containing protease inhibitors (Complete protease inhibitor cocktail tablet, Roche Applied Science) and homogenized for 30 strokes with a Dounce homogenizer. Cellular debris and DNA were removed by centrifugation at 400 × g for 10 min at 4 °C, and the supernatants were collected. Crude plasma membranes were isolated by centrifugation of the supernatants at 150,000 × g for 60 min at 4 °C. The membrane pellets were resuspended in TME buffer. The wtβ2AR and truncation variants were tested for binding with Levo-[Ring, Propyl-3H(N)]-Dihydroalprenolol Hydrochloride [3H]DHA, (81 Ci/mmol, Perkin Elmer Life Sciences). Crude plasma membranes (5 μg of total protein per reaction) were incubated for 30 min at room temperature with serial dilutions of the radioligand (0.15–30 nM). Incubations were rapidly terminated by filtration using a Tomtec Mach III cell harvester (Tomtec) through a 96-well GF/B filter plate (MultiScreen Harvest plate, Millipore Corp.), and rinsed five times with 500 μl of ice-cold buffer (50 mM Tris-HCl, pH 7.4). The harvest plates were dried, and 30 μl of OptiPhase “HiSafe” III scintillation liquid (Perkin Elmer Life Science) were added. The bound radioactivity was measured using a Packard’s TopCounter NTX. Non-specific binding was determined in parallel reactions in the presence of an excess of Alprenolol (100 μM, Sigma-Aldrich), and specific binding was defined as the difference between total and non-specific binding. Protein concentrations were determined with the BCA protein assay (Pierce), using bovine serum albumin as a reference. All incubations were performed in triplicates, and independent experiments were repeated five times. Equilibrium dissociation constants (Kd) and maximal receptor levels (Bmax) were calculated from the results of saturation experiments using GraphPad Prism Software.
Western immunoblot analysis of total extractable material
For β2AR or Cx26 samples, cells from 1 ml aliquots of culture were isolated by centrifugation at 500 × g for 10 min. The supernatant was discarded, and the pellet was resuspended in TBS to a final concentration of 2 × 107 expressing cells/ml. Each aliquot was solubilized with 1% dodecyl maltoside (DDM, Anatrace) in the presence of benzonase for 1 h at 4 °C. The samples were then incubated with SDS-loading buffer, applied to a 4–10% Bis-Tris SDS-PAGE gel (Invitrogen), and separated by electrophoresis at 150 V. Samples were then transferred onto nitrocellulose and probed with an alkaline phosphatase conjugated anti-Flag monoclonal antibody (Sigma) for β2AR, or an alkaline phosphatase conjugated anti-histidine tag monoclonal antibody (Sigma) for Cx26. The resulting bands were exposed colorimetrically and analyzed by pixel density relative to wild type using ImageJ software (β2AR) or scored visually by relative band intensity (Cx26).
Results
Protein expression
The well-to-well variability of β2AR protein expression was assessed by measuring cell surface expression of twelve aliquots of a single Sf9 culture infected at the same recombinant baculoviral stock and MOI. The variability in cell surface expression was determined to be acceptable for an accurate comparison to the total protein yields quantified by Western immunoblotting (data not shown).
Cell surface expression of β2AR
All β2AR constructs were probed with Alexa488-Flag and assayed using a 96-well Guava EasyCyte microcapillary flow cytometer. The raw data from a typical assay (Figure 2A) are represented as a MFI of the expressing population of cells. Markers under which the MFI and %CV were calculated were set by inspection of the histograms for each sample. In cases where the expressing population is not clearly delineated based on the histogram (Figure 2A panel 1), the markers are set to include no less than 70% of the total cell population. In the case where the expressing population is clear based on the histogram (Figure 2A panel 2), the marker is set to include the entire expressing population. Interestingly, the MFI values showed that the shortest truncations (5 to 25 residues) displayed an increase in cell surface expression, whereas the larger truncations (30 to 65 residues) showed a marked decrease (Figure 2B).
