Abstract
Fluorescent proteins have been widely used as genetically encodable fusion tags for biological imaging. Recently, a new class of fluorescent proteins was discovered that can be reversibly light-switched between a fluorescent and a non-fluorescent state. Such proteins can not only provide nanoscale resolution in far-field fluorescence optical microscopy much below the diffraction limit, but also hold promise for other nanotechnological applications, such as optical data storage. To systematically exploit the potential of such photoswitchable proteins and to enable rational improvements to their properties requires a detailed understanding of the molecular switching mechanism, which is currently unknown. Here, we have studied the photoswitching mechanism of the reversibly switchable fluoroprotein asFP595 at the atomic level by multiconfigurational ab initio (CASSCF) calculations and QM/MM excited state molecular dynamics simulations with explicit surface hopping. Our simulations explain measured quantum yields and excited state lifetimes, and also predict the structures of the hitherto unknown intermediates and of the irreversibly fluorescent state. Further, we find that the proton distribution in the active site of the asFP595 controls the photochemical conversion pathways of the chromophore in the protein matrix. Accordingly, changes in the protonation state of the chromophore and some proximal amino acids lead to different photochemical states, which all turn out to be essential for the photoswitching mechanism. These photochemical states are (i) a neutral chromophore, which can trans-cis photoisomerize, (ii) an anionic chromophore, which rapidly undergoes radiationless decay after excitation, and (iii) a putative fluorescent zwitterionic chromophore. The overall stability of the different protonation states is controlled by the isomeric state of the chromophore. We finally propose that radiation-induced decarboxylation of the glutamic acid Glu215 blocks the proton transfer pathways that enable the deactivation of the zwitterionic chromophore and thus leads to irreversible fluorescence. We have identified the tight coupling of trans-cis isomerization and proton transfers in photoswitchable proteins to be essential for their function and propose a detailed underlying mechanism, which provides a comprehensive picture that explains the available experimental data. The structural similarity between asFP595 and other fluoroproteins of interest for imaging suggests that this coupling is a quite general mechanism for photoswitchable proteins. These insights can guide the rational design and optimization of photoswitchable proteins.
Author Summary
Proteins whose fluorescence can be reversibly switched on and off hold great promise for applications in high-resolution optical microscopy and nanotechnology. To systematically exploit the potential of such photoswitchable proteins and to enable rational improvements of their properties requires a detailed understanding of the molecular switching mechanism. Here, we have studied the photoswitching mechanism of the reversibly switchable fluoroprotein asFP595 by atomistic molecular dynamics simulations. Our simulations explain measured quantum yields and excited state lifetimes, and also predict the structures of the hitherto unknown intermediates and of the irreversibly fluorescent state. Further, we find that the proton distribution in the active site of the asFP595 controls the photochemical conversion pathways of the chromophore in the protein matrix. Our results show that a tight coupling between trans-cis isomerization of the chromophore and proton transfer is essential for the function of asFP595. The structural similarity between asFP595 and other fluoroproteins suggests that this coupling is a quite general mechanism for photoswitchable proteins. These insights can guide the rational design and optimization of photoswitchable proteins.
Introduction
Fluorescent proteins have been widely used as genetically encodable fusion tags to monitor protein localizations and dynamics in live cells [1]–[3]. Recently, a new class of green fluorescent protein (GFP)-like proteins has been discovered, which can be reversibly photoswitched between a fluorescent (on) and a non-fluorescent (off) state [4]–[10]. As the reversible photoswitching of photochromic organic molecules such as fulgides or diarylethenes is usually not accompanied by fluorescence [11], this switching reversibility is a very remarkable and unique feature that may allow fundamentally new applications. For example, the reversible photoswitching, also known as kindling, may provide nanoscale resolution in far field fluorescence optical microscopy much below the diffraction limit [12]–[15]. Likewise, reversibly switchable fluorescent proteins will enable the repeated tracking of protein location and movement in single cells [16]. Since fluorescence can be sensitively read out from a bulky crystal, the prospect of erasable three-dimensional data storage is equally intriguing [17].
