Abstract
Objective
To develop an electronic health record that facilitates rapid capture of detailed narrative observations from clinicians, with partial structuring of narrative information for integration and reuse.
Design
We propose a design in which unstructured text and coded data are fused into a single model called structured narrative. Each major clinical event (e.g., encounter or procedure) is represented as a document that is marked up to identify gross structure (sections, fields, paragraphs, lists) as well as fine structure within sentences (concepts, modifiers, relationships). Marked up items are associated with standardized codes that enable linkage to other events, as well as efficient reuse of information, which can speed up data entry by clinicians. Natural language processing is used to identify fine structure, which can reduce the need for form-based entry.
Validation
The model is validated through an example of use by a clinician, with discussion of relevant aspects of the user interface, data structures and processing rules.
Discussion
The proposed model represents all patient information as documents with standardized gross structure (templates). Clinicians enter their data as free text, which is coded by natural language processing in real time making it immediately usable for other computation, such as alerts or critiques. In addition, the narrative data annotates and augments structured data with temporal relations, severity and degree modifiers, causal connections, clinical explanations and rationale.
Conclusion
Structured narrative has potential to facilitate capture of data directly from clinicians by allowing freedom of expression, giving immediate feedback, supporting reuse of clinical information and structuring data for subsequent processing, such as quality assurance and clinical research.
Introduction
Electronic health records have been a major objective of research in biomedical informatics for decades. 1,2,3,4 This research seeks to improve the legibility, accessibility and quality of health records to support patient care. Tremendous effort has been expended in representing health information in highly structured ways that support subsequent computer processing, such as decision support and quality assurance. 5,6,7,8,9,10,11,12 This vision of electronic health records transcends the basic function of communication among providers, and enables computers to support and augment care processes in myriad ways.
Despite these efforts, electronic health records have relatively low penetration into health care delivery. 13,14,15 Acquisition of data directly from clinicians remains one of the largest potential obstacles. 16 As a result, many electronic health records often lack key pieces of documentation, such as progress notes or admission notes. Low utilization is caused in part by the difficulty of capturing data in structured form. 17 While effective in narrow, predictable domains, structured data entry can be quite slow when events are broad in scope and exhibit high variation. 18,19,20,21 Clinicians are pressed for time, and cannot assume the burden of data entry without significant returns for their efforts. 1,22,23 Diffusion of electronic health record technology will remain low until these barriers are addressed.
Clinicians have a long tradition of using paper forms and dictation services, and are beginning to adopt direct entry of text and speech recognition. 24 This abiding preference for narrative data (clinical text written in a natural language such as English) deserves further consideration. Narrative has the advantage of familiarity, ease of use and freedom to express anything the clinician wishes. But more than this, clinicians need a way to interpret raw data, synthesize the facts and weave them into a coherent narrative. 25,26 Natural language provides many mechanisms that augment or enrich simple facts, for example to qualify their severity or degree, convey temporal relationships, indicate patterns of causality, provide rationale, propose hypotheses, and suggest alternatives. 27,28
This paper proposes a new model for electronic health records called structured narrative, in which unstructured text and coded data are fused into a single representation combining the advantages of both. Each major clinical event (e.g., encounter or procedure) is represented as a document to which Extensible Markup Language (XML) is added to indicate gross structure (sections, fields, paragraphs, lists) as well as fine structure within sentences (concepts, modifiers, relationships). This form of representation is known as semi-structured because the gross structure imposes restrictions on the clinician (standard fields for data entry), while allowing freedom of expression within those units (free text paragraphs). However, unlike traditional structured documentation, the free text paragraphs are marked up (using natural language processing) to yield a fine structure that identifies facts within the free text (diseases, medications, procedures, etc.), as well as modifiers (severity, certainty, etc.), and ways of connecting these facts (temporally, causally, etc.). All of these structured elements are associated with standardized codes that enable them to be reused for various computational purposes. At the same time, the extra verbiage in the narrative helps to weave the facts together, conveying temporal, causal, and reasoning relationships among the facts that are essential to contextualize, interpret, and synthesize the information. The structured narrative model is intended to be convenient for data entry, highly synthesized, and amenable to computer processing.
Background
Our interest in developing the structured narrative model for electronic health records is based on a variety of findings in the research literature that contrast the strengths and weaknesses of structured and unstructured clinical data. Van Ginnegan provides an excellent comprehensive review. 29 Several of these studies point toward some kind of hybrid model that could combine advantages of both. The follow sections summarize background in these two areas.