Radioligand saturation binding assay for β2AR
The most reliable method for assessing β2AR expression is a saturation binding assay using a radio-labeled agonist or antagonist [18]. This quantitative method yields the absolute number of receptors per cell or sample, traditionally expressed as picomoles of receptor per milligram of total protein (Figure 3A). There is a general decrease in functional β2AR binding sites as the C-terminus is progressively truncated (Figure 3B; Table 1). However, the binding affinities for the receptor series remained relatively constant, indicating a simple decrease in the amount of functional receptor that was produced. At this time, it is unclear how the C-terminus influences the natural expression of β2AR although receptor signalled recycling is a likely explanation.
Fig. 3.
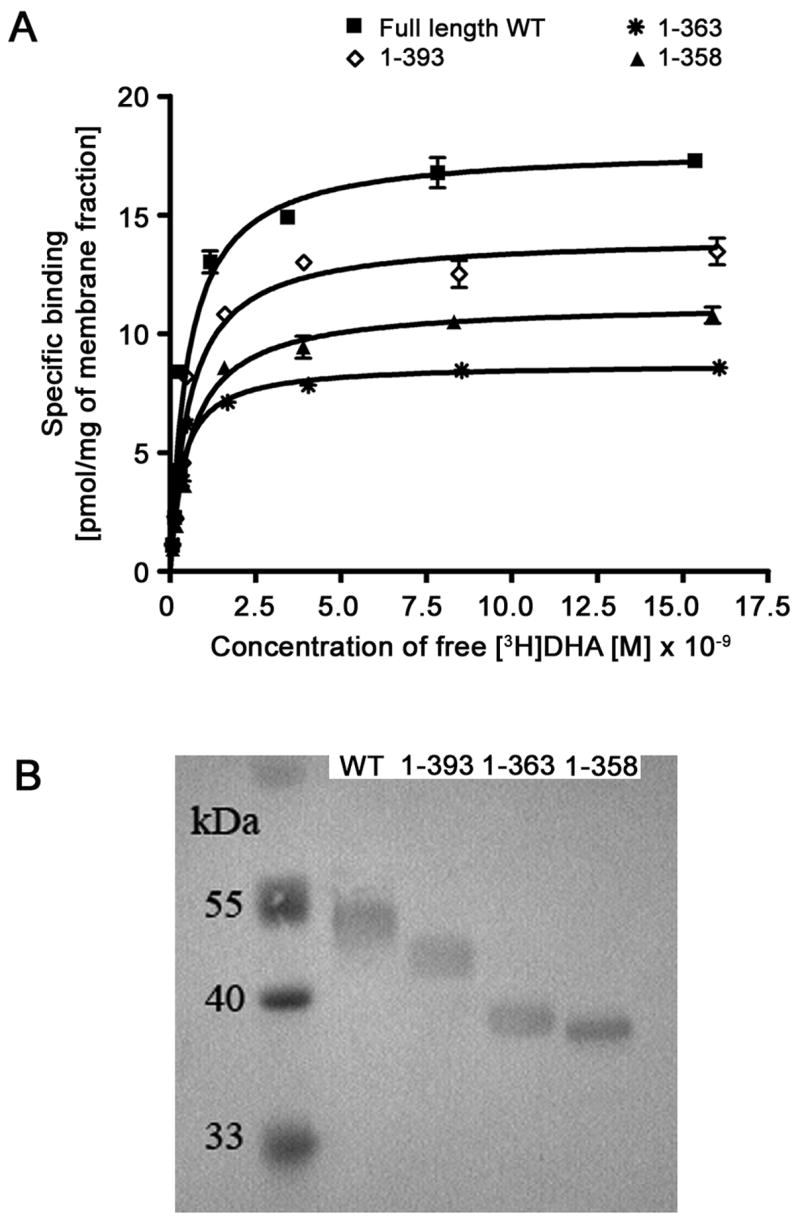
Saturation ligand binding data for β2AR. (A) [3H]DHA isothermal binding curves to a selected set of C-terminal β2AR truncation variants compared to wild-type. The data represent five repetitions of three assay replicates. (B) Protein immunoblot of isolated SF9 cell membranes.
Table 1.
