Abstract
Objective
Explore the automated acquisition of knowledge in biomedical and clinical documents using text mining and statistical techniques to identify disease-drug associations.
Design
Biomedical literature and clinical narratives from the patient record were mined to gather knowledge about disease-drug associations. Two NLP systems, BioMedLEE and MedLEE, were applied to Medline articles and discharge summaries, respectively. Disease and drug entities were identified using the NLP systems in addition to MeSH annotations for the Medline articles. Focusing on eight diseases, co-occurrence statistics were applied to compute and evaluate the strength of association between each disease and relevant drugs.
Results
Ranked lists of disease-drug pairs were generated and cutoffs calculated for identifying stronger associations among these pairs for further analysis. Differences and similarities between the text sources (i.e., biomedical literature and patient record) and annotations (i.e., MeSH and NLP-extracted UMLS concepts) with regards to disease-drug knowledge were observed.
Conclusion
This paper presents a method for acquiring disease-specific knowledge and a feasibility study of the method. The method is based on applying a combination of NLP and statistical techniques to both biomedical and clinical documents. The approach enabled extraction of knowledge about the drugs clinicians are using for patients with specific diseases based on the patient record, while it is also acquired knowledge of drugs frequently involved in controlled trials for those same diseases. In comparing the disease-drug associations, we found the results to be appropriate: the two text sources contained consistent as well as complementary knowledge, and manual review of the top five disease-drug associations by a medical expert supported their correctness across the diseases.
Introduction
Clinical knowledge is constantly evolving as new discoveries are made and practices change. This knowledge is valuable but often buried in text within a range of sources such as journal articles and clinical narratives in the patient record. Through automated approaches, information associated with diseases can be extracted and integrated from these disparate text sources for understanding the various characteristics of diseases (e.g., treatment or symptoms 1–3 ) and how they may change over time. The availability of up-to-date disease profiles may be valuable for a variety of applications including decision support (e.g., recommending treatments), quality assurance (e.g., inter- and intra-institutional review), clinical information needs (e.g., answering clinical questions), information retrieval (e.g., classifying documents), and data mining (e.g., hypothesis discovery).
Literature on randomized controlled trials (RCTs) reports on the results of testing one or more treatments such as drugs, devices, or procedures that are studied for diagnostic, therapeutic, or prophylactic effectiveness. 4 Patient records reflect practices within an institution and provide patient-specific information such as past and present diseases or medications. Given these contrasting roles, these text sources may offer valuable complementary disease-specific knowledge. Natural language processing (NLP) has been shown to facilitate the tasks of extracting information and relations between information captured within text. 5–7 While Medline articles are indexed by manually-assigned MeSH headings to list some important biomedical concepts in the articles, techniques such as NLP could automatically extract biomedical concepts as well as their relations in a high throughput manner.
Towards the goal of acquiring, integrating, and managing disease-specific knowledge from disparate sources, we applied a text mining approach for the automated acquisition of disease-drug associations in Medline articles for RCTs and discharge summaries in the electronic medical record at NewYork-Presbyterian Hospital (NYP). This paper describes the annotation of these text sources by MeSH and two NLP systems (BioMedLEE 7,8 and MedLEE 5,6,9 ) to extract disease and drug entities, and the use of statistical techniques to identify strong disease-drug associations for a subset of eight diseases. While a number of studies have explored the use of text mining methods either for the literature or patient record, to the best of our knowledge, this study presents a first attempt to acquire and compare disease-specific associations from both.
In this paper, we first provide some background on the NLP systems used in this study and related work on knowledge acquisition from biomedical and clinical text. Next, we describe the approach involving NLP and statistical techniques to identify disease-drug associations from RCTs and discharge summaries. We then present a subset of associations derived for each of the eight study diseases and highlight the commonalities and differences. Finally, we end with a discussion about issues encountered and potential solutions, implications of this work, and future directions.
Background
The acquisition and management of disease-drug knowledge is challenging due the continuous growth of clinical knowledge, range of sources offering such knowledge (e.g., human experts, literature, or patient record), and the variety of manual or automated methods that can be applied. Knowledge acquisition techniques using text mining have been adapted and applied for numerous studies in the biomedical domain. 10–14 As part of the text mining process, NLP has come to play an increasing role for knowledge acquisition through its ability to automatically extract entities and relations within documents in a high throughput manner.
Natural Language Processing
Through Natural Language Processing (NLP) techniques, information can be extracted from text for applications including patient management, decision support, quality assurance, and clinical research. To support these various applications, NLP systems have emerged to identify, extract, and encode information within biomedical literature and clinical narratives. These systems include MPLUS, 15 MEDSYNDIKATE, 16 MetaMap 17 and SemRep, 18 and MedLEE and BioMedLEE.
MedLEE (Medical Language Extraction and Encoding) is a natural language processing system at NYP that has been used to extract and encode information in clinical narratives for a number of applications and studies. 5,6,9 BioMedLEE (BioMedical Language Extraction and Encoding) is an adaptation of MedLEE focused on extracting and structuring biomedical entities and relations in biomedical literature, 7,8 including phenotypic and genotypic information. Both NLP systems produce a set of primary findings (e.g., problem, procedure, device, and medication) along with associated modifiers (e.g., certainty, change, body location, and frequency) for a given document (e.g., discharge summary or Medline article). An additional feature of BioMedLEE is the ability to capture relations between findings (e.g., entity1 treats entity2 or entity1 complicated by entity2). In this work, we did not look at the relations and instead explored statistical co-occurrence methods as an initial study.
The output from MedLEE and BioMedLEE is based on frames in the form Type-Value-Modifiers where Type and Value refer to the primary finding followed by Modifiers, which are also frames following the same format, thereby allowing for nesting of modifiers and representing highly specific information. For example, in the sentence “His past medical history is significant for asthma” from a discharge summary, MedLEE extracts asthma as a primary finding with type problem where modifiers include certainty and status with values high certainty and past history, respectively (▶). Codes may be available for the primary finding as well as certain modifiers and are represented as additional modifiers called code. In this work, MedLEE primarily assigns UMLS 19,20 codes to findings (e.g., C0004096 corresponding to asthma) while BioMedLEE assigns codes from the UMLS as well as other sources including the Mammalian Phenotype Ontology 21 and Cell Ontology. 22
Figure 1.
Example NLP XML Output. Simplified MedLEE XML output for asthma in a sentence from a discharge summary.
Knowledge Acquisition from Biomedical and Clinical Text
Several groups have focused on the development of text mining approaches for identifying specific types of co-occurring concepts, particularly concept pairs such as disease-drug, based on MeSH headings and subheadings in Medline. 23–28 Many of these studies involved use of knowledge sources such as the UMLS Metathesaurus (Meta) 19,20 and comparison with established clinical knowledge sources. Srinivasan et al. have focused on the creation of MeSH-based profiles for purposes such as generating hypotheses and providing information useful to health care providers and researchers. 2,29–34 Other studies have used concept-based approaches involving NLP and the Meta. Rindflesch et al. have developed linguistically-based systems such as SemRep and SemGen for identifying relationships between entities extracted by MetaMap (e.g., gene-drug, disease-gene, and disease-drug). 18,35 In a recent study, Hristovski et al. explored the use of SemRep and BioMedLEE for integrating semantic relation extraction with co-occurrence based literature-based discovery systems. 36 In another study by Duda et al., both MeSH and a concept-oriented approach were explored to assist with the classification of drug-drug interaction articles in Medline. 37 Other studies have looked at using knowledge within the UMLS Knowledge Sources such as the MRCOC table that contains the frequency of co-occurring concepts for sources including Medline. 25,38
NLP and text mining have also been applied to clinical documents for a range of applications including detecting clinical conditions and medical errors, coding and billing, tracking physician performance and resource utilization, improving provider communication, and monitoring alternate courses of treatment. 39–43 Heinze et al. have discussed the use of NLP to mine free-text medical records for the creation of disease profiles based on demographic information, primary diseases, and other clinical variables. 44 To validate inferences produced by SemRep about drug treatments for diseases, Rindflesch et al. constructed a repository of drug-disorder co-occurrences based on a large collection of clinical notes from the Mayo Clinic using MetaMap. 1
Previous work has primarily focused on studying either the biomedical literature (particularly, Medline) or the patient record for identifying co-occurring concepts. In a recent survey, Collier et al. observed the advances in text mining applied to molecular biology and biomedical literature, and discussed the need for studies focused on clinical corpora and electronic patient records in order to bring together the domains. 45
Statistical Approaches to Acquiring Disease-specific Associations
In recent studies, Cao et al. used NLP and statistical methods to discover disease-finding associations in discharge summaries for the automated generation of medical problem lists. 3,46 Such associations are generally not explicitly stated in the patient record and the chi-square (χ2) statistic offers a measure of significance for association rules based on co-occurrence. 47 However, large sample sizes create problems in the simple χ2 analysis as the sheer volume of data is sufficient to make any hypothesis test significant thereby observing statistical significance versus scientific significance.
