Abstract
This article describes our system entry for the 2006 I2B2 contest “Challenges in Natural Language Processing for Clinical Data” for the task of identifying the smoking status of patients. Our system makes the simplifying assumption that patient-level smoking status determination can be achieved by accurately classifying individual sentences from a patient’s record. We created our system with reusable text analysis components built on the Unstructured Information Management Architecture and Weka. This reuse of code minimized the development effort related specifically to our smoking status classifier. We report precision, recall, F-score, and 95% exact confidence intervals for each metric. Recasting the classification task for the sentence level and reusing code from other text analysis projects allowed us to quickly build a classification system that performs with a system F-score of 92.64 based on held-out data tests and of 85.57 on the formal evaluation data. Our general medical natural language engine is easily adaptable to a real-world medical informatics application. Some of the limitations as applied to the use-case are negation detection and temporal resolution.
Introduction
Within the Informatics for Integrating Biology and the Bedside (I2B2) initiative (see https://www.i2b2.org/), the First Shared Task on Natural Language Challenges for Clinical Data was organized. 1 Sharing data in the clinical domain is highly restricted to protect patient confidentiality. Hence, it is difficult to produce comparable results, evaluate techniques, and share platforms. Our system tries to address these issues by using an open-source framework, IBM’s Unstructured Information Management Architecture (UIMA) (see http://uima-framework.sourceforge.net/), and text analytics components previously developed by the Mayo Clinic Natural Language Processing (NLP) group. Thus, we show that it is possible to build a shareable system with a modest amount of effort by addressing the I2B2 Natural Language challenge for the identification of the patient smoking status from clinical records.
The goal of text classification is to label a document with a predefined set of categories. Usually the problem is approached as supervised learning where classifiers are learned from examples in an automated way. A fairly recent development in the machine learning world has been the advent of support vector machines (SVMs). 2,3 Joachims 4 explores the use of SVMs for learning text classifiers. He shows that SVMs “acknowledge the particular properties of text: (a) high dimensional feature spaces, (b) few irrelevant features, and (c) sparse instance vectors”. Chen et al. 5 apply SVMs to document classification of biological literature. Brank et al. 6 study the interaction of feature ranking and selection with the learning algorithm, in particular feature selection through linear SVMs, which then are used to train the SVM classifier.
Our objective for this study is to build a high-performance classifier for patient smoking status assignment. Because of theoretical and empirical evidence showing that SVMs are well-suited for text categorization, 4,5 our efforts focus on building such a classifier for the task.
Methods
The challenge of automated patient smoking status discovery was to accurately classify a patient with one of five categories: smoker, current smoker, past smoker, NONSMOKER and unknown based on the patients’ respective medical records. 1 We made the simplifying assumption that the documents could be categorized by accurately classifying the individual sentences within them followed by the final document level assignment based on a simple set of prioritized rules.
To build a sentence-level classifier, we manually identified every sentence that we judged related to the patient’s smoking status in each document and assigned that sentence the smoking status category assigned to the document. All other sentences were labeled as unknown.
Our system was built on IBM’s UIMA, which is a framework that facilitates the construction of reusable text analysis components (see Figure 2, available as a JAMIA on-line data supplement at www.jamia.org). Two UIMA-compliant components within the freely available Mayo Weka/UIMA Integration (MAWUI) project (see http://informatics.mayo.edu/text/) are of particular interest here as they provide a link between UIMA and the machine learning package Weka (see http://www.cs.waikato.ac.nz/ml/weka/). We used the WEKA SVM implementation for our classification task. One of the MAWUI components provides a way to generate Weka-compliant data files suitable for training classifiers from features created by UIMA annotators. The other MAWUI component provides a way to expose a Weka classifier to UIMA components for document classification.
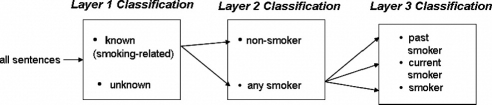
Our system has three layers of sentence classification (▶). Layer 1 classifies sentences as unknown or smoking-related (an umbrella category for nonsmoker, past smoker, current smoker, and smoker). All sentences labeled smoking-related are passed on to Layer 2. Layer 2 applies a negation detection algorithm to find the nonsmoker category. Sentences not marked as nonsmoker by Layer 2 are passed to Layer 3. Layer 3 assigns current smoker, past smoker, and the generic smoker categories by performing temporal resolution.
Figure 1.
High-level architecture for the sentence classifiers.
After each sentence in a document is classified, we apply precedence rules to assign the document level smoking status. The category current smoker is given the highest status, followed by past smoker, smoker, nonsmoker, and unknown.
