Abstract
We describe the architecture of LifeCode® (A-Life Medical, Inc.), a natural language processing system for free-text clinical information extraction, our methodology in applying LifeCode® to the i2b2 smoking challenge, and statistical measures for performance evaluation. Due to the limited test size and the coefficient of variation in the test standard, it is difficult to draw conclusions regarding the relative efficacy of approaches that were applied to this challenge.
Introduction
LifeCode® is a natural language processing (NLP) and medical coding expert system that extracts information from free-text clinical records. First commercially available in 1998, LifeCode® processes documents for radiology, pathology, and emergency medicine.
In this paper, we describe the basic architecture of the LifeCode® system and the methodology of applying CM-Extractor, a LifeCode® engine designed for quality measure abstraction, to the i2b2 First Shared Task for Challenges in NLP for Clinical Data smoking challenge. We discuss issues with sample size and coefficient of variation which make it difficult to draw conclusions from comparisons between NLP systems. The full version of this article is available as a JAMIA on-line data supplement at www.jamia.org.
LifeCode® is a predominantly symbolic natural language processing system that relies on morphological, syntactic, semantic, and pragmatic analysis to extract and synthesize concepts from free-text medical documents. 1
The NLP extraction engine and medical expert system modules are driven by domain specific knowledge bases (KBs). The KBs contain core medical vocabulary concepts, domain specific concepts such as ICD-9-CM diagnoses, and concept logic.
Due to its commercial use, LifeCode® has been optimized for speed. LifeCode® references the KBs an average of 50,000 times per sentence. A table storing partial results during vector analysis allows these calculations to be performed in typically less than one second per page of text.
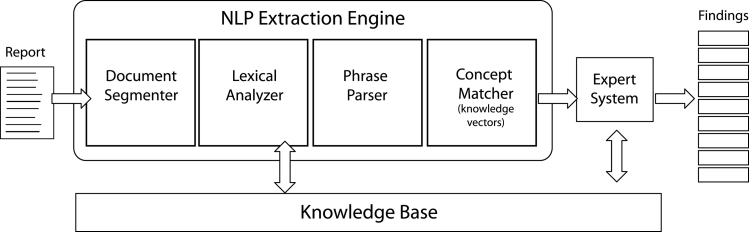
The NLP module of the system is comprised of four components (▶): document segmenter, lexical analyzer, phrase parser, and concept matcher. The document segmenter delimits and categorizes the content of medical notes based on section headings. This process associates information from a note to its contextual origin. The lexical analyzers are a series of processors designed to transform the text into a string of symbols consistent with the KB vocabulary. Functionality includes acronym expansion, morphological reduction and a variety of specialized parsers for additional analysis. The phrase parser employs bottom-up syntactic analysis to chunk the input into phrases. This parsing is tolerant of incorrect grammar and unknown words. Text chunks range from two words to complete sentences, corresponding roughly to the granularity of KB concepts. The concept matcher uses vector analysis to assign meanings (KB concept labels) to each phrase. A second evaluation uses anatomy, medication, and microbiology concept hierarchies and synonyms to improve matches, and syntactic heuristics to join and redistribute words from consecutive phrases and compute the meaning for the combined phrase.
Figure 1.
LifeCode® Architecture. The NLP Extraction Engine breaks reports up into concept-level phrases and matches them to known concepts using knowledge vectors.
The medical expert system module applies general and specialty-specific logic to refine the set of concepts identified by the engine. This logic is used to resolve ambiguous concepts, eliminate redundant information, combine concepts, and implement application specific business rules. Logic rules are developed, in consultation with medical coding experts, through analysis of medical documents and published guidelines.
Methods
For the i2b2 Challenge, we selected A-Life Medical’s (ALM’s) CM-Extractor, a second-generation NLP abstraction system designed for use with JCAHO Core Measures in a multiple document inpatient record application. 2,3 This system includes a module for detecting smoking status, which we adjusted for purposes of the challenge. This involved adjusting the time frames used in categorization, adjusting to single document cases, adding new sections to be scanned, and adding new phrases encountered in the i2b2 data.
In our initial test, our engine matched the i2b2 score in 355 of 399 (89%) training documents. Through an iterative process we revised the KB to better match the i2b2 training data and infer smoking status, as necessary.