The discovery of association rules that reflect the relationships between data items is among the basic data mining tasks. The typical methods used to measure the quality of these rules are support and confidence; however these measures do not take into account both the presence and absence of items in sets. As a result, the χ2 statistic has been proposed as a measure of significance for correlation rules (a form of association rules). 47 Cao et al. proposed a statistical methodology to analyze co-occurrence data from a large sample. 3,46,48 In this work, the χ2 statistic is used to compute an adjustment of its p-value, the volume test adjustment, ɛ(χ2), that measures the distance between the 2×2 contingency table under study from the surface of independence. A graphical device (that adjusts for multiple comparisons) is then used to estimate the number of true null hypotheses. This graphical device, called the p-value plot, is used to calibrate the χ2 statistic by providing a heuristic cutoff point for the adjusted p-values that estimates the number of true associations. For a detailed description of the method see Cao et al. 3,46,48 Here we only note that it is a computationally easy and fast procedure to implement.
The present study is motivated by the work of Cao et al. that was focused on disease-finding associations in the patient record and seeks to build upon this work by applying the combination of NLP and statistical methods for acquiring disease-drug relationships in both the biomedical literature and patient record.
Study Design
Design
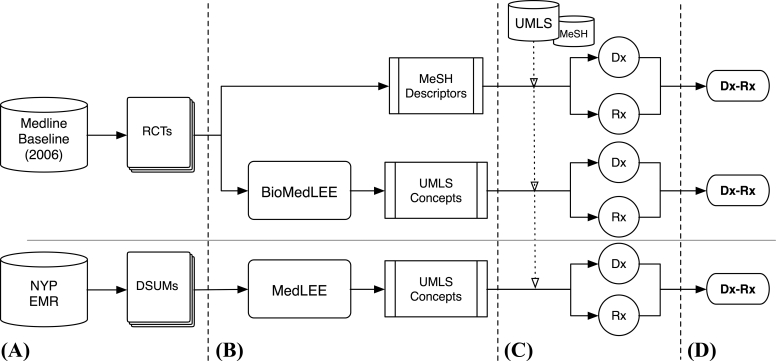
The approach we applied for identifying disease-drug associations from text sources involves four major phases (▶):
a Collect relevant documents from the biomedical literature and patient record (specifically, Medline RCT articles and discharge summaries in this study)
b Process the documents to extract entities
c Filter and normalize disease and drug entities using external knowledge sources
d Apply statistical methods to reveal associations between diseases and drugs
Figure 2.
Overview of Methods. (A) Documents are collected from the 2006 Medline baseline database (i.e., randomized controlled trials) and the patient record at NewYork-Presbyterian Hospital (i.e., 2003 and 2004 discharge summaries). (B) MeSH descriptors are obtained for the RCT articles and UMLS concepts are extracted by the NLP systems, BioMedLEE and MedLEE, for the RCT articles and discharge summaries, respectively. (C) Using external knowledge sources (i.e., MeSH Thesaurus and UMLS Knowledge Sources), disease and drug entities are filtered and normalized from the MeSH- and NLP-based annotations. (D) Co-occurrence statistics are applied to discover associations between the study diseases and drugs.
This approach was applied separately to the Medline RCTs annotated by MeSH, Medline RCTs annotated by BioMedLEE with UMLS concepts, and discharge summaries from 2003 and 2004 annotated by MedLEE with UMLS concepts (henceforth referred to as RCT/MeSH, RCT/UMLS, DSUM/UMLS 2003, and DSUM/UMLS 2004 where DSUM/UMLS will be used as shorthand to represent both years). Comparisons of the disease-drug associations were performed to provide insight on the similarities and differences between the text sources and annotation methods.
Scope
Our main objective for this study was to explore the acquisition of knowledge in biomedical and clinical documents using a combination of text mining and statistical techniques to identify disease-drug associations. The motivation for limiting the scope was to highlight the feasibility and bring to light challenges in applying this automated approach to disparate text sources for acquiring disease-specific knowledge. Additionally, the initial results provide insight on the characteristics of disease-drug knowledge captured within Medline RCT articles and discharge summaries.
At the start of the study, we performed several preliminary analyses of the text sources of interest (i.e., Medline articles and discharge summaries) and available annotations (i.e., MeSH and UMLS concepts extracted by the NLP systems). Several potential challenges in comparing disease-specific knowledge across RCT/MeSH, RCT/UMLS, and DSUM/UMLS became apparent at this time and we therefore limited the scope of this study as follows:
• Though these methods were developed to be generalizable across diseases and to other types of associations, we have initially focused on a subset of diseases and disease-drug associations. Based on results from a previous study that identified the 100 most frequent diseases described in discharge summaries at NYP, a medical expert in our group selected eight diseases representing a range of conditions and body systems for this study: acquired immunodeficiency syndrome, asthma, breast neoplasms, congestive heart failure, diabetes mellitus, Parkinson’s disease, pneumonia, and schizophrenia.
• MeSH annotations for the Medline literature include descriptors (also referred to as main headings), qualifiers, and supplementary concepts. While qualifiers further specify descriptors and supplementary concepts include chemical substances, we initially only used MeSH descriptors.
• MeSH generally includes broader classes whereas MedLEE and BioMedLEE detect more specific concepts. For example, the MeSH hierarchy for Asthma consists of ‘Asthma,’ ‘Asthma, Exercise-Induced,’ and ‘Status Asthmaticus’; the BioMedLEE and MedLEE UMLS codes that are generated include over 50 asthma-related concepts including mild asthma, acute asthma, and asthma childhood. While this indicates the need for disease classes in order to group relevant disease concepts, we initially use the broadest form of the disease by selecting the single highest-level MeSH descriptor to represent the eight diseases. The only exception was Diabetes Mellitus where we found that the more specific descriptors, ‘Diabetes Mellitus, Type 1’ and ‘Diabetes Mellitus, Type 2’, are often used to index RCT articles related to diabetes mellitus. ▶ presents the MeSH descriptor and unique identifier (for the descriptor and its entry terms) from 2006 MeSH for the set of diseases and the corresponding UMLS concepts from both the 2005AA version (used by BioMedLEE and MedLEE at the start of this study) and 2006AB version (available at the start of this study).
• Similar to the granularity issue with diseases, while the MeSH descriptors for RCT articles primarily include drug classes or generic names, UMLS concepts identified by MedLEE for the discharge summaries consist more of trade name (or brand name) drugs because these are frequently mentioned in the patient record. In order to resolve these differences, we leveraged drug knowledge within MeSH and the Meta to characterize the drug entities (e.g., class, generic name, or trade name). For this study, the focus was on mapping trade names to generic names (e.g., Retrovir is a brand name equivalent for the generic drug Zidovudine) in order to facilitate comparisons of generic drugs across RCT/MeSH, RCT/UMLS, and DSUM/UMLS.