Structured and Unstructured Data
• Despite evidence of positive effects such as document completeness30,31 and ease of billing,32 documentation approaches based solely on coded data entry also have significant limitations, including clinician time for note completion,33,34,35 difficulty ascertaining medical relevance,36,37 and loss of information.38
• Narrative is a critical factor in evaluating medical evidence, making management decisions, and communicating medical knowledge39,40,41 and is often more accurate,42 more comprehensive,43 and provides data complementary to other sources.44 Medical narrative captures multiple pieces of data that, when used effectively, can reduce length of stay45 and diminish unnecessary tests.46 Well-written narrative can be easier to comprehend, more edifying, and even more convincing than structured data.47
• Natural language is by far the most expressive carrier of medical information while also permitting fairly rapid data entry.48,49 However, when the entry of text data is completely free, there is potential for omission errors.50 Unlimited freedom of data entry can also lead to inconstancies between different parts of the electronic health record.51
Semi-structured Data
• Imposing a certain degree of structure on documents can improve completeness and accuracy of clinical narrative.52,53,54,55 Physicians prefer reading such standardized documents.56,57,58 Displaying documents in labeled chunks or paragraphs helps to locate data efficiently.59
• While some electronic medical systems have attempted to separate structured data entry from free text entry, several prominent researchers advocate for some kind of combination of the two.60,61,62 A key factor in effectiveness is to allow clinicians to switch smoothly between the two types of entry.63
• When the gross structure of a document (sections, fields, paragraphs, lists, etc.) is represented explicitly, a number of useful functions can be supported that facilitate access to information and communication among providers such as the construction of new documents by reusing information from designated paragraphs of existing documents.64
• Representation of the fine structure (concepts, modifiers, etc.) is crucial to enable computer systems to carry out tasks such as decision support. In addition, representation of the fine structure of documents has several important uses in electronic health records. For example, highlighting can be used to draw attention to particular portions of text, e.g., sentences describing abnormal conditions.65
• XML was developed to support what is known as self-describing data or semi-structured data—data that has an irregular structure not known in advance, and that can change frequently and without notice.66,67 Thus, XML markup is ideal for representing both the gross document structure of documents and the fine structure of medical sentences. XML standards have emerged to represent medical documents.68,69,70,71
• XML representations can be enhanced through markup of individual medical terms using codes from a suitable dictionary72 and through integration of the results of natural language processing with the document structure.73 This feature enables semi-structured documents to be linked with an ontology of document types and sections, enabling subsequent processing.74
Formulation Process
The structured narrative model presented below is part of a larger program of research on electronic health records at Columbia University Medical Center (CUMC). A few years ago, our existing electronic web-based clinical information system (WebCIS) 75 provided an abundance of structured clinical data (e.g., laboratory results), but a dearth of narrative data (in particular, admission and progress notes). At the same time, our studies of medical narrative revealed an incredible richness of information. To close this gap, we decided to explore new paradigms for data capture, with an eye toward increasing direct entry by physicians and nurses.
We developed the structured narrative model described here through two parallel efforts: extension of our existing production system and development of an entirely new prototype system.
Beginning in 2003, one of the authors (PDS) led an effort to extend our electronic health records system (WebCIS). They developed and deployed a documentation module that allows physicians to write admission notes, progress notes and discharge summaries. Physicians at CUMC began using the module in February 2004 and have generated over 100,000 notes.
During the same period, another author (SBJ) led an effort to experiment with new techniques not available on our current platform. We convened a interdisciplinary biomedical informatics research group with core investigators consisting of a linguist, nurse, physician, and system analyst, with significant participation by another nurse, a surgeon, two internists, and multiple software experts. We conducted an extensive review of the literature to identify key studies of narrative and its role in patient care. The model of structured narrative emerged through a long series of white board discussions.
These two efforts are highly synergistic. New design ideas suggested by the model can be tested in a system which real providers use for real patients. Observing the use of the production system by real users in turn helps to inform and enhance the model. The templates used by physicians in the production system adhere to the guiding principles of structured narrative as outlined in the Introduction above, but do not yet incorporate all aspects of the structured narrative model. For example, the production system does not yet embed natural language processing capability. A series of related studies of these users 76 and the production system 77 confirmed several of our design principles for the structured narrative model.
Model Description
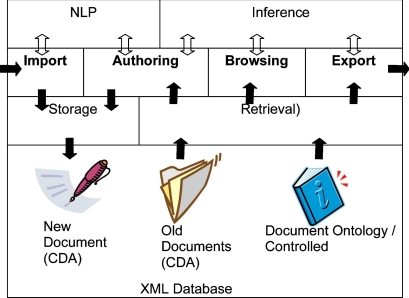
Structured Narrative is a model for electronic health records designed to capture and manage documents from many different kinds of clinicians (physicians, nurses, therapists, social workers, etc.). ▶ depicts a general system architecture using structured narrative, which comprises several distinct modules. The arrows in the figure indicate how information flows between the modules. The main functions performed by the system appear in bold: import (acquisition of documents from an external system), authoring (creation of new documents by a clinician), browsing (search and presentation of documents in a patient’s electronic health record), and export (transmission of documents to an external system). The sections below provide a concise description of each of these modules; this is followed by an extended example illustrating how the system is designed to be used by clinicians.
Figure 1.
An electronic health record using structured narrative consists of four primary modules that communicate with supporting modules (arrows indicate the flow of information): Import (acquisition of documents from an external system); Authoring (creation of new documents by a clinician); Browsing (search and presentation of documents in a patient’s electronic health record); and Export (transmission of documents to an external system). Import and Authoring use the Storage module to create new clinical documents, and use NLP to semantically annotate the document. Authoring, Browsing and Export use the Retrieval module to extract documents from a patient’s electronic health record. These modules also use Retrieval to access concepts in the document ontology and the Inference module to reason about concepts.
Clinical Document Database and Structured Narrative
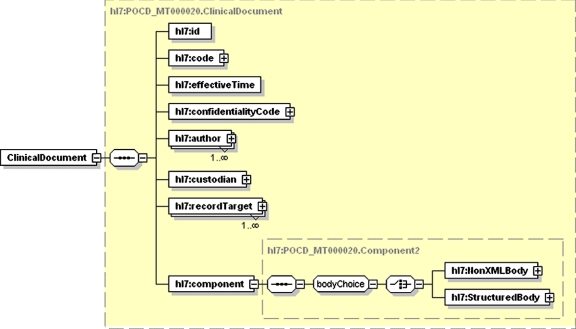
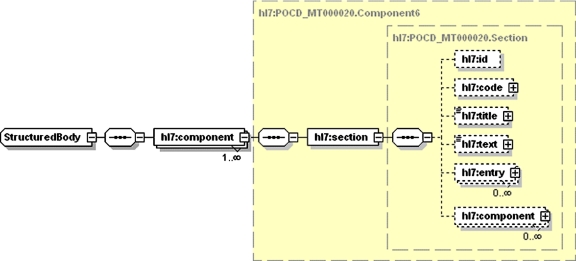
All data in the structured narrative model are represented as documents in XML. Clinical documents are represented using the Health Level Seven (HL7) clinical document architecture (CDA) standard. 78 The proposed model uses CDA for clinical notes (e.g., progress notes), clinical reports (e.g., radiology and pathology reports), as well as for non-narrative data (e.g., laboratory or pharmacy data). Each document comprises structured information giving circumstances of the document and a body containing the narrative information. The structured information (▶) includes a code specifying the type of document, effective time (date and time of creation), author, and record target (patient). The representation of the body (▶) is called structured narrative because the text is divided into named sections that contain reusable units of clinical information. Each section has a code that specifies its type, a title that describes its content, text containing one or more paragraphs and zero or more entry items that convey structured data. In addition, a section may contain one or more sections, to any level of nesting.