There were two problems involving document sections. First, because “history of” statements are not codeable from a diagnosis section in inpatient visits, our engine ignores such statements. Second, one of the training documents had no sections. Our system is set to route such documents to a human reviewer.
We disagreed with the classification of several training documents. We chose not to adapt our system to these examples, as we thought that might negatively impact our handling of other documents.
In our final run against the training data, our engine matched the i2b2 score in 371 of 399 documents (93%). We then halted development, and downloaded and ran the 104 i2b2 test documents. We were pleased with our system’s performance (absent an i2b2 scoring key), but did note a few mistakes due to novel formatting not present in the training data. While our system is linguistically robust, it does rely on the standard formatting and segmentation generally found on medical documents.
Observations
Analyzing the results of language processing tasks can be challenging in that there is often a subjective aspect to scoring. This is present in healthcare coding and abstracting, where variation between human “experts” is not uncommon. ALM has developed a set of statistical tools for the purposes of quality assurance and performance auditing. 4 After reporting the i2b2 results, we include examples that demonstrate the need for further analysis, briefly discuss the pertinent techniques from our production analysis tool suite, and provide the analytic results as derived from the raw test scores.
The i2b2 confusion matrix for ALM’s challenge performance, and the precision, recall, and F-measure scores (rounded to 4 decimal places) for all categories are reproduced in ▶ ▶ below.
Table 1.
Table 1 A-Life Medical (Reported Matrix)
| U | N | P | S | C | i2b2 |
|---|---|---|---|---|---|
| 63 | 0 | 0 | 0 | 0 | U |
| 1 | 15 | 0 | 0 | 0 | N |
| 1 | 2 | 4 | 1 | 3 | P |
| 0 | 2 | 0 | 0 | 1 | S |
| 0 | 1 | 3 | 0 | 7 | C |
Table 2.
Table 2 A-Life Medical (Reported Scoring)
| Precision | Recall | F-Meas | |
|---|---|---|---|
| Unknown | 0.9690 | 1.0000 | 0.9840 |
| Non-Smoker | 0.7500 | 0.9375 | 0.8333 |
| Past-Smoker | 0.5714 | 0.3636 | 0.4444 |
| Smoker | 0.0000 | 0.0000 | 0.0000 |
| Current-Smoker | 0.6364 | 0.6364 | 0.6364 |
The weighted F-measure for our system was 0.8388. The mean across all participants in the i2b2 challenge was 0.7957.
Overall we agreed with the i2b2 scoring of the documents. Systematic issues we recognized in our review included cases in which our engine failed to recognize novel formatting, and cases in which our engine’s production design prevented information extraction, such as the “history of” issue with diagnosis sections.
We also found five cases which we believe were miscategorized. While we recognize that human coders may disagree on the coding of a document, 4 we felt that the coding for these five cases was clear. Three examples are detailed below:
• “SOCIAL HISTORY: Widowed since 1972, no tobacco, no alcohol, lives alone.” (ALM “non-smoker”, i2b2 “past-smoker”).
• Social History: “No alcohol use and quit tobacco greater than 25 years ago with a 10-pack year smoking history.” (ALM “past-smoker”, i2b2 “current-smoker”).
• “He is a heavy smoker and drinks 2–3 shots per day at times.” (ALM “current-smoker”, i2b2 “past-smoker”).
Given the small size of the test set, rescoring using our judgments on these five documents provides a significant increase in our scores (▶). ▶
Table 3.
Table 3 A-Life Medical (Revised Matrix)
| U | N | P | S | C | i2b2 |
|---|---|---|---|---|---|
| 63 | 0 | 0 | 0 | 0 | U |
| 1 | 16 | 0 | 0 | 0 | N |
| 1 | 1 | 5 | 0 | 2 | P |
| 0 | 2 | 0 | 1 | 0 | S |
| 0 | 1 | 2 | 0 | 9 | C |
Table 4.
Table 4 A-Life Medical (Revised Scoring)
| Precision | Recall | F-Meas | |
|---|---|---|---|
| Unknown | 0.9692 | 1.0000 | 0.9844 |
| Non-Smoker | 0.8000 | 0.9412 | 0.8649 |
| Past-Smoker | 0.7143 | 0.5556 | 0.6250 |
| Smoker | 1.0000 | 0.3333 | 0.5000 |
| Current-Smoker | 0.8182 | 0.7500 | 0.7826 |
The weighted F-measure for our system given the revised document scoring was 0.8965.