• In order to perform direct comparisons between RCT/MeSH and RCT/UMLS, we used the same set of Medline RCT articles (although the BioMedLEE annotations may have identified additional documents containing the pertinent diseases).
Table 1.
Table 1 MeSH and UMLS Concepts for Study Diseases
| MeSH ID and Descriptor (2006) | UMLS CUI and Term (2005AA and 2006AB) |
|---|---|
| D000163 Acquired immunodeficiency syndrome | C0001175 Acquired Immunodeficiency Syndrome |
| D001249 Asthma | C0004096 Asthma |
| D001943 Breast neoplasms | C0006142 Malignant neoplasm of breast |
| C0006149 Breast Neoplasms (2005AA only) | |
| C1257930 Mammary Carcinoma, Human | |
| C1257931 Mammary Neoplasms, Human | |
| C1458155 Mammary Neoplasms (2006AB only) | |
| D003920 Diabetes Mellitus | C0011849 Diabetes Mellitus |
| D003922 Diabetes Mellitus, Type 1 | C0011854 Diabetes Mellitus, Insulin-Dependent |
| C0205734 Diabetes, Autoimmune | |
| D003924 Diabetes Mellitus, Type 2 | C0011860 Diabetes Mellitus, Non-Insulin-Dependent |
| D006333 Heart failure, congestive | C0018802 Congestive heart failure |
| D010300 Parkinson disease | C0030567 Parkinson Disease |
| D011014 Pneumonia | C0032285 Pneumonia |
| C0887898 Experimental Lung Inflammation | |
| D012559 Schizophrenia | C0036341 Schizophrenia |
Materials and Methods
Collecting Document Sets
Documents were collected from two major sources: Medline and the electronic medical record at NYP. For this study, we focused on randomized controlled trials (RCTs) and discharge summaries from the respective sources.
Medline
Medline is the United States National Library of Medicine’s biomedical bibliographic database containing citations dating back to the mid-1960s. 49 For this study, we used the 2006 Medline baseline files consisting of 15,433,668 articles. The PubMed Clinical Query for ‘therapy’ and ‘narrow, specific search’ was adapted to identify PubMed identifiers (PMID) for RCTs pertaining to drug therapy resulting in the following query (performed in June 2006): 50
“(drug therapy[sh]) AND (randomized controlled trial[Publication Type] OR (randomized[Title/Abstract] AND controlled[Title/Abstract] AND trial[Title/Abstract]))”
Only PMIDs retrieved by this query that were also in the 2006 Medline baseline were maintained resulting in a document set of 81,828 RCT articles. For each PMID, the following information was extracted from the baseline XML files: publication month and year, title, abstract, and MeSH annotations (in this study, we focused on the MeSH descriptors). 51
Patient Record
The clinical data repository at NYP maintains a wide range of clinical narratives for patient care and research including discharge summaries, operative reports, and reports from numerous ancillary services (e.g., radiology and pathology). 52 Patient-specific information, such as diseases or conditions and related medications, can be present within a single section or across sections; however, most importantly, the relationships between these types of information are generally not explicitly stated. For this study, we obtained IRB approval and used de-identified discharge summaries from the years 2003 and 2004 totaling 22,609 and 25,751 reports, respectively.
Extracting Entities from Documents
Two methods were applied for identifying entities (specifically those referring to diseases and drugs) in the RCT articles. First, all MeSH descriptors associated with each article were extracted. Second, BioMedLEE was applied for extracting and encoding both entities and relations within the title and abstract of each article (this study focuses on only the extracted entities). ▶ presents the MeSH descriptors and UMLS concepts assigned to an article with the respective methods. For the discharge summaries, MedLEE was used to extract and encode clinical information. Findings were extracted from the XML output of both NLP systems and filtered based on primary type (i.e., problem, finding, med, and substance), certainty modifier (e.g., values not indicating a negated finding such as no), and status modifier for MedLEE output (e.g., values not indicating a past finding like past history) leaving only present disease and drug findings.
Figure 3.
MeSH and NLP Annotations for RCT Article. (A) Excerpt from Medline citation that includes title and abstract (obtained from PubMed in January 2007). The source of the citation is: LeWitt PA, Ward CD, Larsen TA, Raphaelson MI, Newman RP, Foster N, Dambrosia JM, Calne DB. Comparison of pergolide and bromocriptine therapy in parkinsonism. Neurology. 1983 Aug; 33(8):1009-14. 56 (B) MeSH descriptors assigned to the article (with subheadings and ‘*’ indicating a major topic) identified as diseases, drugs, or other based on UMLS semantic type (specified in italics for other). (C) BioMedLEE UMLS concepts with problem, finding, or substance as the primary type differentiated as diseases, drugs, or other based on UMLS semantic type (specified in italics for others).
Filtering and Normalizing Extracted Entities
Identifying Diseases and Drugs
UMLS semantic types for each of the entities extracted in the previous phase were identified in order to select disease and drug entities. Disease entities were considered those with the following semantic types: Disease or Syndrome (T047), Mental or Behavioral Dysfunction (T048), or Neoplastic Process (T191); the semantic types Pharmacologic Substance (T121) or Antibiotic (T195) were used to identify drug entities.
Characterizing Drug Entities
Knowledge in MeSH and the Meta were used as an initial attempt to characterize each drug entity as a class, generic name, and trade name. While MeSH was primarily used to infer drug classes through its knowledge of pharmacologic actions, several sources in the Meta were used to map trade name drugs to their generic names. In the latter case, the tradename_of and has_tradename relationships in the 2006AB version were leveraged for RxNorm (RxNorm Vocabulary at the National Library of Medicine), 53 NCI (National Cancer Institute), and PDQ (Physician Data Query). An example mapping for the generic name Zidovudine would result in the trade name drugs, Combivir, Retrovir, and Trizivir. In cases where there is a one-to-many mapping between trade name and generic name, all mappings are used. For example, Sinemet is the combination of Carbidopa and Levodopa and therefore will map to both these generic name drugs. Where possible, all trade name drugs were mapped to their generic names leaving primarily drug classes and generic name drugs. In cases where a mapping could not be found through the techniques used (e.g., loviride), the drug entity was marked as unknown.
Generating Disease–Drug Associations
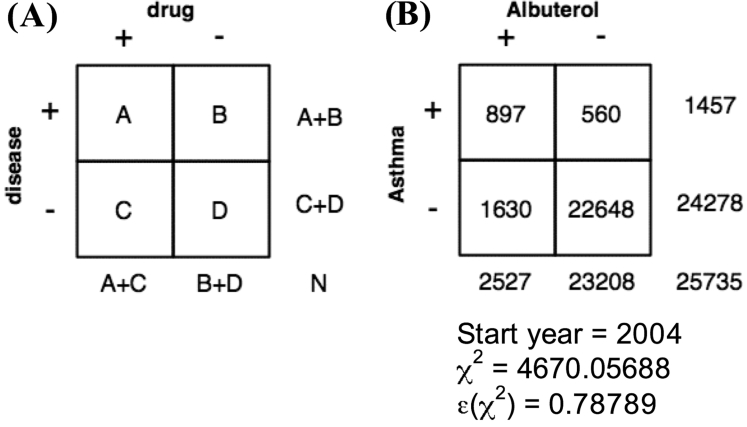
The definition from the study by Cao et al. for diseases and findings was adapted to disease-drug associations to produce the following characterization. 3 For a document set, a disease and drug are considered to co-occur if they appear in the same document (and both are considered present findings based on the status and certainty modifiers extracted by the NLP systems). Disease-drug pairs can be characterized by S = {start year, sample size, frequency of disease, frequency of drug, frequency of disease and drug co-occurrence}, where start year refers to the year the drug first appeared in the document set (for discharge summaries, this would either be 2003 or 2004) and sample size is the number of documents since the start year. Based on this definition, 2×2 tables for each disease-drug pair can be generated where the disease is either present or absent and the drug is similarly present or absent. ▶ presents a graphical representation of the 2×2 table definition and example table for asthma and albuterol for DSUM/UMLS. To test the hypothesis of no association between the disease and drug, the χ2 statistic and the adjustment to its p-value, ɛ(χ2) are used and appropriate cutoff identified. 3,46 A method for cutoff identification for “true” disease-drug associations is described by Cao et al.; 3,46 the present study uses a variation of this method where we fit, on sequentially defined ranges of the ɛ(χ2) values, a no-intercept regression model and stop the algorithm when the highest R2 value is observed.