Figure 2.
The CDA Header (shown as an XML Schema diagram) contains structured information about a document, including (but not limited to) a code specifying the type of document, effective time (date and time of creation), author, and record target (patient).
Figure 3.
The CDA Body (shown as an XML Schema diagram) is divided into named sections, each of which contains a code that specifies its type, a title that describes its content, text containing one or more paragraphs, and zero or more entry items that convey structured data. A section may contain one or more sections, to any level of nesting.
Document Ontology and Controlled Terminology
The codes for documents and their sections are managed in a classification system or document ontology. The HL7/ Logical Observation Identifiers, Names, and Codes (LOINC) document ontology is used to classify clinical documents along several axes, including subject matter domain (e.g., surgery), professional role (e.g., resident), type of service (e.g., evaluation and management) and setting (e.g., inpatient). 79 In addition to the axes of the HL7/LOINC document ontology, our document ontology also manages the codes for sections, and indicates whether they contain text (e.g., physical exam or impression) or structured data (date of birth or blood pressure), as well as whether they have subsections (e.g., physical exam may contain sections for different body systems). Each document has an associated template, which specifies the order of sections and their properties (e.g., required during data entry). In this model a section can be defined once and used many times in different types of documents. For example, Family History may appear in the attending physician’s admission note and in the nurses’ admission document.
In the structured narrative model, the document ontology is part of a larger controlled terminology system, a knowledge base for integrating standard and local controlled terminologies (our local implementation uses the Medical Entities Dictionary). 80 This strategy allows each document and section code to be associated with any number of coding systems, whether locally defined (e.g., vendor-specific codes), or standardized (e.g., LOINC). It also allows a degree of independence when adapting to changes in these terminologies over time. In addition, other codes are required in documents to represent structured data elements, whether entered by the user or identified through natural language processing.
The controlled terminology (which includes the document ontology) can also be represented in XML, for example, using OWL (Web Ontology Language). 81 This enables the system to use common XML technologies for both patient-specific and ontology information.
Natural Language Processing
In the structured narrative model, text can be marked up with coded information at any level of detail, down to sentences, phrases, and words. This function is performed by the natural language processing (NLP) module, which takes text as input and returns XML, marking up medical concepts and their modifiers. The principal innovation of the proposed system is to apply NLP in real time, and use it to improve the entry of documents. The NLP system analyzes medical text and identifies semantic structures consisting of core concepts (e.g., demographics, diseases, symptoms, medications, and procedures) and their modifiers (e.g., anatomic location, time, frequency, degree, and certainty). In exploring our model, we focused on a system called MedLEE (Medical Language Extraction and Encoding), 82 but similar systems can be used.
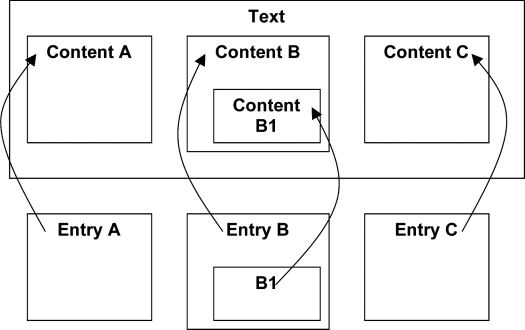
MedLEE can produce output in XML format, which is easily transformed into HL7 CDA. Significant phrases in the text are marked by a content tag, which is given a unique identification number. Following the text, one or more entry elements are used to define concepts and link them back to the marked items; these may be nested to any depth. ▶ illustrates this linked structure: three regions in the text are identified as A, B, and C, with B having a subregion identified as B1; the three entry items refer back to the text using these same identifiers. Entries can be nested to represent complex semantic structures (a detailed example appears below). Each entry has a code that links it with standardized coding schemes, e.g., Unified Medical Language System (UMLS), International Classification of Diseases 10 (ICD-10) or SNOMED CT. Using this approach, documents can represent coded information at the gross structural level (sections and fields), as well as the fine structural level (medical concepts and their modifiers).
Figure 4.
The CDA model allows any stretch of text (e.g. a phrase) to be marked by a content tag (here identified as A, B and C), which may be nested to any depth (B contains a stretch of text B1). Following the text, one or more entry elements are used to define concepts and link them back to the marked items using these identifiers (references indicated by arrows).
Inference Engine
The inference module enables the system to reason about controlled terminology elements. A separate module for this purpose helps to simplify the software functions for importing, authoring, browsing and exporting by enabling the system to reason about types of documents, sections and coded entries identified by NLP. Basic inference functions include translating from one controlled terminology to another (e.g., between our local terminology and national standards), and determining whether concepts are members of given classes. For example, to determine whether the document mentions antibiotics, medication codes can be extracted from the text using MedLEE, located in the ontology, and tested to see whether they are descendents of the antibiotic class or not.
Import and Export
The structured narrative model is designed to interoperate with other clinical information systems, such as ancillaries (radiology, pathology, laboratory, pharmacy, etc), health information management systems (e.g., chart deficiency), order entry, and billing. The model represents all clinical information in the form of documents. Structured data acquired from systems such as laboratory and pharmacy data can be managed using the same techniques as those used for reports and notes. For example, a laboratory panel (e.g., complete blood count) can be represented as a document, with sections for each laboratory test (e.g., red blood cell count). This approach makes it easy to incorporate structured data into our structured narrative documents.