The GFP-like protein asFP595, isolated from the sea anemone Anemonia sulcata, is a prototype for a reversibly switchable fluorescent protein. The protein can be switched from its non-fluorescent off state to the fluorescent on state by green light of 568 nm wavelength [5],[6],[18],[19]. From this so-called kindled on state, the same green light elicits a red fluorescence emission at 595 nm. Upon kindling, the intensity of the absorption maximum at 568 nm diminishes, and an absorption peak at 445 nm appears. The kindled on state can be promptly switched back to the initial off state by this blue light of 445 nm. Alternatively, the off state is repopulated through thermal relaxation within seconds. In addition, if irradiated with intense green light over a long period of time, asFP595 can also be irreversibly converted into a fluorescent state that cannot be quenched by light any more [5]. The nature of this state is hitherto unknown.
The switching cycle of asFP595 is reversible and can be repeated many times without significant photobleaching. These properties render asFP595 a promising fluorescence marker for high-resolution optical far-field microscopy, as recently demonstrated by Hofmann and coworkers [20]. Currently, however, with its low fluorescence quantum yield (<0.1% and 7% before and after activation, respectively [6],[16]) and rather slow switching kinetics, the photochromic properties of asFP595 need to be improved. To systematically exploit the potential of such switchable proteins and to enable rational improvements to the properties of asFP595, a detailed molecular understanding of the photoswitching mechanism is mandatory.
The aim of this study is to obtain a detailed mechanistic picture of the photoswitching mechanism of asFP595 at the atomic level, i.e., to understand the dynamics of both the activation process (off-to-on switching) and the de-activation process (on-to-off switching).
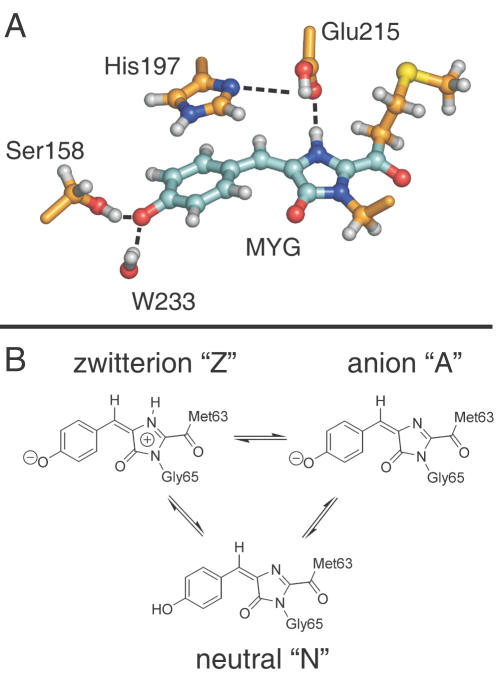
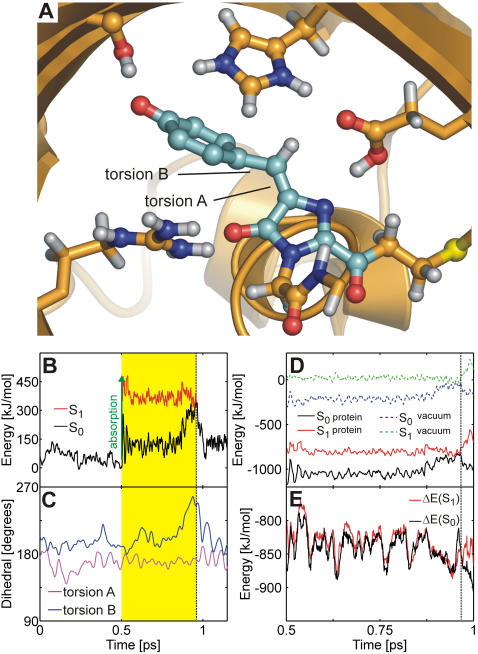
High-resolution crystal structures of the wild-type (wt) asFP595 in its off state [19],[21],[22], of the Ser158Val mutant in its on state [19], and of the Ala143Ser mutant in its on and off states [19] were recently determined. Similar to GFP, asFP595 adopts a β-barrel fold enclosing the chromophore, a 2-acetyl-5-(p-hydroxybenzylidene)imidazolinone (Figure 1). The chromophore is post-translationally formed in an autocatalytic cyclization-oxidation reaction of the Met63-Tyr64-Gly65 (MYG) triad. As compared to the GFP chromophore, the π-system of MYG is elongated by an additional carbonyl group [23].