Figure 4.
Disease-Drug 2×2 Table. (A) General disease-drug 2×2 contingency table where cells represent the number of documents in which the disease is either present or absent and the drug is similarly present or absent. (B) Table for asthma and albuterol generated from UMLS concepts extracted by MedLEE for discharge summaries from 2004 (DSUM/UMLS). In this table, A=897 represents the number of documents including both asthma (C0004096) and albuterol (C0001927), start year is the year the disease is first mentioned in the document set, and χ2 and ɛ(χ2) are calculated from the table and indicate the strength of the association between asthma and albuterol.
All relevant information including PMID, publication year, MeSH descriptors, and UMLS concepts was used to create the 2×2 tables, and co-occurrence statistics were applied to calculate the strength of association between the eight diseases under investigation (represented by the MeSH descriptors and UMLS concepts in ▶) and respective normalized drugs based on the document collection. The cutoffs for “true” disease-drug associations derived for RCT/MeSH, RCT/UMLS, and DSUM/UMLS were identified and corresponding associations compiled for further analysis.
Comparing Disease–Drug Associations
As part of a preliminary assessment, associations acquired from RCT/MeSH, RCT/UMLS, and DSUM/UMLS were compared to gain insight on the similarities and differences. Based on the cutoffs applied to the sets of associations, in addition to understanding overall agreement, several analyses can be performed to compare and contrast: MeSH and NLP-extracted UMLS annotations for the biomedical literature, findings in the biomedical literature and patient record, and associations for different time periods of the patient record.
One question of interest is whether the approaches are in agreement over all diseases examined in this study and the extent to which they agree in terms of the number of most associated drugs. To answer this question, we formulated the hypothesis of no agreement and computed Kendall’s coefficient of concordance. 54 Another question pertains to whether the proportion of common drugs that are identified by the various approaches is significantly different. In total, there are 4 different sources of 2×2 tables, thus the number of discrete pairs is 6 for: (RCT/MeSH, RCT/UMLS), (RCT/MeSH, DSUM/UMLS 2003), (RCT/MeSH, DSUM/UMLS 2004), (RCT/UMLS, DSUM/UMLS 2003), (RCT/UMLS, DSUM/UMLS 2004), and (DSUM/UMLS 2003, DSUM/UMLS 2004). A drug that is identified by both sources is called common. To compute the proportion of common drugs in each source, and thus to measure agreement between the sources, we count the number of common drugs in each source and divide these numbers by the total number of drugs identified by each source. For example, if the pair (RCT/MeSH, RCT/UMLS) produces lists of 22 and 20 drugs, respectively and the number of drugs common to both sources is 13, then the proportion of common drugs in RCT/MeSH is 13/22=0.59 and in RCT/UMLS is 13/20=0.65.
Results
Descriptive Statistics
The document collection used in this study included a total of 81,828 RCT articles from Medline and 48,360 discharge summaries from patient records at NYP. Within the set of RCT articles and discharge summaries, those pertaining to the eight diseases of interest were identified (based on the highest-level MeSH descriptor or UMLS concept). ▶ presents the total number of documents, number of 2×2 tables generated representing the disease-drug associations (excluding drug classes), and cutoffs representing the number of “true” associations considered for further analysis. ▶ highlights the 5 most associated generic name drugs for each disease as determined by RCT/MeSH, RCT/UMLS, and DSUM/UMLS based on sorting by ɛ(χ2) adjustment values in descending order.
Table 2.
Table 2 Distribution of Documents and Disease-Drug Associations
| Disease | Source/Annotation | Total Documents | Disease-Drug Associations | “True” Disease-Drug Associations |
|---|---|---|---|---|
| Acquired immunodeficiency syndrome | RCT/MeSH | 270 | 75 | 20 (22) |
| RCT/UMLS | 270 | 106 | 20 | |
| DSUM/UMLS 2003 | 685 | 724 | 35 | |
| DSUM/UMLS 2004 | 805 | 755 | 37 | |
| Asthma | RCT/MeSH | 3,349 | 215 | 13 (18) |
| RCT/UMLS | 3,349 | 425 | 29 | |
| DSUM/UMLS 2003 | 1,332 | 889 | 23 | |
| DSUM/UMLS 2004 | 1,457 | 956 | 18 | |
| Breast neoplasms | RCT/MeSH | 1,931 | 191 | 8 (8) |
| RCT/UMLS | 1,931 | 210 | 2 | |
| DSUM/UMLS 2003 | 350 | 610 | 4 | |
| DSUM/UMLS 2004 | 391 | 679 | 8 | |
| Congestive heart failure | RCT/MeSH | 1,521 | 246 | 10 (11) |
| RCT/UMLS | 1,521 | 433 | 16 | |
| DSUM/UMLS 2003 | 1,817 | 1,157 | 13 | |
| DSUM/UMLS 2004 | 1,916 | 1,212 | 22 | |
| Diabetes mellitus | RCT/MeSH | 2,202 | 172 | 26 (27) |
| RCT/UMLS | 2,202 | 241 | 47 | |
| DSUM/UMLS 2003 | 3,926 | 874 | 4 | |
| DSUM/UMLS 2004 | 4,407 | 894 | 7 | |
| Parkinson’s disease | RCT/MeSH | 494 | 80 | 10 (11) |
| RCT/UMLS | 494 | 135 | 5 | |
| DSUM/UMLS 2003 | 211 | 450 | 5 | |
| DSUM/UMLS 2004 | 275 | 525 | 11 | |
| Pneumonia | RCT/MeSH | 273 | 116 | 37 (40) |
| RCT/UMLS | 273 | 198 | 105 | |
| DSUM/UMLS 2003 | 1,610 | 962 | 31 | |
| DSUM/UMLS 2004 | 1,794 | 1,036 | 30 | |
| Schizophrenia | RCT/MeSH | 1,098 | 186 | 8 (10) |
| RCT/UMLS | 1,098 | 241 | 10 | |
| DSUM/UMLS 2003 | 213 | 479 | 23 | |
| DSUM/UMLS 2004 | 232 | 463 | 24 |
This table presents several statistics for the text sources and annotation methods with respect to the eight diseases under investigation. “Total Documents” represents the number of disease-specific documents, “Disease-Drug Associations” refers to the number of 2×2 tables generated for each disease and respective generic name drugs, and “True Disease-Drug Associations” are the number of associations above the identified cutoff used for comparison.
The same set of documents was used for RCT/MeSH and RCT/UMLS.
For comparison, entities in RCT/MeSH represented by MeSH identifiers were mapped to UMLS concepts. Due to one-to-many mappings, two numbers are presented in the “True Disease-Drug Associations” column – one before mapping and one after.
Table 3.