Bmax and Kd for β2AR wild type and truncations
| Truncation | Kd (nM) | Bmax (pmol/mg) | ||
|---|---|---|---|---|
| SEM | SEM | |||
| WT | 0.61 | ± 0.06 | 19.75 | ± 1.17 |
| 1–393 | 0.71 | ± 0.10 | 16.81 | ± 0.76 |
| 1–363 | 0.50 | ± 0.07 | 9.57 | ± 0.72 |
| 1–358 | 0.50 | ± 0.04 | 12.44 | ± 0.47 |
Semi-quantitative Western immunoblot analysis of β2AR
β2AR was solubilized by treatment of crude membrane fractions with 1% DDM. The solubilized protein was then analyzed by SDS-PAGE. For this purpose, non-specific aggregation was minimized by not heating the sample prior to loading. There is a clear decrease in the total intensity of the bands relative to WT with all of the truncations (Figure 3). This trend becomes more clear upon pixel quantitation analysis with ImageJ (Figure 4). Due to the ability of protein immunoblot analysis to detect both surface and internal receptor, and the Guava EasyCyte analysis detecting cell surface receptor only, one should get a better approximation of total expression. However, this is often not the case as the protein immunoblot technique suffers from a number of technical limitations, including differential transfer rates to nitrocellulose and non-quantitative staining. In fact, these shortcomings are evident by examining the correlation coefficient (CC; Table 2) relative to saturation ligand binding of the normalized data (Figure 4) for both cell surface expression as measured by flow cytometry (CC: 0.93) and total extractable material as measured by protein immunoblot (CC: 0.74).
Fig. 4.
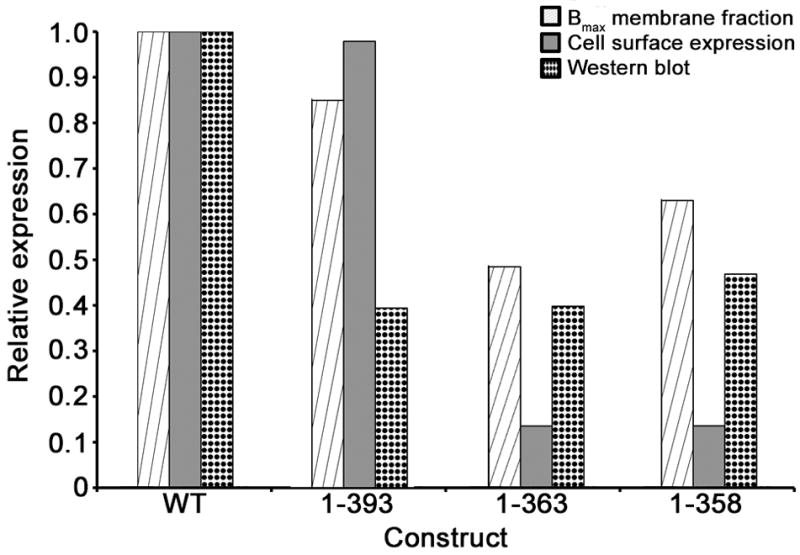
Expression comparison of β2AR truncations relative to wild type. (A) Relative expression levels of three selected truncation variants of β2AR relative to wild-type as represented by the maximal receptor expression levels (Bmax) of the crude membrane fraction (striped), cell surface fluorescence (gray) and by protein immunoblot analysis (dotted).
Table 2.
Correlation table for relative expression levels
| Ligand binding | Cell surface expression | Protein immunoblot | |
|---|---|---|---|
| Ligand binding | 1.00 | 0.93 | 0.74 |
| Cell surface expression | 1.00 | 0.54 | |
| Protein immunoblot | 1.00 | ||
Connexin truncation series
Our studies examined the expression of the 26 kDa connexin, Cx26, which has one of the shortest C-termini in the family. Since the N- and C-termini reside within the cytoplasm, the Guava EasyCyte could not be used to assess cell surface expression with immunoaffinity tags. Instead the total protein expression level was qualitatively assessed relative to wild type by Western immunoblot analysis with antibodies directed against the 6-histidine C-terminal tag (Figure 6). Overall 96 truncation or deletion variants were attempted, consisting of single amino acid serial truncations on both the N- and C- termini, as well as a series of deletion variants in each extracellular and cytoplasmic loop (Figure 1B; Supplementary Figure 1). These were each expressed in the baculovirus expression system. Wild type yields are approximately 1.5 mg per liter of culture by traditional shake flask growth methods. The highest expressing of the mutants in the Vertiga-IM system yielded similar levels to wild-type, Of particular importance, several trends emerged with deletions at the N- or C-termini, and to a lesser extent the EL1 loop which resulted in no recombinant virus or a reduction of expression in comparison. Truncation of the cytoplasmic M2–M3 loop IL and EL2 loops were surprisingly well tolerated (Figure 5).