Layer 1: Classifying Unknown and Smoking-related Sentences
Feature Selection
A subcorpus containing all sentences that were labeled a category other than unknown was created from the sentence-level training data described above. All features for the Layer 1 classifier were drawn from this subcorpus. The features were normalized words that did not appear in a stopword list, e.g., a, the, on, in. For normalization, the National Library of Medicine’s Lexical Variant Generation library (LVG) was used (see http://SPECIALIST.nlm.nih.gov). Words that appeared only once in the subcorpus were removed from the set of features. Features were chosen only from the smoking-related subcorpus to reduce those unrelated to smoking. This assumes that sentences labeled unknown did not share useful features for classification, but simply lack the features that describe sentences labeled smoking-related. The feature selection decisions we made had a marked positive impact on the efficiency of the model building step over choosing features from the entire corpus (5,312 features if entire corpus were used versus 98 features extracted from the smoking-related subcorpus).
SVM Classifier
We built a linear SVM sentence classifier from the entire corpus with the features described above with an unordered bag-of-words representation using the Weka SVM implementation.
Layer 2: Classifying Nonsmoker Sentences
We customized NegEx, a negation detection algorithm developed by Chapman et al. 7 for the Layer 2 classification of the nonsmoker category. NegEx takes a sentence and an anchor word and determines if that anchor word has been negated according to a set of negation rules. The anchor words that we used were from a small dictionary we created that contained the top 10 features as ranked by the weights in the SVM model built for Layer 1 and included words like smoke, smoker, tobacco, and cigarette. If a sentence contained a word in this dictionary, then negation detection was applied to the matched word. If the word was determined to be negated, then we labeled the sentence with nonsmoker. All sentences that were not labeled nonsmoker were passed to the Layer 3 classifier.
Layer 3: Classifying Current Smoker, Past Smoker, and Smoker Sentences
Feature Selection
A subcorpus containing all sentences that were labeled current smoker or past smoker was created from the sentence-level training data described above. An initial set of features for the Layer 3 classifier included words that did not appear in our stopword list but did appear at least twice in the subcorpus. Normalization was not applied to the features in order to retain verb tense information, which is important for temporal resolution. These initial features were used to build a linear SVM model for Layer 3 classification. The features were then ranked by their weights. We retained only the features that were important for temporal resolution indicated by higher weights, e.g., verb tense indicators (for now the nonnormalized individual tensed words) and lexical items such as day, year, ago. We refer to this set of features as temporal resolution features.
SVM Classifier
The corpus comprising of sentences indicating past smoker, current smoker, and smoker was represented as vectors using the temporal resolution features. We built a linear SVM classifier from that corpus using Weka. The Layer 3 classifier labels each sentence into current smoker, past smoker, and smoker.
Final Resolution: Discovering Smoking Status at the Document Level
After each sentence in a document was classified into one of the unknown, past smoker, current smoker, smoker, or nonsmoker categories, we applied additional logic to assign the final document-level smoking status. Current smoker has the highest precedence, followed by past smoker, smoker, nonsmoker, and unknown (see Final resolution: Discovering smoking status at the document level available as a JAMIA on-line data supplement at www.jamia.org).
Data Sets and Evaluation
The I2B2 challenge organizers released three data sets (▶). 1 Set 1 and Set 2 were made available three months before the formal competitive evaluation. Set 3 was used during the formal competition.
Table 1.
Table 1 I2B2 Data Sets by Category (in Number of Documents)
| Set | Past Smoker | Current Smoker | Smoker | Nonsmoker | Unknown | Total |
|---|---|---|---|---|---|---|
| Set 1 | 25 | 23 | 7 | 45 | 164 | 264 |
| Set 2 | 11 | 14 | 2 | 21 | 88 | 136 |
| Set 3 | 10 | 11 | 4 | 16 | 63 | 104 |
During the three-month period before the formal evaluation, we used Set 1 as a development and training set and Set 2 as our test set. After we determined the best configuration parameters on the training/development set, we built our models from Set 1. Testing was performed on Set 2. This is our informal evaluation set up for which we report results.
For the final competitive evaluation, we trained our models on the data from Set 1 and Set 2 with the best configurations as determined from our informal evaluation experimentation. Formal competition results were run on Set 3. We submitted three sets of results with our top performing models, which we describe in the next section.
We used precision, recall, and F-score as our evaluation metrics: Because our system makes assignments to every document that is processed, totalNumberOfDocumentsClassifiedByTheSystem and numberOfDocumentsInTestSet are the same. Hence, precision, recall, and F-score are the same. A simple baseline is to assign the most frequent category to each report (Unknown). The 95% confidence intervals for each metric are reported computed by the method of Clopper and Pearson. 8
Results and Discussion
▶ summarizes our results (see also Table 3 and Table 4 available as a JAMIA on-line data supplement at www.jamia.org). The best F-score from our informal evaluation is 92.64. The F-score baseline for this set is 63.31. We limited our runs on Set 2 to three to avoid overtraining/overfitting on the test data. The three runs differed slightly in the features used to build the Layer 3 model for classifying past smoker, current smoker, and smoker instances, and in the features for negation detection used in the Layer 2 model for discovering nonsmoker instances. However, the F-scores turned out similar.