Table 3 Top 5 Disease–Drug Associations
| RCT/MeSH |
RCT/UMLS |
DSUM/UMLS 2004 |
||||||
|---|---|---|---|---|---|---|---|---|
| Disease | Drug | Start Year | ɛ(χ2) | Drug | Start Year | ɛ(χ2) | Drug | ɛ(χ2) |
| Acquired immunodeficiency syndrome | zidovudine | 1987 | 0.63153 | zidovudine | 1987 | 0.90564 | ritonavir | 0.74390 |
| didanosine | 1992 | 0.25971 | didanosine | 1992 | 0.53224 | lamivudine | 0.74145 | |
| zalcitabine | 1989 | 0.25482 | lamivudine | 1994 | 0.50146 | lopinavir | 0.64139 | |
| foscarnet | 1986 | 0.22152 | stavudine | 1993 | 0.47512 | lopinavir-ritonavir | 0.63180 | |
| pentamidine | 1976 | 0.19434 | ritonavir | 1995 | 0.42316 | zidovudine | 0.63027 | |
| Asthma | albuterol | 1972 | 0.80320 | albuterol | 1971 | 0.64084 | albuterol | 0.78789 |
| budesonide | 1980 | 0.50067 | salmeterol | 1988 | 0.47005 | montelukast | 0.55013 | |
| beclomethasone | 1973 | 0.48950 | methacholine | 1976 | 0.46152 | montelukast sodium | 0.54505 | |
| terbutaline | 1973 | 0.43036 | budesonide | 1980 | 0.43908 | ipratropium | 0.41305 | |
| methacholine chloride | 1983 | 0.42112 | montelukast | 1996 | 0.40051 | fluticasone | 0.40265 | |
| Breast neoplasms | tamoxifen | 1976 | 0.90979 | tamoxifen | 1973 | 0.71016 | tamoxifen | 0.40440 |
| cyclophosphamide | 1966 | 0.58549 | cyclophosphamide | 1966 | 0.36420 | anastrozole | 0.34439 | |
| fluorouracil | 1967 | 0.55386 | fluorouracil | 1969 | 0.30104 | letrozole | 0.30108 | |
| methotrexate | 1965 | 0.39540 | anastrozole | 1996 | 0.25993 | trastuzumab | 0.23170 | |
| epirubicin | 1984 | 0.38076 | toremifene | 1990 | 0.23456 | exemestane | 0.22265 | |
| Congestive heart failure | digoxin | 1971 | 0.31366 | digoxin | 1971 | 0.28106 | frusemide | 0.74348 |
| enalapril | 1982 | 0.26417 | frusemide | 1965 | 0.17439 | carvedilol | 0.50234 | |
| captopril | 1979 | 0.25010 | dobutamine | 1979 | 0.17096 | digoxin | 0.49833 | |
| furosemide | 1965 | 0.23694 | enalapril | 1982 | 0.16701 | lisinopril | 0.38617 | |
| milrinone | 1985 | 0.22112 | captopril | 1979 | 0.15419 | spironolactone | 0.32166 | |
| Diabetes mellitus | insulin | 1965 | 1.00000 | insulin | 1965 | 0.95470 | metformin | 0.69717 |
| glyburide | 1974 | 0.51144 | glyburide | 1970 | 0.44653 | insulin isophane | 0.63515 | |
| metformin | 1969 | 0.44713 | metformin | 1970 | 0.40722 | metformin hydrochloride | 0.49561 | |
| insulin, isophane | 1982 | 0.35043 | insulin, regular, human | 1982 | 0.35550 | glipizide | 0.47934 | |
| glucagon | 1970 | 0.31428 | rosiglitazone | 1999 | 0.34800 | glyburide | 0.43595 | |
| Parkinson’s disease | levodopa | 1971 | 1.00000 | levodopa | 1970 | 1.00000 | carbidopa | 1.00000 |
| carbidopa | 1975 | 0.62779 | carbidopa | 1973 | 0.58552 | levodopa | 1.00000 | |
| selegiline | 1980 | 0.53362 | entacapone | 1994 | 0.46421 | pramipexol | 0.36856 | |
| bromocriptine | 1976 | 0.42983 | selegiline | 1987 | 0.45679 | entacapone | 0.29354 | |
| benserazide | 1976 | 0.32870 | pramipexol | 1995 | 0.42478 | ropinirole | 0.27994 | |
| Pneumonia | cefamandole | 1978 | 0.19196 | ceftazidime | 1983 | 0.19061 | azithromycin | 0.61668 |
| cefoperazone | 1983 | 0.15708 | azithromycin | 1990 | 0.18618 | ceftriaxon | 0.48426 | |
| ceftazidime | 1983 | 0.14577 | telithromycin | 2000 | 0.16826 | ipratropium | 0.34349 | |
| erythromycin | 1966 | 0.14514 | ceftriaxon | 1981 | 0.16452 | albuterol | 0.34127 | |
| roxithromycin | 1990 | 0.13789 | imipenem | 1983 | 0.15372 | ipratropium bromide | 0.33408 | |
| Schizophrenia | haloperidol | 1970 | 0.88813 | haloperidol | 1970 | 0.75683 | benztropine | 0.32158 |
| risperidone | 1989 | 0.62936 | olanzapine | 1996 | 0.75337 | benzatropine methanesulfonate | 0.30715 | |
| clozapine | 1978 | 0.58340 | risperidone | 1992 | 0.65631 | olanzapine | 0.27719 | |
| fluphenazine | 1967 | 0.46367 | clozapine | 1978 | 0.59956 | risperidone | 0.24228 | |
| pirenzepine | 1980 | 0.37131 | quetiapine | 1996 | 0.37114 | fluphenazine hydrochloride | 0.23775 | |
This table presents the generic name drugs most associated with the diseases as identified in Medline RCT articles (annotated by MeSH and UMLS concepts with BioMedLEE) and discharge summaries (annotated by UMLS concepts with MedLEE). For RCT/MeSH and RCT/UMLS, the start year indicates the year the drug first appeared in the literature based on the respective annotations; for DSUM/UMLS, the start year is the year of discharge summaries (in this case, 2004).
Overall Agreement Between RCT/MeSH, RCT/UMLS, and DSUM/UMLS
The measurement of agreement over all diseases results in a value of 0.653 (p = 0.02), which indicates that the null hypothesis is rejected. Thus, RCT/MeSH, RCT/UMLS, and DSUM/UMLS agree in the number of drugs associated more closely with the different diseases.
The number of drugs identified by RCT/MeSH, RCT/UMLS, and DSUM/UMLS varies. For example, 75 drugs were identified by RCT/MeSH, 106 by RCT/UMLS, and 724 and 755 by DSUM/UMLS for the two years as mentioned together with acquired immunodeficiency syndrome. From those, 22 medications were identified as highly associated by RCT/MeSH, 20 by RCT/UMLS, 35 by DSUM/UMLS for 2003, and 37 by DSUM/UMLS for 2004. These lists were used to compute the proportions that appear in ▶. Given that the confidence intervals for the median differences in the proportions all include 0, the hypothesis of equality over all diseases cannot be rejected; however, there may be disease-dependent differences. In looking at the Spearman correlations 54 for each pair of approaches reported in ▶, a high degree of agreement is noticed between (RCT/MeSH, RCT/UMLS) and (DSUM/UMLS 2003, DSUM/UMLS 2004) as expected while the agreement between the other pairs is weaker.
Table 4.