Fig. 5.
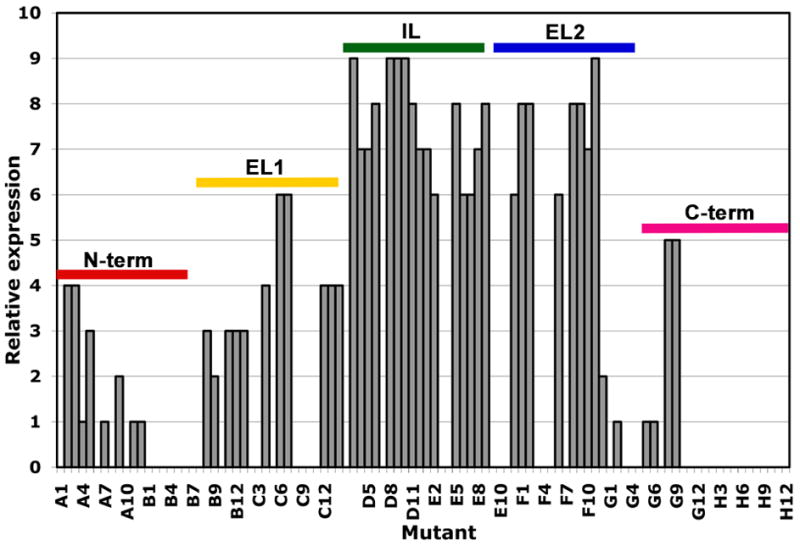
Cx26 parallel expression in SF9 cells using the Vertiga-IM. Estimated relative expression levels of expressed variants on a relative scale of 1 to 10, as determined by western blot analysis. The associated regions of the protein as described in Figure 1B are indicated. Full definition of the constructs are described in supplementary figure 1.
Discussion
Until recently, small scale expression trials in the baculovirus system were commonly carried out in small volume shake flasks or adherent cell cultures [6,9]. While these formats have the advantage of simplicity and familiarity, it can be prohibitively cumbersome to process multiple samples in parallel, scale-up to larger production volumes can be inconsistent, and this approach is wasteful as most of the cell culture is not needed for downstream analysis [6,9]. To obviate these problems, we developed the Vertiga-IM for growing small volumes of cell cultures in suspension. The key to the success of this instrument was careful optimization of the shaker’s orbital radius. If the throw-arm length was too small, cells were damaged by shear forces, whereas a throw-arm that was too large resulted in settling of the cells and suboptimal aeration (data not shown). In this particular instance, we utilized Sf9 insect cells for expression, which were maintained in suspension in 24-well blocks with a 1/2” throw arm at 300 rpm. The length of the throw arm and the rotation speed can be optimized for other cell types or culture vessels.
Development of protocols for baculovirus microexpression that use the Vertiga-IM was carried out using fourteen C-terminal truncation variants created from the full length β2AR gene. A parallel expression process was designed with this device in conjunction with the Guava EasyCyte microcapillary flow cytometer. The latter being used to assess cell surface targeting of heterologously expressed FLAG epitope tagged receptor (Figure 2B). Three truncations mutants were then chosen for scale up to medium scale shake flasks along with wtβ2AR for the purpose of comparing cell surface expression data to two other commonly used metrics: saturation ligand binding and protein immunoblot.
Results from medium scale expression studies showed high correlation between cell surface expression and the two more traditional metrics (Table 2), providing sufficient confidence so that cell surface expression data may be used as a means to screen out low expressing constructs in small scale expression trials prior to further characterization and scale up. We have further been able to use MFI of cell surface expression as a metric for predictive scale up in Wave bioreactors (data not shown). Although the correlation in positive expression data helps to select a construct for scale up and biophysical analysis, there are inconsistencies in the data (e.g. expression levels and Bmax values) that may be due to a higher limit of detection for the FACS analysis or a difference between the amount of functional protein being produced and the amount of total protein trafficked successfully to the cell surface. Indeed, we have observed an apparent early saturation of cell surface expression levels for other constructs in this system, while total functional protein levels continue to increase. This problem is much more pronounced when the target is expressed under the polyhedrin late stage promoter, rather than the GP64ie1 fusion promoter (Novagen) presumably due to the adverse effect of viral infection on the endogenous secretory machinery of the cell. Nevertheless, the ability to screen constructs in a high-throughput manner not only conserves reagents, but may also eliminate the need to generate high-titer stocks of each construct being screened, as the P2 viral passages may contain enough virus under favourable conditions to infect small scale expression cultures for screening purposes.