The import module acquires documents from external sources using standard messaging services. NLP is applied to the text to identify gross structural elements (e.g., header information and section names), and to markup the fine structure (e.g., important medical concepts and their modifiers). Different kinds of document formats are converted into CDA. The export module retrieves designated types of documents from the database, and transforms them from CDA into formats required by other systems. The inference module may be needed to translate between controlled terminologies or to simplify the logic for certain complex transformation operations.
Storage and Retrieval
In our model, XML documents are managed by a database system that enables storage of XML in its native form and retrieval through the XQuery language (an emerging standard for XML queries). In exploring our model, we selected Tamino, 83 which can support very large collections of documents, but other systems with similar functionality could be used. By managing native XML in this manner, the space overhead of the markup is minimized. Designated elements can be indexed to make retrieval rapid. For example, all documents are indexed by medical record number to support browsing and authoring documents of a single patient.
Browsing
The purpose of browsing is to assist a clinician in locating information within the vast collection of documents in a patient’s electronic health record, usually by narrowing down to a single document and then focusing on a particular section or content item (terms in bold refer to elements of the CDA model in ▶, and axes of the LOINC document ontology). Browsing involves querying the database of existing documents using the CDA elements recordTarget (patient), effectiveTime (when the document was written), and code (the type of document). The document ontology and inference module are used to filter the document code according to its properties domain (area of health care), service (clinical activity being documented), and setting (location of activity).
The order in which these features are specified determines the model of browsing. As each feature is restricted, the set of relevant documents narrows for the clinician, allowing selection of the information of interest. These conceptual models can be rendered graphically in a variety of ways (see examples below). For temporal browsing, the order of document features is recordTarget, effectiveTime, and domain. For content browsing, the order is recordTarget, domain, and effectiveTime. Either sequence of narrowing can be then optionally followed by further restrictions on service and setting.
Once a document is selected, the clinician needs assistance in locating appropriate sections and content items. To drill down into documents, the codes associated with these elements are interpreted using the inference module and document ontology. Different sections and content codes have different display characteristics governing amount of screen space allocated, emphasis, layout, highlighting, etc.
Authoring
The purpose of the authoring function in the structured narrative model is to assist the clinician in creating new documents. The first challenge is to narrow down the type of document to be created from the thousands of potential types. The same document features used in browsing are employed, but the sequence of features used to narrow the space differs. For document authoring, the primary order is recordTarget (patient) and professional level. This can be further restricted by a combination of domain and service. In many cases, professional level and domain are fixed characteristics of the user that can be retrieved from the user profile.
Once a document type is selected, the template associated with the document is retrieved from the document ontology, which specifies the display and entry characteristics of sections, e.g., how big the entry box is for a section and whether it requires data or not. Another important property of a section is whether it is automatically filled with text extracted from old documents (pre-fill rules). The rules used to pre-fill are implemented with database queries similar to those described above for document browsing, but typically pull information from individual sections (or parts of sections) rather than whole documents.
Sections may also have associated post-fill rules, which specify processing actions that occur after the clinician has finished entering text into the section. These include natural language parsing that marks up the text with semantic content codes, and validation rules that check the content codes accordingly to pre-specified logic.
Validation through Example
The primary function of the structured narrative model is to facilitate the capture of rich clinical data directly from clinicians. Accordingly, this section will illustrate the model described above by focusing on the authoring function and how the other modules support it. Our principal concern is to make the acquisition of data as natural and rapid as possible.
We will use as an example a common clinical scenario—admitting a patient to the hospital. The steps articulated below will illustrate how the structured narrative model might augment existing workflow for reviewing and authoring clinical documents. Consider the following use case:
A general medicine attending physician working in the hospital receives notification of a new admission from the Emergency Room. The patient is being admitted for an asthma exacerbation. The attending physician accepts the admission and begins to work up the patient.
A typical workflow for this scenario is as follows:
1 Review the patient’s history in the electronic health record, focusing on summary-level descriptions of the patient.
2 Visit with the patient, take the history, and conduct the physical examination.
3 Review available clinical data in more detail using the electronic health record.
4 Author a new note documenting the initial encounter, assessment, and plan.
The clinician uses the Browsing function in the structured narrative model to complete Steps 1 and 3 and the Authoring function to create a new note (Step 4).
Browsing
After logging into the system, the user’s profile is retrieved, which provides some basic knowledge about the user related to the document ontology, such as professional level, domain, and setting (in this use case, the values are attending, internal medicine, and inpatient, respectively). For some users these characteristics will be fixed for long periods, others may change over time (e.g., a resident rotating through different services), while some may have multiple roles at a given time. Next, the clinician selects a patient to focus on, by specifying a medical record number or name (recordTarget).
The document ontology and inference engine support multiple ways to browse the patient’s collection of documents. The clinician first browses from the temporal view (effectiveTime) to determine how frequently the patient has been admitted over the last year with asthma exacerbations (▶). She next browses by domain to retrieve the most recent pulmonology note. In addition, she browses the document collection by content to determine if the patient’s asthma was ever severe enough to warrant intubation and mechanical ventilation support.
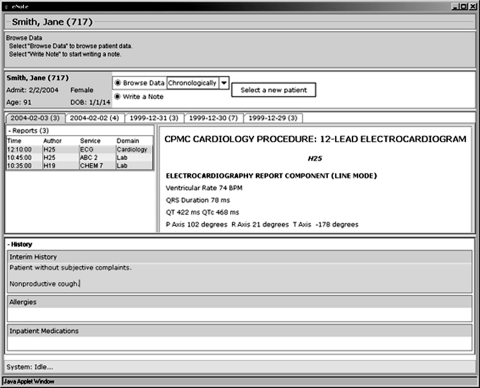
Figure 5.
Screen shot of structured narrative in use shows demographic information for a patient and panels for browsing and authoring. The user has chosen to browse documents by time, and has selected an ECG to view. The authoring panel shows part of a history section (other sections are not currently in view) for the current note, with some text entered in the interim history subsection.