Table 4 Pairwise Comparison and Agreement of Disease–Drug Associations
| Disease | (RCT/MeSH, RCT/UMLS) | (RCT/MeSH, DSUM/UMLS 2004) | (RCT/UMLS, DSUM/UMLS 2004) | (DSUM/UMLS 2003, DSUM/UMLS 2004) |
|---|---|---|---|---|
| Acquired immunodeficiency syndrome | (0.59, 0.65) | (0.45, 0.27) | (0.65, 0.35) | (0.94, 0.89) |
| Asthma | (0.72, 0.45) | (0.22, 0.22) | (0.28, 0.44) | (0.78, 1.00) |
| Breast neoplasms | (0.25, 1.00) | (0.25, 0.25) | (0.50, 0.12) | (1.00, 0.50) |
| Congestive heart failure | (0.66, 0.38) | (0.15, 0.57) | (0.01, 0.07) | (1.00, 0.57) |
| Diabetes mellitus | (0.72, 0.50) | (0.45, 0.23) | (0.43, 0.32) | (1.00, 0.60) |
| Parkinson’s disease | (0.27, 0.60) | (0.36, 0.36) | (1.00, 0.45) | (1.00, 0.45) |
| Pneumonia | (0.83, 0.31) | (0.20, 0.27) | (0.11, 0.40) | (0.74, 0.77) |
| Schizophrenia | (0.40, 0.40) | (0.60, 0.25) | (0.70, 0.29) | (0.78, 0.75) |
| Spearman correlation across diseases | ρ = 0.9345 | ρ = 0.2678 | ρ = 0.2619 | ρ = 0.9524 |
(a, b) = (proportion of common drugs in a, proportion of common drugs in b) where a and b = (RCT/MeSH, RCT/UMLS, DSUM/UMLS for 2003, or DSUM/UMLS for 2004); a common drug is one that is identified by both a and b for the disease.
In total, there are 4 different sources of 2×2 tables, giving 6 discrete pairs—e.g, (RCT/MeSH, RCT/UMLS) or (RCT/UMLS, DSUM/UMLS 2004). A drug that is identified by both sources is called common. To compute the proportion of common drugs in each source to measure agreement between the sources, the number of common drugs in each source is counted and divided by the total number of drugs identified by each source. This table presents 4 of the 6 pairs—for (RCT/MeSH, DSUM/UMLS 2003), ρ = 0.4226 and (RCT/UMLS, DSUM/UMLS 2003), ρ = 0.4226.
Characterizing Associations, Annotations, and Text Sources
With regard to characteristics of the data in the document collection, this study deals with a large data set. The smallest sample size corresponding to the RCTs equals 3,394 (i.e., articles since 2005, the latest start year identified for drugs in this study) and the largest sample size is 81,828 (i.e., all articles in the document set). Thus, p-values associated with χ2 are not informative and an adjustment, ɛ(χ2), can be used to address this. A problem of imbalance in 2×2 tables (created if the marginal row and column probabilities are away from 0.5) is usually met when one attempts to analyze tables generated from large samples. 3,46 The imbalance associated with the tables for five of the diseases (acquired immunodeficiency syndrome, asthma, breast neoplasms, congestive heart failure, and diabetes mellitus) is negligible. In contrast, a greater amount of imbalance was observed for the other three diseases: Parkinson’s disease, pneumonia, and schizophrenia. Certain degrees of imbalance can be addressed by modification of χ2 and use of this modified value to compute ɛ(χ2). Further analyses, however, are needed when the degree of imbalance is extreme.
Through individual ɛ(χ2) adjustment values, characteristics of certain disease-drug associations may be revealed. For some diseases, these values come closer to 1 indicating highly associated drugs (e.g., values for Parkinson’s disease and Carbidopa range from 0.59 to 1.00 in ▶). While for others, the values are relatively distant from 1 (e.g., values for pneumonia and Ceftazidime range from 0.15 to 0.19 in ▶). This finding could indicate that the former types of drugs are particular to a disease, while the latter are more general drugs and can be given for a variety of diseases (Ceftazidime is an antibiotic for treating infections and possibly used to treat commonly co-occurring conditions).
In reviewing specific drugs identified by RCT/MeSH and RCT/UMLS, the top drug was found to be the same across all diseases (e.g., levodopa for Parkinson’s disease). Examination of disease-drug associations from the RCT articles compared with those from discharge summaries reveals several interesting findings. DSUM/UMLS (for both 2003 and 2004) appears to include more recent drugs than the literature. In some cases, an association was found only in the literature or only in the patient record. For example, in the RCT articles, methacholine is found to be associated with asthma and benserazide with Parkinson’s disease. This former finding can be explained by the fact that methacholine is used to diagnose asthma (which is specified in some citations with the MeSH subheading diagnostic use) and the discharge summaries have a focus on stating drugs used to treat a disease. Alternatively, in looking at results for pneumonia, ipratropium and albuterol appear as associated drugs for pneumonia in the patient record, but not in the literature. These drugs are typically given for other lung diseases such as asthma, thus indicating the presence of other diseases or frequent co-morbidities within the discharge summaries leading to these potentially “false positives.” These findings support the complementary nature of the two text sources emphasizing the focus of RCT literature on testing of therapies over a long time span and the discharge summaries conveying current practice with respect to prescribing medications for certain conditions.
Discussion
Our overall goal was to explore the feasibility of the automated detection and validity of disease-specific knowledge from the biomedical literature and patient record. As an initial step, this study examines an approach involving NLP and statistical techniques for identifying disease-drug associations in Medline RCT articles and discharge summaries. The results presented demonstrate the potential value of using NLP to enhance existing annotations (e.g., MeSH for Medline) as well as the consistent and complementary nature of the biomedical literature and patient record. Physician review of the top five disease-drug associations also determined that they were appropriate.
Limitations and Future Work
Analysis of the disease-drug associations obtained in this study demonstrates that the text sources offer complementary knowledge and that the combination of NLP and statistical techniques could play a valuable role in extracting relevant information within these sources. Several issues were encountered and initial attempts were made to resolve those related to drug names, but further work is needed to thoroughly interpret the findings (e.g., studying all associations compared with those above the cutoff as done in this study), refine the techniques (e.g., exploring other methods for identifying the cutoff), learn other aspects of diseases (e.g., comorbidities or symptoms), handle the various challenges to integration (e.g., concepts at different levels of granularity), and explore other sources and techniques for extracting disease-specific knowledge (e.g., association rule mining). Subsequent studies could include using established clinical knowledge sources (e.g., Micromedex and UMLS), knowledge bases (e.g., QMR and PharmGKB), and NLP systems (e.g., MetaMap) for comparison and verification. Additional statistical techniques (e.g., sensitivity and specificity) may also be used to further assess the effectiveness of the text mining approach.
The comparison and integration of knowledge from disparate text sources such as the biomedical literature and patient record presents several challenges. Given these challenges, we limited the scope at the start of this study to address issues related to the varying levels of granularity among entities where MeSH provides more general terms and the NLP systems studied present more specific UMLS concepts. These limitations included using only the MeSH descriptor and certain relationships in the Meta for drug knowledge, which resulted in some of the inconsistent associations observed among the text sources and annotations. The results presented in this study should thus be viewed as a lower bound for agreement among the approaches and further work is needed to address the challenges and increase the power of the findings. The following sections summarize the limitations and potential next steps with respect to text sources, annotations, and disease and drug entities.
Exploring Text Sources
The document collection used in this study consisted of titles and abstracts for RCT articles focused on drug therapy and two years of discharge summaries. One key difference between the document sets was the use of all RCT articles in Medline (spanning 1960s-present), but discharge summaries for particular years. The rationale behind this selection was that Medline articles for particular diseases might be sparse within a single year. Other experiments would involve processing articles at specific time intervals (e.g., every 3 or 5 years) and also analyzing discharge summaries in these intervals or in aggregate (i.e., all discharge summaries available at NYP) to determine how different timeframes may affect the associations identified. Other criteria could be used for creating the document sets such as considering literature with particular characteristics (e.g., publication type or MeSH subheading) and reviewing other types of clinical narratives (e.g., radiology reports and cardiology reports) to understand their impact on acquiring disease-drug associations as well as other types of disease-specific knowledge.
