Abstract
The Clinical Language Understanding group at Nuance Communications has developed a medical information extraction system that combines a rule-based extraction engine with machine learning algorithms to identify and categorize references to patient smoking in clinical reports. The extraction engine identifies smoking references; documents that contain no smoking references are classified as UNKNOWN. For the remaining documents, the extraction engine uses linguistic analysis to associate features such as status and time to smoking mentions. Machine learning is used to classify the documents based on these features. This approach shows overall accuracy in the 90s on all data sets used. Classification using engine-generated and word-based features outperforms classification using only word-based features for all data sets, although the difference gets smaller as the data set size increases. These techniques could be applied to identify other risk factors, such as drug and alcohol use, or a family history of a disease.
Introduction
Medical reports contain a wealth of data about diagnoses, medications, procedures, etc., expressed primarily as narrative text. This information is difficult to access in free text. A technology that identifies and extracts such data from text makes it possible to build applications that perform tasks such as population of computerized electronic records, report summarization for physicians, support for medical coding, and government/insurance reporting.
The Clinical Language Understanding group at Nuance Communications has built an engine that extracts targeted data from the text of electronic medical reports. In addition to determining that a particular span of text refers to a diagnosis, procedure, or medication, the Nuance medical extraction system analyzes linguistic context to determine the relevance or status of medical entities. Thus, a medication may be current or discontinued, a diagnosis may be confirmed or denied, etc. Several research groups 1-6 have addressed the task of clinical data extraction in recent years. a The Nuance system follows in this tradition, but is distinguished by its emphasis on determining the status of medical entities.
i2b2 Smoking Challenge
In 2006, Nuance participated in the “Smoking Challenge,” a natural language processing shared task competition sponsored by Informatics for Integrating Biology and the Bedside (i2b2). The work done for the i2b2 Smoking Challenge served as a targeted trial that led to more general experiments presented in this paper.
The Smoking Challenge required the automatic classification of patients with respect to smoking status, based on clinical reports. I2b2 defined five smoking categories: 7 PAST SMOKER, CURRENT SMOKER, SMOKER, NON-SMOKER, and UNKNOWN.
The Nuance medical extraction system can recognize text that refers to patient smoking behavior, distinguish statements denying smoking from statements asserting or implying smoking, and distinguish expressions that indicate current time, recent past, and distant past. We submitted three test results to the Smoking Challenge, and they were the winning submissions. b
Since we used a supervised machine learning approach for the Smoking Challenge, we augmented the training data that was supplied by the challenge organizers. Our hypothesis that using a larger training set would lead to better learning models was verified experimentally in the course of the challenge. Linguistic information provided by our extraction engine also improved results significantly. Since the Challenge, we have investigated in more detail the contributions that data set size, data set homogeneity, and the use of linguistically-based classification features make to document classification accuracy.
Methods
Data
Data provided by i2b2 for analysis and training includes a training set of 398 documents with smoking classification labels, including documents classified as UNKNOWN. After the Challenge, i2b2 released a test set of 104 documents labeled similarly.
We collected 4,292 additional medical reports from three healthcare facilities representing a variety of medical specialties and report types, and annotated them using the five smoking category labels. We applied the same category labels to each smoking reference extracted from the documents, in order to support a two-step document classification approach starting at the smoking reference (or mention) level. We hypothesized that classifying individual smoking references using local evidence might allow for a more straightforward feature representation of the problem, and that we could then categorize documents by reasoning over the set of mention-level labels. We created three training sets: one (“i2b2”) contained 502 i2b2 documents; another (“Nuance”) contained 4,292 documents; a third set (“Combo”) combined Nuance and i2b2 documents for a total of 4,794 documents. Differences between Nuance and i2b2 data with respect to writing style and vocabulary made the Combo data set less homogeneous.
Identification of Smoking References
The production version of the Nuance medical extraction engine, which we used to analyze all documents, identifies document structure (sections, paragraphs, sentences), expressions referring to medical entities (medications, problems, procedures, and allergens), and the referential status of those entities; it also normalizes problem (diagnosis) references. For the Smoking Challenge and for subsequent experiments presented here, the engine identified smoking references as problems, and assigned each one a status category indicating whether smoking was asserted or denied, for patient or family. The extraction engine also identified additional smoking-related information, including anti-smoking medications and treatments. Sentences containing phrases indicative of smoking were then extracted from the documents.
We assigned smoking status UNKNOWN to documents in which the extraction engine found no potential references to a patient’s smoking behavior. These documents did not undergo subsequent steps.
Instance Creation and Classification
A document, and even a single sentence, may contain several smoking mentions. The assignment of a smoking category label to a document should be based on all smoking mentions in the document. This can be done by (1) classifying the mentions first and reconciling them into a single document-level judgment (mediated approach), or (2) classifying the document in one step (direct approach).
To collect mention-level details used in both approaches, we extracted all smoking-related references from documents that mentioned smoking, along with information that might characterize the status of the smoking, including predicates (e.g., “quit”); temporal expressions (e.g., “20 years ago”); and normalized representations of section headings (e.g., “DIAGNOSIS”). Each smoking reference, together with its status, code, section heading, heading normalization, negated and current flags, and sentence words and bigrams, became an instance for classification in the mediated approach.