Authoring
The clinician selects Admission Note from a list that has been filtered based upon her professional level (Attending), domain (Internal Medicine), and setting (Inpatient) as represented in the user profile. Using these default constraints, the resulting list will largely reflect the different kinds of services that the clinician can document (in this case, initial visit—evaluation and management). All of these values are default restrictions that the user can alter (e.g., if documenting a service in a different setting or domain).
Once the clinician selects the type of document, the header portion of the document is filled with information about the context of creation, such as the patient’s identifying information, the date and time, information about the author and institution. The content of the body is determined by a document template represented in CDA (▶). The empty patient-specific document is then rendered into graphical objects with which the clinician interacts. Section names are arranged in the window, with input boxes sized accordingly; these expand automatically when nearly full of text. The clinician adds ad hoc sections and subsections as needed.
Figure 6.
XML representation of a CDA document showing header information: id (unique identifier), code (type of document), recordTarget (patient information), author and custodian (institution). The structured body shown here has one section which has a code (type of section) and its text (narrative data).
Through pre-fill rules enabled by XQuery (▶), the laboratory data that the clinician ordered in the emergency room are automatically integrated into the note. She adds narrative to annotate the abnormal values. In addition, she browses relevant documents and highlights certain acts to import them into the current document, saving considerable time through reducing data entry, while ensuring the continuity of information across documents.
Figure 7.
Example of XQuery used to pre-fill the laboratory results section of a progress note. The query looks for documents with id matching a given medical record number (MRN), where the type of document (code) is defined in the medical entities dictionary (MED) as having a LOINC domain of laboratory. The creation time of the document (effectiveTime) must be within the last 24 hours. This query can be refined to extract the test values from the laboratory reports in order to embed them into the current note.
After she has written a section and moved on to the next, a variety of post-fill functions process and validate the content. Short fields containing relatively structured data (e.g., blood pressure, temperature, birth date) are validated to make sure they are in range and sensible. In addition, misspelled words and inappropriate abbreviations are highlighted.
Natural language processing is invoked as soon as the clinician has completed a field, and carried out without interfering with her workflow. ▶ shows how the text There is a history of asthma is marked up to encode a disease (asthma), a modifier (history of) and the composite concept (history of asthma). In general, this detailed semantic markup is entirely transparent to users. However, the availability of rich clinical data in real time makes possible a wide range of functions that interpret the data, and interact with the user in various ways. A simple example is the use of highlighting to identify particular kinds of information, such as clinical problems. A more interactive use is to trigger various decision support rules to provide alerts, warnings, etc. A novel use is to critique the document in various ways to improve quality. For example, abnormal lab results that are not mentioned in the assessment section may have been missed. Similarly, problems identified in the assessment section that are not addressed in the plan section may be untreated.
Figure 8.
CDA representation of past medical history section with NLM markup for the text There is history of asthma. The phrase history of asthma is marked as content with identifier p1, history of as p2 and asthma as p3. The text is followed by an entry for p1 given a UMLS code for the phrase, with value p3 and modifier p2.
Discussion
Significance
Our model proposes a new vision of electronic health records as collections of rich, interrelated narratives rather than lists of isolated facts. We believe that this representation is more in accord with the cognitive models of clinicians, and will therefore serve as a more accurate reflection of a patient’s health and a more effective source of knowledge for clinical decision making. This model provides a platform from which to test important, yet unanswered, questions related to provider data entry and reuse. First, we will be able to assess whether a more natural mode of data input (narrative) is more efficient for providers. Additionally, this model enables the assessment of whether capture of coded data through natural language processing facilitates subsequent analysis and interpretation operations. For example, the use of NLP may allow a much more precise method to carry text forward from previous notes, which may alleviate some problems caused by uncontrolled cutting and pasting of text.
The long-term objective of this work is to enhance the electronic health record by capturing documentation of clinician’s thought processes and decisions. The Structure Narrative model supports the following functions to reach that goal:
Maintaining continuity of the electronic health record: A lengthy electronic health record requires significant time to review and digest. Many facts from past narratives remain true in the present or persist with minor changes. By automatically bringing these facts forward into the current narrative, the system can reduce the time to enter the document. There is also potential to improve the completeness of documentation by maintaining continuity of what is known about a patient.
Integrating the electronic health record: Electronic health records contain vast amounts of data. However, most data are raw facts. By helping the clinician to connect, interpret and summarize these facts, the system can improve the usefulness of the information in the electronic health record. There is also potential to reduce the time to enter documents by performing some syntheses automatically.
Harmonizing the electronic health record: The multidisciplinary nature of health care creates the potential for the differing perspectives and interpretations in the electronic health record, and even contradictions. By bringing possible discrepancies to the attention of the responsible clinician, the system can help resolve or at least document the inconsistencies.
Through its facilitation of these electronic health record goals, the structured narrative model has the potential to support delivery of health care that is “safe, effective, patient-centered, timely, efficient, and equitable.” 84
Limitations
Our development team is large in comparison to most informatics research projects, but it is small by the standards of software companies engaged in this area. We can therefore develop only a limited number of interface features and assess their impact on clinician workflow. In addition, all members of the team are present at the same institution, and familiar only with the local information systems, practice patterns and clinical culture. Although paper narrative records are universal, it remains to be seen whether their electronic form will be embraced widely. Most crucially, the project is founded on the assumption that natural language processing can eventually identify the fine structure of clinical narratives. While no existing NLP system has complete coverage, we believe that a real-time approach that enables the immediate intervention of the clinician can address the inadequacies of this approach. For example, the codes assigned by NLP might trigger an alert inappropriately. The clinician could respond by rejecting the alert, which could provide feedback to the developers, or with more experience, could rephrase text to enable a correct parse to be obtained. In any case, the semi-structured representation allows fine-tuning of the data entry templates, permitting the use of structured entry when automated methods fail.