Enhancing Annotations
The comparison of associations derived from MeSH and BioMedLEE revealed both similarities and differences indicating that NLP could play a valuable role in supporting and supplementing MeSH indexing. Based on analysis of some of the differences, possible solutions for resolving differences in associations include incorporating MeSH supplementary concepts that provide chemical substances and subheadings that refine the main heading. The use of subheadings may also be valuable for identifying the type of association (e.g., drug used to treat a disease or drug that causes an adverse effect). Additionally, the ability of NLP systems to extract relations may remove the need to perform additional techniques (in this case, co-occurrence statistics) for identifying associations or may prove to be a supplementary method. For example, the BioMedLEE output for articles related to Parkinson’s disease identified several types of relationships with levodopa (e.g., associated with and treats) indicating a “treats” relation, although finding relations was not the focus of the current study.
The findings from this study indicate that while MeSH could be used for identifying the primary disease and drug entities in a paper, NLP could enhance the existing MeSH annotations to find associations with other types of entities such as symptoms or procedures, which are less likely to be associated with a MeSH annotation. For example, in a small experiment, we compared MeSH descriptors in articles with UMLS concepts obtained using BioMedLEE classified as a sign or symptom by semantic type, and found that BioMedLEE on average found four times more concepts related to signs or symptoms (or findings) for each disease.
Resolving Issues with Disease and Drug Entities
With respect to disease entities, the MeSH hierarchy is relatively shallow for the diseases in this study (containing 0 to 10 descendants) while the NLP systems produced a range of both general and specific concepts for particular diseases. Due to these varying levels of granularity and challenges with performing accurate mapping, we focused on the broadest MeSH descriptor and UMLS concept in this initial study (as depicted in ▶), thereby not taking advantage of the specific disease concepts extracted by the NLP systems and producing associations limited to these high-level entities. The use of hierarchical relationships present in the Meta (e.g., has narrower relationship [RN]) or particular vocabulary sources such as SNOMED could be explored for building disease classes that would allow the specific concepts to be grouped and used for determining disease-drug associations at a class-level.
For drug entities, we briefly explored drug-specific knowledge within the Meta and MeSH to characterize the entities as trade name drugs, generic name drugs, or drug classes. In some cases, a drug could not be mapped indicating the need for additional techniques (e.g., use of other relationships in RxNorm such as ingredient_of) or other drug sources. For example, while Advair was found to be a highly associated drug for asthma and is a known trade name drug for this disease, its generic names could not be identified with the techniques used. Another issue arises from the one-to-many mapping of some trade name drugs and their generic names. For instance, kaletra used to treat HIV infections is a combination of two drugs and therefore maps to multiple concepts (i.e., lopinavir, ritonavir, and lopinavir-ritonavir). In this study, all mappings were made; however, filtering techniques could be applied to find the “best” match. Finally, while the focus was on generic name drugs, we were able to use knowledge from MeSH to identify drug classes and generate co-occurrence statistics (e.g., Anti-Asthmatic Agents and Bronchodilator Agents are highly associated with asthma). Further work on combining related drugs (e.g., methacholine and methacholine chloride or montelukast and montelukast sodium for asthma) and merging drugs into classes would be valuable to understanding what drug classes are associated with particular diseases.
Next steps include using sophisticated concept mapping methods 1,55 for both disease and drug entities to facilitate the identification of associations at different levels of granularity (i.e., trade name, generic name, and classes for drugs and specific and general classes for diseases) and comparing the associations at each level.
Implications
The analysis of disease-drug associations from RCT/MeSH, RCT/UMLS, and DSUM/UMLS provides some insight on the potential power of the combined use of the literature and patient record, types of comparisons possible, and implications of the findings. When the same disease-drug association is found in both the literature and patient record, it increases confidence in the validity of the association, and it may be possible to accept it in a completely automated process. Other inferences could also be possible based on assumptions about the patient record. For example, although the statistical method we used for the literature did not determine the type of association (e.g., treat, side effect, or diagnosis), it is more likely that when a patient is prescribed a particular drug for a particular disease over a sustained time period, the patient is being treated with the drug or being given the drug for preventative measures. Further studies are needed to fully quantify the similarities and differences between the text sources and annotation methods, explore use of their juxtaposition, and analyze the acquired knowledge. Additionally, manual review of the associations by clinical experts is needed to interpret the clinical significance of each disease-drug pair; for this study, a practicing physician who reviewed the top 5 associations in ▶ found them to have relatively good face validity and a next step includes having multiple experts review all disease-drug associations above the cutoff.
Disease-specific knowledge can be found within a range of text sources offering different aspects of a disease. This work presents an initial step to acquiring disease-drug knowledge from the literature and patient record using a combination of text mining and statistical techniques (i.e., MeSH, NLP, and co-occurrence statistics). A framework for discovering pairwise associations was developed and applied to a subset of diseases concentrating on disease-drug pairs. This framework could be applied to additional sources and diseases, other types of two-way associations such as disease-disease and drug-drug, and eventually expanded to higher dimensional associations (e.g., disease-drug-symptom for learning symptoms that may result from giving a drug for a particular disease). Additionally, time series analysis or trend analysis can be used to capture changes in diseases over time and allow for time-oriented comparisons (e.g., studying emerging or disappearing drugs).
Conclusions
Text sources such as biomedical literature and clinical narratives in the patient record are rich resources for learning and tracking disease-specific knowledge (e.g., what drugs are associated with a particular disease and how associations change over time). In this study, we applied an automated approach involving NLP and statistical techniques for identifying disease-drug associations within these text sources. Comparison of the associations demonstrates that Medline RCT articles and discharge summaries offer consistent and complementary knowledge. Additionally, the top five disease-drug associations were found to be appropriate based on physician review. Given these findings, the use of automated annotations through NLP techniques appears to enhance the perspective offered by existing annotations such as MeSH. This study also reveals the challenges in the comparison and integration of disease-drug knowledge from disparate sources, and implications for extending this approach towards the creation of comprehensive disease profiles that reflect both current and historical knowledge. The results achieved by the methodology described in this paper demonstrate for the first time the feasibility of automated acquisition of medical knowledge by capture of information from both the biomedical literature and patient record.
Acknowledgments
The authors thank Hui Cao for sharing information and programs for the co-occurrence statistics, Lyudmila Shagina for technical assistance with BioMedLEE and MedLEE, and Peter Stetson for reviewing the results obtained in this study.
Marianthi Markatou, PhD and Carol Friedman, PhD contributed equally to this work.
Footnotes
This work is supported in part by grants LM007659, LM008635, and LM006910 from the National Library of Medicine. Dr. Markatou is supported by NSF DMS-0504957.
References
- 1.Rindflesch TC, Pakhomov SV, Fiszman M, Kilicoglu H, Sanchez VR. Medical facts to support inferencing in natural language processing AMIA Annu Symp Proc 2005:634-638. [PMC free article] [PubMed]
- 2.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE Proc AMIA Symp 2002:722-726. [PMC free article] [PubMed]
- 3.Cao H, Markatou M, Melton GB, Chiang MF, Hripcsak G. Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics AMIA Annu Symp Proc 2005:106-110. [PMC free article] [PubMed]
- 4.Tsay MY, Yang YH. Bibliometric analysis of the literature of randomized controlled trials J Med Libr Assoc 2005;93(4):450-458. [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language J Am Med Inform Assoc 1999;6(1):76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing J Am Med Inform Assoc 2004;11(5):392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing Medinfo 2004;11(Pt 2):758-762. [PubMed] [Google Scholar]
- 8.Lussier Y, Borlawsky T, Rappaport D, Liu Y, Friedman C. PhenoGO: Assigning phenotypic context to gene ontology annotation with natural language processing Pac Symp Biocomput 2006:64-75. [PMC free article] [PubMed]
- 9.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology J Am Med Inform Assoc 1994;1(2):161-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Fuller S, Friedman C, Hersch W. Medical informatics: Knowledge management and data mining in biomedicineNew York: Springer-Verlag; 2005.