Although we prefer these revised scores, they are neither more nor less indicative of relative system performance than the original scores. We discuss this point through consideration of two important measures:
• An estimated coefficient of variation (CV) based on observed variation between human “experts.”
• A confidence interval and precision level based on test size.
Because medical coding and abstracting are, to a degree, subjective and prone to error, we argue there is no straightforward ‘gold standard’ for evaluation. A measure of coefficient of variation (CV) should be used in evaluating test scores. 4 For medical coding/abstracting, the CV is a measure of the observed (or expected) variation in performance from one test to the next, or a measure of the variation between qualified coders/abstractors performing the same test. Observed CV on medical coding tasks can be very large when measured on production coders. When measured on qualified auditors, a CV of 0.05 (5%) is not unusual and a CV less than 0.03 (3%) is highly unlikely.
Based on a sample of only two (ALM’s judgments and i2b2’s judgments), combined with large-scale observations of production medical coding and auditing, we believe that a CV of 0.05 (5%) is minimal for the i2b2 smoking history challenge. The challenge organizers noted that on an individual basis the physicians who created the test standard only agreed 80% of the time. This variation seems to argue for measuring the test results on the basis of inter-rater agreement. 5
Appropriate confidence and precision levels are dependent in part on test set size. The US Department of Health & Human Services, Office of the Inspector General (OIG) provides the Rat-Stats 6 calculator for determination of minimum test set sizes for auditing purposes. Applying the Rat-Stats unrestricted sample calculator to the parameters of this challenge produces sample sizes as shown in ▶:
Table 5.
Table 5 Rat-Stats Sample Sizes
| Confidence Level | |||||
|---|---|---|---|---|---|
| Precision Level | 80% | 90% | 95% | 99% | |
| 1% | 4089 | 6718 | 9512 | 16317 | |
| 2% | 1025 | 1688 | 2395 | 4130 | |
| 5% | 164 | 270 | 384 | 663 | |
| 10% | 41 | 68 | 96 | 166 | |
| 15% | 18 | 30 | 43 | 74 | |
Making an approximate fit of the i2b2 smoking history 104 document test set to the Rat-Stats sample size yields only an approximate 10% precision level at 95% confidence. Alternatively, at a less than 80% confidence level, the precision level rises to 5%.
We next apply these CV and confidence-precision level measures in our analysis of the probability distributions of the lowest performing, highest performing, and ALM systems.
The following defines the parameters and formulas for selecting an unrestricted random sample fpc * n from a population (universe) of size N. Defect number x is recalculated to provide X which is the defect number modified to account for the expected subjectivity and error of the auditor according to the formula X = x − (CV * P * fpc * n). The rationale for this formula is that if the error level of the auditor is CV and the auditee is expected or observed to make proportion P errors, then the number of correct auditee codes that were incorrectly judged to be errors by the auditor is CV * P * fpc * n. This value is subtracted from the raw defect number x. The complete set of values and equations is as follows.
• CVis the expected or observed judgment subjectivity/error proportion of the auditor.
• CL is the desired confidence level as a percent where
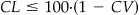 is preferred.
is preferred.• Z is the area under the tails of the distribution for the desired CL.
• H is the half width of the desired confidence interval where
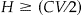 and
and 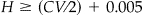 is preferred.
is preferred.• P is the expected or observed proportion of errors on the part of the person/process under test.
• N is the size of the population (universe) of documents to be sampled.
• n is the unadjusted sample size where
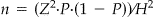 .
.• fpc is the finite population correction factor where
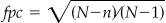 .
.•
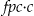 is the finite population adjusted sample size.
is the finite population adjusted sample size.• x is the observed defect/error number.
• X is the defect/error number adjusted for the auditor error rate where
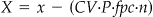 .
.
Note that the desired confidence interval should be greater than CV, i.e., no matter how large the sample, we cannot be more confident of our test results than we are of our scorer. Increasing the sample size, which is the practical effect of decreasing H, will not truly improve precision once H ≤ CV/2. H ≥ (CV/2) + 0.005 is recommended. We also recommend CL ≤ 100 * (1 − CV) because, similar to H, we cannot expect to achieve a confidence level in the test that is greater than the maximum accuracy that the scorer can achieve.