Table 2.
Table 2 Best Results. Informal evaluation set up: training on Set 1; testing on Set 2.Formal evaluation set up: training on Set 1 and Set 2; testing on Set 3. (Numbers in brackets are 95% exact confidence intervals)
| Training Set | Test Set | No. Documents Correctly Classified by System | Total No. Documents Classified by System | No. Documents in Test Set | Precision | Recall | F-score | Baseline |
|---|---|---|---|---|---|---|---|---|
| Set 1 | Set 2 | 126 | 136 | 136 | 92.64 (86.89-96.42) | 92.64 (86.89-96.42) | 92.64 (86.89-96.42) | 63.31 (56.05-72.70) |
| Set 1 and Set 2 | Set 3 | 89 | 104 | 104 | 85.57 (77.33-91.70) | 85.57 (77.33-91.70) | 85.57 (77.33-91.70) | 60.58 (50.51-70.02) |
For the formal evaluation and the final i2b2 submission, our models were built from Set 1 and Set 2 and were run on the official I2B2 test set (Set 3). We submitted three sets of results run with models that differed slightly as described in the preceding paragraph. Our best F-score is 85.57 for this final formal evaluation (most frequent category baseline is 60.58). If we remove the unknown category and consider 2 categories (current smoker and noncurrent, which includes smoker, past, and nonsmoker), precision, recall and F-score for current are 53.33, 72.72, and 61.53 respectively; and for noncurrent are 88.46, 76.66, and 82.14 respectively.
Our error analysis uncovered several areas for improvement. Currently, our negation detection does not account for nonnegated lexical items indicating nonsmoker status, e.g., nonsmoker, nonsmoker. Also, phrases such as “nor does she smoke” are not flagged as negated.
Our temporal resolution component does not include an explicit one-year rule for distinguishing between past smoker and current smoker, but relies on the features and labeled data to learn the differences. The most challenging category for our system to classify is past smoker. Our system’s upper bound for this category is 78% when training and testing is performed on the same data. Potential enhancements are the inclusion metadata information as features, e.g., section headings, and experimenting with higher order SVMs, especially for temporal resolution.
We also noticed interesting cases such as the following report, which contained the sentence “He does drink alcohol three drinks per day, denies any current tobacco use.” The final classification as provided by the challenge organizers is unknown despite the fact that based on the above sentence one would be tempted to assign the nonsmoker label. Assigning smoking status by human experts might include information over the entire report and involve some inference based on the facts, medical and otherwise, as present in the entire record, which requires processing beyond sentence classification.
Conclusion
In this article, we described our system for identifying patient smoking status as part of the First Shared Task on Natural Language Challenges for Clinical Data. We reduce the problem of document classification to a sentence classification task to discover the relevant smoking information from which the final document level assignment is derived. The system uses a series of components developed by the Mayo Clinic NLP group within IBM’s UIMA. The total effort invested in this project was approximately 160 hours, which includes manual sentence-level annotation, code development of the project’s UIMA components, model building, experimentation, and discussions.
Acknowledgments
The authors thank Lesa Rohde for manual annotations and the JAMIA reviewers for their critiques.
References
- 1.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records J Am Med Inform Assoc. 2008;15:xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortes C, Vapnik V. Support-vector networks Machine Learning 1995;10:273-297. [Google Scholar]
- 3.Witten IH, Frank E. Data Mining: Practical Machine-Learning Tools and Techniques with Java ImplementationsSan Francisco, CA: Morgan Kaufmann Publishers; 2000.
- 4.Joachims T. Text categorization with support vector machines: learning with many relevant features Machine Learning: ECML-98. 10th European Conference on Machine Learning. Heidelberg, Germany: Springer-Verlag; 1998. pp. 137-142.
- 5.Chen D, Muller H-M, Sternberg PW. Automatic document classification of biological literature BMC Bioinformatics 2006;7:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brank J, Grobelnik M, Milic-Frayling N, Mladenic D. Feature selection using linear support vector machines. Microsoft Research Technical report MSR-TR-2002-63. Available at http://research.microsoft.com/research/pubs/view.aspx?id=580&type=Technical+Report&0sr=a. Accessed October 25, 2007.
- 7.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries J Biomed Inform 2001;34:301-310. [DOI] [PubMed] [Google Scholar]
- 8.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial Biometrica 1934;26:404-413. [Google Scholar]