In the direct approach, we combined features, generated as described above, from all smoking mentions in a document into one data instance. Mention-level linguistic features were converted to binary features, indicating that a particular attribute occurred with a particular value. This permitted us to handle cases where the same attribute had different values in subsequent mentions in the same document.
For both approaches, we assigned smoking categories using a Support Vector Machine 8 trained with a sequential minimal optimization algorithm. 9 SVMs have been shown empirically to give very good generalization performance on a variety of tasks, including some in text domains. c
In the direct approach, this completed the task. In the mediated approach, classification assigned document-level smoking categories to each smoking mention separately. We then used a simple heuristic to derive a document classification from the combination of categories obtained for all smoking instances in the document. The heuristic selected the most specific of the mention categories to be the document category, where UNKNOWN is less specific than SMOKER or NON-SMOKER, and SMOKER is less specific than either CURRENT SMOKER or PAST SMOKER. Conflicts between categories of equal specificity were resolved by taking into account the number of instances in each category and generalizing from examples in the training data.
Results
Filtering UNKNOWNs
UNKNOWN was the most frequent label in the data set. Of the 502 i2b2 training and test documents, 315 (63%) were labeled UNKNOWN by i2b2 annotators. In the Nuance data set, 3,452 out of 4,292 documents (80%) were marked UNKNOWN after no smoking mentions were found in them by the engine. d
The accuracy of filtering UNKNOWNs using the extraction engine was 100% on i2b2 test data. We cannot report accuracy of engine filtering on the other data sets, because we did not examine the filtered documents to see if they actually did contain smoking mentions. However, the effectiveness of the filtering on i2b2 test data, and additional spot checks, indicated that this was probably a low source of error in our system. Note that documents that were not filtered by the engine could still subsequently be classified as UNKNOWN based on the nature of their associated features.
The extraction engine filtering of UNKNOWNs contributed significantly to the accuracy of document classification. The high degree of accuracy for this category raised our overall classification accuracy.
Effect of Data Set Size on Document Classification Accuracy
We restricted our investigation of the effect of data set size on document classification accuracy to the classification step applied to the set of documents with almost all UNKNOWNs filtered out, as described above. (Note that our accuracy for this step is significantly lower than the overall accuracy we report for the full sets, which include all documents.) For each data set (i2b2, Nuance, and Combo), we created a sequence of subsets of increasing size by random sampling. Each experiment was repeated 10 times, and our results are averaged over these trials. In each trial, we used 10-fold cross-validation to estimate classification accuracy.
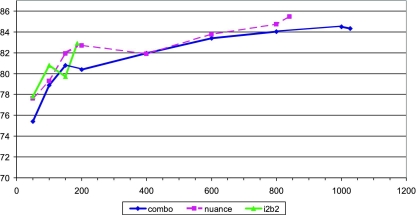
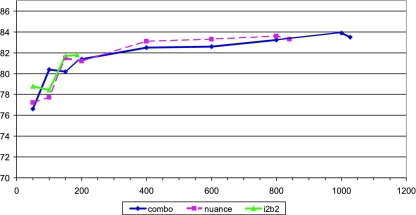
For direct and mediated approaches, the accuracy increases sharply between 50 and 200 document subsets (see ▶). ▶ Above 200, the accuracy increases more slowly. We appear to be approaching the point where additional data would be of little help.
Figure 1.
Effect of data set size on classification accuracy—direct approach
Figure 2.
Effect of data set size on classification accuracy—mediated approach
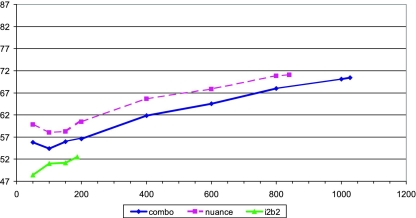
We conducted the same experiment without using our extraction engine to mark mentions and assemble their features. All (filtered) documents were used in their entirety (not just sentences containing mentions as in the directed and mediated approaches), and the features collected were only words and bigrams used in the documents.
The accuracy for all subsets is significantly lower (▶). More interestingly, there is no big increase between 50 and 200 document subsets, and the accuracy keeps rising steadily to the end of the scale. We expect that we would observe a considerable increase still if we could train our models on significantly larger amounts of data. It is not clear, though, whether the accuracy obtained by this approach could ever match the accuracy obtained using our extraction engine-generated features, and if so, how large the data set would need to be.
Figure 3.
Effect of data set size on classification accuracy—no engine features (note a different scale from ▶)
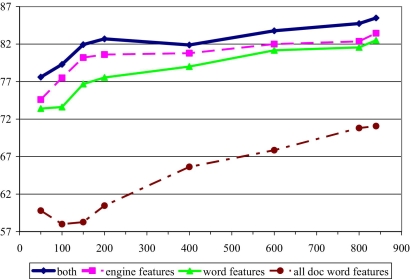
Using informative features provided by our medical fact extraction engine allows our system to learn complicated concepts with much smaller data sets. This point is made even more clearly by a direct comparison of models trained with various feature subsets (▶).