- 11.van Bemmel JH, van Mulligen EM, Mons B, van Wijk M, Kors JA, van der Lei J. Databases for knowledge discoveryExamples from biomedicine and health care. Int J Med Inform 2006;75(3–4):257-267. [DOI] [PubMed] [Google Scholar]
- 12.de Bruijn B, Martin J. Getting to the (c)ore of knowledge: mining biomedical literature Int J Med Inform 2002;67(1–3):7-18. [DOI] [PubMed] [Google Scholar]
- 13.Shatkay H, Feldman R. Mining the biomedical literature in the genomic era: an overview J Comput Biol 2003;10(6):821-855. [DOI] [PubMed] [Google Scholar]
- 14.Krallinger M, Erhardt RA, Valencia A. Text-mining approaches in molecular biology and biomedicine Drug Discov Today 2005;10(6):439-445. [DOI] [PubMed] [Google Scholar]
- 15.Christensen L, Haug P, Fiszman M. MPLUS: a probabilistic medical language understanding system Proceedings of the Workshop on Natural Language Processing in the Biomedical Domain 2002:29-36.
- 16.Hahn U, Romacker M, Schulz S. MEDSYNDIKATE—a natural language system for the extraction of medical information from findings reports Int J Med Inform 2002;67(1–3):63-74. [DOI] [PubMed] [Google Scholar]
- 17.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program Proc AMIA Symp 2001:17-21. [PMC free article] [PubMed]
- 18.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text J Biomed Inform 2003;36(6):462-477. [DOI] [PubMed] [Google Scholar]
- 19.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System Methods Inf Med 1993;32(4):281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology Nucleic Acids Res 2004;32(Database issue):D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bard J, Rhee SY, Ashburner M. An ontology for cell types Genome Biol 2005;6(2):R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information Genome Biol 2005;6(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hristovski D, Stare J, Peterlin B, Dzeroski S. Supporting discovery in medicine by association rule mining in Medline and UMLS Medinfo 2001;10(Pt 2):1344-1348. [PubMed] [Google Scholar]
- 24.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships Bioinformatics 2004;20(3):389-398. [DOI] [PubMed] [Google Scholar]
- 25.Burgun A, Bodenreider O. Methods for exploring the semantics of the relationships between co-occurring UMLS concepts Medinfo 2001;10(Pt 1):171-175. [PMC free article] [PubMed] [Google Scholar]
- 26.Weeber M, Vos R, Klein H, De Jong-Van Den Berg LT, Aronson AR, Molema G. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide J Am Med Inform Assoc 2003;10(3):252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimino JJ, Barnett GO. Automatic knowledge acquisition from MEDLINE Methods Inf Med 1993;32(2):120-130. [PubMed] [Google Scholar]
- 28.Mendonca EA, Cimino JJ. Automated knowledge extraction from MEDLINE citations Proc AMIA Symp 2000:575-579. [PMC free article] [PubMed]
- 29.Srinivasan P, Libbus B. Mining MEDLINE for implicit links between dietary substances and diseases Bioinformatics 2004;20(Suppl 1):I290-I296. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan P, Hristovski D. Distilling conceptual connections from MeSH co-occurrences Medinfo 2004;11(Pt 2):808-812. [PubMed] [Google Scholar]
- 31.Srinivasan S, Rindflesch TC, Hole WT, Aronson AR, Mork JG. Finding UMLS Metathesaurus concepts in MEDLINE Proc AMIA Symp 2002:727-731. [PMC free article] [PubMed]
- 32.Srinivasan P. MeSHmap: a text mining tool for MEDLINE Proc AMIA Symp 2001:642-646. [PMC free article] [PubMed]
- 33.Srinivasan P. Text mining: generating hypotheses from MEDLINE J Am Soc Information Sci and Technol 2004;55:396-413. [Google Scholar]
- 34.Srinivasan P, Wedemeyer M. Mining concept profiles with the vector model or where on earth are diseases being studied? Proceedings of Text Mining Workshop. Third SIAM International Conference on Data Mining. 2003.
- 35.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature Pac Symp Biocomput 2000:517-528. [DOI] [PMC free article] [PubMed]
- 36.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery AMIA Annu Symp Proc 2006:349-353. [PMC free article] [PubMed]
- 37.Duda S, Aliferis C, Miller R, Statnikov A, Johnson K. Extracting drug-drug interaction articles from MEDLINE to improve the content of drug databases AMIA Annu Symp Proc 2005:216-220. [PMC free article] [PubMed]
- 38.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS Proc AMIA Symp 1998:568-572. [PMC free article] [PubMed]
- 39.Mullins IM, Siadaty MS, Lyman J, Scully K, Garrett CT, Greg Miller W, et al. Data mining and clinical data repositories: Insights from a 667,000 patient data set Comput Biol Med 2006;36(12):1351-1377. [DOI] [PubMed] [Google Scholar]
- 40.Hahn U, Romacker M, Schulz S. Creating knowledge repositories from biomedical reports: the MEDSYNDIKATE text mining system Pac Symp Biocomput 2002:338-349. [PubMed]
- 41.Babic A. Knowledge discovery for advanced clinical data management and analysis Stud Health Technol Inform 1999;68:409-413. [PubMed] [Google Scholar]
- 42.Hripcsak G, Bakken S, Stetson PD, Patel VL. Mining complex clinical data for patient safety research: a framework for event discovery J Biomed Inform 2003;36(1–2):120-130. [DOI] [PubMed] [Google Scholar]
- 43.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports Radiology 2002;224(1):157-163. [DOI] [PubMed] [Google Scholar]
- 44.Heinze DT, Morsch ML, Holbrook J. Mining free-text medical records Proc AMIA Symp 2001:254-258. [PMC free article] [PubMed]
- 45.Collier N, Nazarenko A, Baud R, Ruch P. Recent advances in natural language processing for biomedical applications Int J Med Inform 2006;75(6):413-417. [DOI] [PubMed] [Google Scholar]
- 46.Cao H, Hripcsak G, Markatou M. A statistical methodology for analyzing cooccurrence data from a large sample Journal of Biomedical Informatics 2007;40(3):343-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunham M. Data Mining Introductory and Advanced TopicsNew Jersey: Pearson Education, Inc; 2003.
- 48.Diaconis P, Efron B. Testing for independence in a two-way table: new interpretations of the chi-square statistic The Annals of Statistics 1985;13(3):845-874. [Google Scholar]
- 49.Available at:http://www.nlm.nih.gov/pubs/factsheets/medline.html 1985. Accessed July 5, 2007.
- 50.Available at:http://www.pubmed.gov 1985. Accessed July 5, 2007.
- 51.Available at:http://www.nlm.nih.gov/mesh/ 1985. Accessed July 5, 2007.
- 52.Johnson SB, Hripcsak G, Chen J, Clayton P. Accessing the Columbia Clinical Repository Proc Annu Symp Comput Appl Med Care 1994:281-285. [PMC free article] [PubMed]
- 53.Liu S, Ma W, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange IT Professional 2005;7(5):17-23. [Google Scholar]
- 54.Conover W. Practical nonparametric statistics2nd edition. New York: Wiley; 1980.
- 55.Sun Y. Methods for automated concept mapping between medical databases J Biomed Inform 2004;37(3):162-178. [DOI] [PubMed] [Google Scholar]
- 56.LeWitt PA, Ward CD, Larsen TA, Raphaelson MI, Newman RP, Foster N, Dambrosia JM, Calne DB. Comparison of pergolide and bromocriptine therapy in parkinsonism Neurology 1983;33(8):1009-1014. [DOI] [PubMed] [Google Scholar]