One of the primary goals of the proposed model is to reduce the effort required by physicians to enter data. A key mechanism in this model is the use of pre-fill rules that carry forward information from previous documents. A potential risk of this technique is that data that is incorrect or no longer valid might be propagated. To reduce this risk, the model supports the use of document templates that define which fields are pre-filled and which require new entry. Additional constraints can be implemented in the model using NLP, allowing a much more precise identification of text to carry forward. However, these methods are experimental and require further evaluation.
Conclusion
Electronic health records currently offer two major representations of clinical data with complementary strengths. Narrative data (captured through typing, transcription services or speech technology) is flexible and expressive but cannot be reused for other computational purposes. Coded data (captured through template systems or acquired automatically from devices) permits subsequent processing but lacks ease of use and the ability to record key aspects of clinician thought processes. The structured narrative model integrates these approaches: guiding clinician entry with broad templates, allowing full freedom of expression within sections and applying natural language processing to analyze this text for further computer processing. The coded information obtained through this model may enable efficient reuse of narrative material in subsequent documents to reduce clinician effort, and the ability to intervene during the authoring process to improve the quality of clinical documentation and patient care processes.
Acknowledgments
The authors acknowledge important conceptual contributions by Justin Starren, MD, PhD, and the involvement of Jason Shapiro, MD and Genevieve Melton, MD.
Footnotes
References
- 1.Barnett GO. Computer-Stored Ambulatory Record (COSTAR)Department of Health, Education and Welfare; 1976. Report No.: DHEW (HRA) 76–3145.
- 2.McDonald C, Overhage J, Tierney W. The Regenstrief Medical Record System: a quarter of a century experience Int J Med Inf 1999;54:225-253. [DOI] [PubMed] [Google Scholar]
- 3.Rector AL, Nowlan WA. The Galen ProjectGALEN Consortium. Computer Methods and Programs in Biomedicine 1994;45:75-78. [DOI] [PubMed] [Google Scholar]
- 4.van der Lei J, Duisterhout J, Westerhof H, van der Does E, Cromme P, Boon W, et al. The Introduction of Computer-based Patient Records in the Netherlands Ann Intern Med 1993;119(10):1036-1041. [DOI] [PubMed] [Google Scholar]
- 5.Barnett GO, Winickoff RN, Morgan MM, Zielstorff RD. A computer-based monitoring system for follow-up of elevated blood pressure Medical Care 1983;21(4):400-409. [DOI] [PubMed] [Google Scholar]
- 6.Brown PJB, Sonksen P. Evaluation of the quality of information retrieval of clinical findings from a computerized patient database using a semantic terminological model J Am Med Inform Assoc 2000;7(4):392-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chute CG, Cohn SP, Campbell JR, ANSI Healthcare Informatics Standards Board Vocabulary Working Group and the Computer-based Patient Records Institute Working Group on Codes and Structures A framework for comprehensive terminology systems in the United States: Development guidelines, criteria for selection, and public policy implications J Am Med Inform Assoc 1998;5(6):503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ. From data to knowledge through concept-oriented terminologies: experience with the Medical Entities Dictionary J Am Med Inform Assoc 2000;7(3):288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huff SM, Rocha RA, McDonald CJ, De Moor GJE, Fiers T, Bidgood Jr. WD, et al. Development of the LOINC (Logical Observation Identifier Names and Codes) Vocabulary J Am Med Inform Assoc 1998;5(3):276-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology Artif Intell Med 1997;9:139-171. [DOI] [PubMed] [Google Scholar]
- 11.McDonald CJ. Protocol-based computer reminders, the quality of care and the non-perfectibility of man New Engl J Med 1976;295:1351-1355. [DOI] [PubMed] [Google Scholar]
- 12.Spackman KA, Campbell KE, Cote RA. SNOMED RT: A Reference Terminology for health care Proc Am Med Inform Assoc Symp 1997:640-644. [PMC free article] [PubMed]
- 13.Anderson JG. Clearing the way for physicians’ use of clinical information systems Comm ACM 1997;40(8):83-90. [Google Scholar]
- 14.Amatayakul M. The state of computer-based patient record J AHIMA 1998;69:34-36. [PubMed] [Google Scholar]
- 15.Berner ES, Detmer DE, Simborg D. Will the Wave Finally Break?A Brief View of the Adoption of Electronic Medical Records in the United States. JAMIA 2005;12:3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loomis GA, Ries JS, Saywell RM, Thakker NR. If electronic records are so great, why aren’t family physicians using them? J Family Practice 2002;51(7):636-641. [PubMed] [Google Scholar]
- 17.McDonald CJ. The Barriers to Electronic Medical Record Systems and How to Overcome Them J Am Med Inform Assoc 1997;4:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powsner SM, Wyatt JC, Wright P. Opportunities for and challenges of computerization Lancet 1999;352:1617-1622. [DOI] [PubMed] [Google Scholar]
- 19.Symes DR. Physician data entry is the solution Health Management Technology 1999;20:34. [PubMed] [Google Scholar]
- 20.Van Ginneken AM. The computerized patient record: balancing effort and benefit IJMI 2002;65:97-119. [DOI] [PubMed] [Google Scholar]
- 21.Poissant L, Pereira J, Tamblyn R, Kawasumi Y. The impact of electronic health records on time efficiency of physicians and nurses: a systematic review J Am Med Inform Assoc 2005;12(5):505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan B. Reducing barriers to physician data entry for computer-based patient records Topics in Health Information Management 1994;15(1):24-34. [PubMed] [Google Scholar]
- 23.Gilbert JA. Physician data entry: providing options is essential Health Data Management 1998;6(9):84-86. [PubMed] [Google Scholar]
- 24.Rind DM, Safran C. Real and imagined barriers to an electronic medical record AMIA 1993:74-78. [PMC free article] [PubMed]
- 25.Van Ginneken AM. The physician’s flexible narrative Methods of Information in Medicine 1996;35(2):98-100. [PubMed] [Google Scholar]
- 26.Rector AL, Nowlan WA, Kay S. Foundations for an electronic medical record Methods Inform Med 1991;30:179-186. [PubMed] [Google Scholar]
- 27.Sager N, Friedman C, Lyman MS. Medical Language Processing: Computer Management of Narrative DataReading, MA: Addison-Wesley; 1987.