Therefore, using an estimated CV of 0.05 and P of 0.2 (based on the mean F-measure of the challenge participants) and fpc * n of 104 (the actual sample size), then for all scores, X = x − (0.05 * 0.2 * 104). In other words, the raw number of errors (defects) should be reduced by approximately 1, for all participants. Also, the optimal sample size would be approximately 400 documents which would yield a confidence level of about 95% at about a 5% precision level.
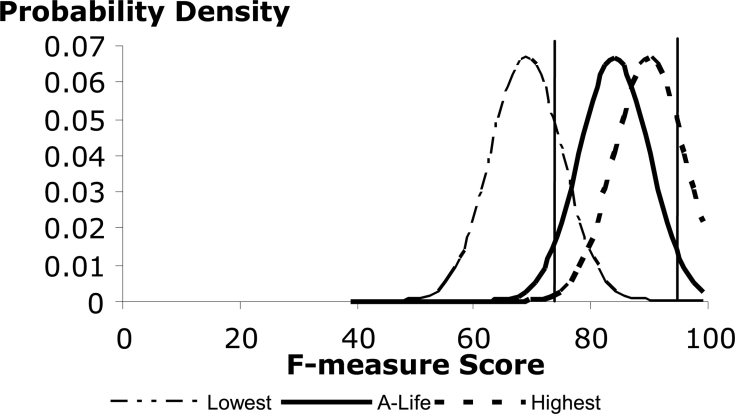
Based on this analysis (95% confidence level and 10% precision level) and the reported scores from the i2b2 smoking history challenge adjusted according to X, the probability distributions for the lowest score, ALM’s score and the best score would be approximately as shown in ▶. The 95% confidence interval for the ALM score is marked by vertical bars. Note that within these bars there is significant system overlap, particularly between ALM’s system and the best performing system, but also including the lowest performing system as well.
Figure 2.
Test Result Probability Distributions.
Although we do not have ready access to the full results from each participant, it appears that a judgment of statistical significance between the majority of the scores is not warranted given the test set size and the variation in judgment regarding the test standard.
Discussion
The development effort for this project, including document review and performance evaluation, totaled approximately 20 hours. Our engine performed with reasonable consistency across the training and test data, with a weighted F-measure of 0.8388 for the test data as originally scored and 0.8965 given ALM’s scoring.
A ’gold standard’ for this task is elusive because of ambiguity in the sources and evaluator disagreement. Due to important issues of patient privacy, it is also often difficult, as in this study, to obtain a sufficient number of documents to justify statistical significance. With the current sample size and CV, it is difficult to draw significant conclusions about the relative efficacy of the various approaches represented in this challenge. We do believe, however, that these types of evaluations provide for meaningful benchmarks and serve as a useful forum for the exchange of ideas and approaches.
References
- 1.Heinze D, Morsch M, Sheffer R, Jimmink M, Jennings M, Morris W, Morsch A. LifeCode: A Deployed Application for Automated Medical Coding AI Magazine 2001;22(2):76-88. [Google Scholar]
- 2.Morsch M, Vengco J, Sheffer R, Heinze D. CM-Extractor: An Application for Automating Medical Quality Measures Abstraction in a Hospital Setting Proc 18th Conf Innov Appl Artif Intell. 2006. July 16-20; Boston, Massachusetts.
- 3.Joint Commission on Accreditation of Healthcare Organizations Specification Manual for National Implementation of Hospital Core Measures Version 2.0Available at:http://www.jointcommission.org/PerformanceMeasurement/PerformanceMeasurement/Current+NHQM+Manual.htm 2006. Accessed Sept 18, 2006.
- 4.Heinze D, Feller P, McCorkle J, Morsch M. Computer Assisted Auditing for High Volume Medical Coding. AHIMA Workshop on Software Standards for Computer Assisted Coding. 2006. September.
- 5.Morris WC, Heinze DT, Warner Jr HR, Primack A, Morsch AE, Sheffer RE, et al. Assessing the Accuracy of an Automated Coding System in Emergency Medicine Proc AMIA Annu Symp 2000:595-599. [PMC free article] [PubMed]
- 6.Department of Health and Human Services—OIG Office of Audit Services Rat-Stats 2007 Companion ManualAvailable at:http://oig.hhs.gov/organization/OAS/ratstats/CompManual%202007.pdf 2000. Accessed March 02, 2007.