Figure 4.
Effect of feature set on classification accuracy—Nuance data set, direct approach
Classification using entire documents with only word-based features (“all doc word features”) performs worst of all. We observe a significant improvement when we base the classification on sentences containing mentions only (all other curves). Among these, using engine-generated linguistic features (“engine features”) from these sentences produces better results than using only word-based features (“word features”), and the combination of engine-generated features and word-based features (“both”) performs the best. We report our results on the Nuance data set; the results on other sets were similar.
Finally, in most of our experiments, more homogenous data sets (Nuance, i2b2) have higher accuracy than the joint data set (the experiment with no engine-generated features reported in ▶ is an exception). We suspect that homogeneity is beneficial, although its effect is small. e
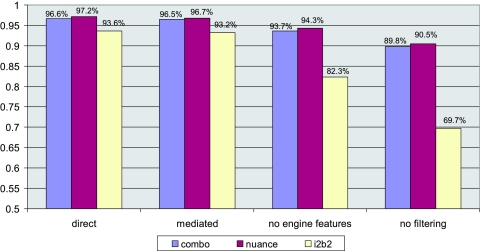
Overall Accuracy Including Filtered UNKNOWNs
The experiments presented in the previous section concern classification accuracy for the subset of documents remaining after a filtering step. For completeness, we report our overall accuracy here. The only data set for which we can report it with full confidence is the i2b2 data set, where all the documents were inspected manually. For Nuance and Combo datasets, we rely on the accuracy of the filtering step, which was tested only on a small subset of the documents.
As can be seen in ▶, mediated and direct approaches have very similar overall accuracy scores (with direct slightly better). The approach using entire documents and no extraction engine-generated features (“no engine features”) is clearly inferior, although the difference here is mitigated by the effect of adding perfectly classified filtered documents. The last category, “no filtering,” is the result of an attempt to classify all documents, including those that were filtered for other approaches. In this experiment, the extraction engine was not used for any part of the process, and the documents were used in their entirety. Features were defined as words and bigrams present in documents.
Figure 5.
Overall accuracy (including filtered documents) assuming 100% accuracy of the filtering step
This last experiment clearly shows the benefit of restricting one’s attention to relevant documents and their relevant fragments. Classification of entire documents requires much more data; classification with a large proportion of data including no relevant features (no smoking related comments), requires more data still. This is particularly evident with the i2b2 data set, which, due to its small size, improves the most with the addition of preprocessing by the extraction engine.
Discussion
The Smoking Challenge task represents a realistic clinical application that requires natural language understanding and reasoning that takes into account the structure of a medical document. We hypothesized that the Nuance medical extraction engine could provide the information needed to categorize a patient’s smoking status. The success of the techniques we used to predict smoking categories supports this hypothesis. We feel that our combination of linguistic analysis and machine learning is the key to our success.
The use of intermediate linguistic analysis can offset the disadvantages of using a small training set in a document categorization task. We believe that the techniques we have used to identify and classify smoking could also be applied to identify other risk factors, such as drug and alcohol use, or even the family history of a disease or disorder.
Footnotes
A description of related work is included in the full paper published as the online data supplement at www.jamia.org.
Details of Smoking Challenge results are included in the full paper published as the online supplement at www.jamia.org.
The learning models for the challenge and our own experiments were created using the Waikato Environment for Knowledge Analysis (WEKA) system. 10
Please see Table 1, available in the full paper published as a JAMIA online data supplement at www.jamia.org.
A discussion of per-class accuracy and class confusions is available in the full paper published as the online data supplement at www.jamia.org.
References
- 1.Long W. Extracting Diagnoses from Discharge Summaries Proc AMIA Symp 2005:470-474. [PMC free article] [PubMed]
- 2.Chapman WW, Dowling JN, Wagner MM. Fever detection from free-text clinical records for biosurveillance J Biomed Inform 2004:120-127. [DOI] [PMC free article] [PubMed]
- 3.Morsch ML, Vengo JL, Sheffer RE, Heinze DT. CM-Extractor: An Application for Automating Medical Quality Measures Abstraction in a Hospital Setting AAAI 2006:1814-1821.
- 4.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing J Am Med Inform Assoc 2004;11(5):392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng Q, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system BMC Med Inform Dec Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sordo M, Zeng Q. On sample size and classification accuracy: a performance comparisonLecture Notes in Computer Science, 3745 2005.
- 7.Uzuner O, Szolovits P, Kohane I. i2b2 Workshop on Natural Language Processing Challenges for Clinical RecordsAvailable athttp://people.csail.mit.edu/ozlem/amia-workshop.pdf 2005. Accessed October 26, 2007.
- 8.Vapnik V. Estimation of Dependences Based on Empirical DataSpringer-Verlag; 1982.
- 9.Platt J. Fast Training of Support Vector Machines using Sequential Minimal OptimizationIn: Schoelkopf B, Burges C, Smola A, editors. Advances in Kernel Methods—Support Vector Learning. MIT Press; 1998.
- 10.Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques2nd Edition. San Francisco: Morgan Kaufmann; 2005.