In our second case study of human Cx26, the cytoplasmic orientation of the N- and C-tails precluded the use of the cell surface expression assay, and there is no straightforward method to quantitatively measure channel activity. Therefore, we used the Vertiga-IM for parallel expression in a Thomson 24-well plate format, monitoring the expression level using semi-quantitative protein immunoblot analysis. The expression level varied greatly with the location and extent of truncation (Figure 5). In general, truncations of the termini and the E1 extracellular loop are not viable or result in much lower protein expression. However, truncations of the IL and E2 loops were surprisingly well tolerated and therefore reasonable to pursue further. This study has provided a large number of potentially useful constructs, as well as important guidance to where and to what extent truncations can be made to the molecule in the future.
Conclusions
Over the last few years considerable effort has been devoted to the systematic, simultaneous analysis of a large number of genes and gene products. While there has been impressive progress in the analysis of soluble protein targets from bacterial sources, there is a lag in the analysis of eukaryotic membrane protein targets where expression levels are typically low and micro-purification procedures are complicated by the need to solubilize folded protein from the cell membrane with detergents. Seemingly minor changes in amino acids or the location and number of affinity tags (e.g. His, Flag, etc.) can have a striking effect on the expression level. For instance, the smaller C-terminal truncations of β2AR showed an increase in cell surface fluorescence compared with wild type, whereas the larger truncations showed a decrease in fluorescence (Figure 2B). Likewise, the expression level of Cx26 was markedly reduced with truncation of the EL1 extracellular loop but less so for the EL2 loop (Figure 5).
The main benefit of the cell surface assay is its ability to monitor expression of a tagged membrane protein on an individual cell level rather than in bulk solution. Because of this feature comparisons between many viral stocks and constructs are simplified, as one may skip the step of normalizing viral load between cultures and compare cellular expression levels directly. Both protein immunoblots and ligand binding analysis use bulk expression levels of the culture and are limited in accuracy by the viral titer and normalization procedures. By analyzing the change in MFI in real time over the course of an expression experiment, one can optimize conditions relative to a known standard, such as ligand binding, and maintain those conditions despite fluctuating cell culture conditions. Translating expression from small shaker flasks to 5 ml cultures enables a more systematic analysis of multiple constructs and expression conditions. The fact that the trends are maintained in both formats and by different metrics portends that this approach will be of general utility.
Given the capricious nature of membrane protein expression, identifying a stable truncation or variant that expresses to a high level and is stable is largely empirical. Thus, it is important to develop high throughput approaches for rapidly and economically screening the expression and folding of many isoforms and variants of a eukaryotic membrane protein. To this end, we have successfully implemented and have described here an efficient method for analyzing the expression of the human membrane proteins (e.g. β2AR and Cx26) in the insect cell-based baculovirus system. We described techniques for maintaining cultured insect cells in a compact 24-well SBS plate format during expression and have demonstrated the utility of cell surface expression measurements as a gauge of the relative expression efficiency of membrane proteins in insect cell cultures. This approach has recently been successfully tested in our laboratory with a number of GPCR proteins, producing samples for crystallization studies.
Supplementary Material
Supplementary Fig 1. Listed are the Cx26 truncations pursued by region of the protein. The mutant identification is followed by the amino acid sequence of the extra-membrane region with the removed residues in green and the remaining residues in red.
Acknowledgments
This work was supported by NIH Roadmap award P50 GM073197 (RCS, PK and MY). KAB was supported by NIH training grant (HL007695). The authors thank Angela Walker for assistance with manuscript preparation.