- 28.Friedman C, Johnson SB. Natural Language and Text Processing in BiomedicineIn: Shortliffe EH, Cimino JJ, editors. Biomedical informatics: computer applications in health care and biomedicine. New York: Springer; 2006.
- 29.Van Ginneken AM. The physician’s flexible narrative Methods Inform Med 1996;35(2):98-100. [PubMed] [Google Scholar]
- 30.Bell DS, Greenes RA. Evaluation of UltraSTAR: performance of a collaborative structured data entry system Proc Annu Symp Comp Appl Med Care 1994:216-222. [PMC free article] [PubMed]
- 31.Gouveia-Oliveira A, Raposo VD, Salgado NC, Almeida I, Nobre-Leitao C, de Melo FG. Longitudinal comparative study on the influence of computers on reporting of clinical data Endoscopy 1991;23(6):334-337. [DOI] [PubMed] [Google Scholar]
- 32.Marill KA, Gauharou ES, Nelson BK, Peterson MA, Curtis RL, Gonzalez MR. Prospective, randomized trial of template-assisted versus undirected written recording of physician records in the emergency department Annals Emerg Med 1999;33(5):500-509. [DOI] [PubMed] [Google Scholar]
- 33.Bates DW, Boyle DL, Teich JM. Impact of computerized physician order entry on physician time AMIA 1994:996. [PMC free article] [PubMed]
- 34.Tierney WM, Miller ME, Overhage JM, McDonald CJ. Physician inpatient order writing on microcomputer workstations—Effects on resource utilization JAMA 1993;269(3):379-383. [PubMed] [Google Scholar]
- 35.Payne TH. The transition to automated practitioner order entry in a teaching hospital: the VA Puget Sound experience AMIA 1999(1–2):589-593. [PMC free article] [PubMed]
- 36.Stein HD, Nadkarni P, Erdos J, Miller PL. Exploring the degree of concordance of coded and textual data in answering clinical queries from a clinical data repository JAMIA 2000;7:42-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jollis JG, Ancukiewicz M, DeLong ER, Pryor DB, Muhlbaier LH, Mark DB. Discordance of databases designed for claims payment versus clinical information system Annals Intern Med 1993;11:844-850. [DOI] [PubMed] [Google Scholar]
- 38.Hohnloser JH, Fisher MR, Koning A, Emmerich B. Data quality in computerized patient records—Analysis of a hematology biopsy report database Clin Monit Comput 1994;11(4):233-240. [DOI] [PubMed] [Google Scholar]
- 39.Atkinson P. Rhetoric as skill in a medical settingIn: Bloor N, Taraborrelli P, editors. Qualitative studies in health medicine. Aldershot: Avebury; 1994.
- 40.Atkinson P. Medical talk and medical work: the liturgy of the clinicLondon: Sage; 1995.
- 41.Hunter K. Doctor’s stories: the narrative structure of medical knowledgePrinceton: Princeton University Press; 1991.
- 42.Fox KM, Reuland M, Hawkes WG, Hebel JR, Hudson J, Zimmerman SI, Kenzora J, Magaziner J. Accuracy of medical records in hip fracture J Am Geriatr Soc 1998;46(6):745-750. [DOI] [PubMed] [Google Scholar]
- 43.Katz DL, Mazhari R, Kalus R, Nawaz H. Preventable inpatient time: adequacy of electronic patient information systems Am J Pub Health 1999;89(12):1885-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein HD, Nadkarni P, Erdos J, Miller PL. Exploring the degree of concordance of coded and textual data in answering clinical queries from a clinical data repository Journal of the American Medical Informatics Association 2000;7(1):42-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz DL, Mazhari R, Kalus R, Nawaz H. Preventable inpatient time: adequacy of electronic patient information systems American Journal of Public Health 1999;89(12):1885-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandler G. Costs of unnecessary tests British Medical Journal 1979;2(6181):21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cawsey AJ, Webber BL, Jones RB. Natural language generation in health care Journal of the American Medical Informatics Association 1997;4(6):473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charlet J, Bachimont B, Brunei V, el Kassar S, Zweigenbaum P, Boisvieux JF. Hospitexte: towards a document-based hypertextual electronic medical record AMIA Symposium 1998:713-717. [PMC free article] [PubMed]
- 49.van Mulligen EM, Stam H, van Ginneken AM. Clinical data entry Proc AMIA Symp 1998:81-85. [PMC free article] [PubMed]
- 50.Kuhn K, Swobodnik W, Johannes RS, Zemmler T, Stange EF, Ditschuneit H, Classen M. The quality of gastroenterological reports based on free text dictation: an evaluation in endoscopy and ultrasonography Endoscopy 1991;23(5):262-264. [DOI] [PubMed] [Google Scholar]
- 51.Zweigenbaum P, Bouaud J, Bachimont B, Charlet J, Seroussi B, Boisvieux JF. From text to knowledge: a unifying document-centered view of analyzed medical language Meth Inform Med 1998;37(4–5):384-393. [PubMed] [Google Scholar]
- 52.Sleszynski SL, Glonek T, Kuchera WA. Standardized medical record: a new outpatient osteopathic SOAP note form: validation of a standardized office form against physician’s progress notes J Am Osteopath Assoc 1999;99(10):516-529. [DOI] [PubMed] [Google Scholar]
- 53.Aghili H, Mushlin RA, Williams RM, Rose JS. Progress notes model Proc AMIA Annu Fall Symp 1997:12-16. [PMC free article] [PubMed]
- 54.Van Ruiswyk J, Erdmann M. Improving the quality and ease of tracking invasive procedures Med Ed 1993;27(1):86-90. [DOI] [PubMed] [Google Scholar]
- 55.Metcalfe DH. J Med Engin Technol 1984;8(2):53-55. [DOI] [PubMed] [Google Scholar]
- 56.Ho LM, McGhee SM, Hedley AJ, Leong JC. The application of a computerized problem-oriented medical record system and its impact on patient care Int J Med Inform 1999;55(1):47-59. [DOI] [PubMed] [Google Scholar]
- 57.van Walraven C, Duke SM, Weinberg AL, Wells PS. Standardized or narrative discharge summariesWhich do family physicians prefer?. Can Fam Phys 1998;44:62-69. [PMC free article] [PubMed] [Google Scholar]
- 58.van Walraven C, Laupacis A, Seth R, Wells G. Dictated versus database-generated discharge summaries: a randomized clinical trial CMAJ 1999;160(3):319-326. [PMC free article] [PubMed] [Google Scholar]
- 59.Tange HJ, Schouten HC, Kester AD, Hasman A. The granularity of medical narratives and its effect on the speed and completeness of information retrieval Journal of the American Medical Informatics Association 1998;5(6):571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tange HJ, Hasman A, de Vries Robbe PF, Schouten HC. Medical narratives in electronic medical records IJMI 1997;46:7-29. [DOI] [PubMed] [Google Scholar]
- 61.McDonald CJ. The barriers to electronic medical records and how to overcome them JAMIA 1997;4:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilbert JA. Physician data entry: providing options is essential Health Data Management 1998;6:84-86. [PubMed] [Google Scholar]
- 63.van Mulligen EM, Stam H, van Ginneken AM. Clinical data entry Proc AMIA Symp 1998:81-85. [PMC free article] [PubMed]
- 64.Lovis C, Baud RH, Revillard C, Pult L, Borst F, Geissbuhler A. Paragraph-oriented Structure for Narratives in Medical DocumentationIn: Patel VL, Rogers R, Haux R, editors. MEDINFO; 2001. pp. 638-642. [PubMed]
- 65.de Estrada WD, Murphy S, Barnett GO. Puya: a method of attracting attention to relevant physical findings Proc AMIA Annu Fall Symp 1997:509-513. [PMC free article] [PubMed]
- 66.Suciu D. Proc Intern Conf Found Data Org, Kobe, Japan, November. Semistructured data and xml 1998.
- 67.Deutsch A, Fernandez M, Florescu D, Levy A, Maier D, Suciu D. Querying XML data Data Eng Bull 1999;22(3):10-18. [Google Scholar]
- 68.Dolin RH, Alschuler L, Behlen F, Biron PV, Boyer S, Essin D, Harding L, Lincoln T, Mattison JE, Rishel W, Sokolowski R, Spinosa J, Williams JP. HL7 document patient record architecture: an XML document architecture based on a shared information model Proc AMIA Annu Symp 1999:52-56. [PMC free article] [PubMed]
- 69.Sokolowski R, Dudeck J. XML and its impact on content and structure in electronic health care documents Proc AMIA Annu Symp 1999:147-151. [PMC free article] [PubMed]
- 70.Chueh HC, Raila WF, Berkowicz DA, Barnett GO. An XML portable chart format Proc AMIA Annu Symp 1998:730-734. [PMC free article] [PubMed]
- 71.Tessier C, Waegemann CP. The continuity of care recordOrganizations unite to create a portable, electronic record for use at transfer points in patients’ care. Healthc Inform 2003;20(10):54-56. [PubMed] [Google Scholar]
- 72.Altmann U, Tafazzoli AG, Noelle G, Huybrechts T, Schweiger R, Wachter W, Dudeck JW. Combining dictionary techniques with extensible markup language (XML)—requirements to a new approach towards flexible and standardized documentation Proc AMIA Annu Symp 1999:12-16. [PMC free article] [PubMed]
- 73.Zweigenbaum P, Bouaud J, Bachimont B, Charlet J, Seroussi B, Boisvieux JF. From text to knowledge: a unifying document-centered view of analyzed medical language Methods Inform Med 1998;37(4–5):384-393. [PubMed] [Google Scholar]
- 74.Shapiro JS, Bakken S, Hyun S, Melton GB, Schlegel C, Johnson SB. Document ontology: supporting narrative documents in electronic health records Proc AMIA Annu Symp 2005:684-688. [PMC free article] [PubMed]
- 75.Hripcsak G, Cimino JJ, Sengupta S. WebCIS: large scale deployment of a Web-based clinical information system Proc AMIA Annu Symp 1999:804-808. [PMC free article] [PubMed]
- 76.Haas J, Bright TJ, Bakken S, Stetson P, Johnson SB. Clinician perceptions of the usability of eNote Proc AMIA Symp 2005:973. [PMC free article] [PubMed]
- 77.Bright TJ, Bakken S, Johnson SB. Heuristic evaluation of eNote: an electronic notes system AMIA Annu Symp Proc 2006:864. [PMC free article] [PubMed]
- 78.Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shabo Shvo A. HL7 Clinical Document Architecture, Release 2 J Am Med Inform Assoc 2006;13(1):30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, Forrey A, Mercer K, DeMoor G, Hook J, Williams W, Case J, Maloney P. LOINC, a universal standard for identifying laboratory observations: a 5-year update Clin Chem 2003;49(4):624-633. [DOI] [PubMed] [Google Scholar]
- 80.Cimino JJ. From data to knowledge through concept-oriented terminologies: experience with the Medical Entities Dictionary J Am Med Inform Assoc 2000;7(3):288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Web Ontology Language (OWL). Available at: www.w3.org/TR/owl-features/. Accessed.
- 82.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing J Am Med Inform Assoc 2004;11(5):392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Software AG. Available at: www.softwareag.com. Accessed.
- 84.Committee on Quality of Health Care in America Crossing the Quality Chasm: A New Health System for the 21st CenturyWashington, D.C: Institute of Medicine, National Academy of Sciences; 2001. 164.