Abbreviations
- β2AR
β2 adrenergic receptor
- Cx26
connexin 26
- PCR
polymerase chain reaction
- Sf9
Spodoptera frugiperda
- EL
extracellular loop
- IL
intracellular (cytoplasmic) loop
- IP
infectious particles
- TBS
Tris-buffered saline
- MOI
multiplicity of infection
- MFI
mean fluorescence intensity
- DDM
dodecyl maltoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards AM, Arrowsmith CH, Christendat D, Dharamsi A, Friesen JD, Greenblatt JF, Vedadi M. Protein production: feeding the crystallographers and NMR spectroscopists. Nat Struct Biol. 2000;7(Suppl):970–2. doi: 10.1038/80751. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert M, Albala JS. Accelerating code to function: sizing up the protein production line. Curr Opin Chem Biol. 2002;6:102–5. doi: 10.1016/s1367-5931(01)00291-5. [DOI] [PubMed] [Google Scholar]
- 3.Stevens RC. Design of high-throughput methods of protein production for structural biology. Structure. 2000;8:R177–85. doi: 10.1016/s0969-2126(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 4.Geisse S, Gram H, Kleuser B, Kocher HP. Eukaryotic expression systems: a comparison. Protein Expr Purif. 1996;8:271–82. doi: 10.1006/prep.1996.0101. [DOI] [PubMed] [Google Scholar]
- 5.Possee RD. Baculoviruses as expression vectors. Curr Opin Biotechnol. 1997;8:569–72. doi: 10.1016/s0958-1669(97)80030-4. [DOI] [PubMed] [Google Scholar]
- 6.Hunt I. From gene to protein: a review of new and enabling technologies for multi-parallel protein expression. Protein Expr Purif. 2005;40:1–22. doi: 10.1016/j.pep.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Page R, Moy K, Sims EC, Velasquez J, McManus B, Grittini C, Clayton TL, Stevens RC. Scalable high-throughput micro-expression device for recombinant proteins. BioTechniques. 2004;37:364–370. doi: 10.2144/04373BM05. [DOI] [PubMed] [Google Scholar]
- 8.Peti W, Page R, Moy K, O’Neil-Johnson M, Wilson IA, Stevens RC, Wüthrich K. Towards miniaturization of a structural genomics pipeline using micro-expression and microcoil NMR. J Struct Funct Genomics. 2005;6:259–67. doi: 10.1007/s10969-005-9000-x. [DOI] [PubMed] [Google Scholar]
- 9.Chambers SP, Austen DA, Fulghum JR, Kim WM. High-throughput screening for soluble recombinant expressed kinases in Escherichia coli and insect cells. Protein Expr Purif. 2004;36:40–7. doi: 10.1016/j.pep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 10.McCall EJ, Danielsson A, Hardern IM, Dartsch C, Hicks R, Wahlberg JM, Abbott WM. Improvements to the throughput of recombinant protein expression in the baculovirus/insect cell system. Protein Expr Purif. 2005;42:29–36. doi: 10.1016/j.pep.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Bahia D, Cheung R, Buchs M, Geisse S, Hunt I. Optimisation of insect cell growth in deep-well blocks: development of a high-throughput insect cell expression screen. Protein Expr Purif. 2005;39:61–70. doi: 10.1016/j.pep.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi J, Takarada T, Yunoki S, Kikuchi Y, Maeda M. FRET-based monitoring of conformational change of the β2 adrenergic receptor in living cells. Biochem Biophys Res Commun. 2006;343:1191–6. doi: 10.1016/j.bbrc.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 13.Wenzel-Seifert K, Lee TW, Seifert R, Kobilka BK. Restricting mobility of GSα relative to the β2-adrenoceptor enhances adenylate cyclase activity by reducing GSα GTPase activity. Biochem J. 1998;334:519–24. doi: 10.1042/bj3340519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris AL. Emerging issues of connexin channels: biophysics fills the gap. Quart Rev Biophys. 2001;34:325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- 15.Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–80. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- 16.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 17.Mulvania T, Hayes B, Hedin D. A flow cytometric assay for rapid, accurate determination of baculovirus titers. BioProcess J. 2004;3:47–53. [Google Scholar]
- 18.Hein L, Kobilka BK. Adrenergic receptor signal transduction and regulation. Neuropharmacology. 1995;34:357–366. doi: 10.1016/0028-3908(95)00018-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig 1. Listed are the Cx26 truncations pursued by region of the protein. The mutant identification is followed by the amino acid sequence of the extra-membrane region with the removed residues in green and the remaining residues in red.