Figure 1.
(A) Chromophore (MYG) in the trans conformation and adjacent amino acid side chains. Ser158, Glu215, and the crystallographic water molecule W233 are hydrogen-bonded to MYG (dashed lines). His197 is π-stacked to the MYG phenoxy-moiety and forms a hydrogen bond to Glu215 (dashed line). The carbon skeleton of the quantum mechanical (QM) subsystem is shown in cyan, and the carbon atoms modelled by molecular mechanics (MM) are shown in orange. (B) Schematic drawings of the different MYG protonation states considered in this work.
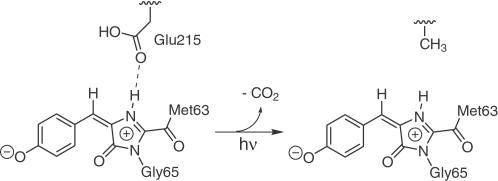
Reversible photoswitching of asFP595 was possible even within protein crystals, and x-ray analysis showed that the off-on switching of the fluorescence is accompanied by a conformational trans-cis isomerization of the chromophore [19]. In a recent study [24], we have shown that the isomerization induces changes of the protonation pattern of the chromophore and some of the surrounding amino acids, and that these changes account for the observed shifts in the absorption spectrum upon kindling. Based on the comparison between measured and calculated absorption spectra, the major protonation states in the ground state have been assigned to the zwitterion (Z) and the anion (A) for the trans conformer, whereas the neutral (N) chromophore is dominant for the cis conformation (Figure 1B).
Here, we study the photochemical behavior of each of the previously identified protonation states. We have addressed the following questions: How does light absorption induce the isomerization of the chromophore within the protein matrix, and how do the different protonation states affect the internal conversion mechanism? Which is the fluorescent species, and how can the fluorescence quantum yield be increased? To address these questions, we have carried out nonadiabatic molecular dynamics (MD) simulations using a hybrid quantum-classical QM/MM approach. This approach includes diabatic surface hopping between the excited state and the ground state. The forces acting on the chromophore were calculated using the CASSCF [25],[26] multi-reference method, which, although not always yielding highly accurate excitation and fluorescence energies, has shown to be a reliable method for mechanistic studies of photochemical reactions involving conical intersections [27].
A number of approaches for modeling nonadiabatic dynamics have been described in the literature, such as Tully's fewest switches surface hopping [28], and multiple spawning [29]. For recent reviews, see [30],[31]. In the context of QM/MM simulations, the surface hopping approach to photobiological problems has been pioneered by Warshel and coworkers [32],[33]. The diabatic surface hopping approach used in this work differs from the other approaches in two main respects. First, in our approach a binary decision (hop or no hop) is made at each integration time step of the trajectory, based only on the current wavefunctions of the ground and excited states. Second, hopping is only allowed at the conical intersection (CI) seam, where hopping probability approaches unity. This could in principle underestimate the crossing probabilty, because we do not allow for transitions in regions of strong coupling but no real crossing. However, for ultra-fast photochemical reactions in large polyatomic systems, decay predominantly takes place at the CI seam, as also shown by others [31]. Thus, most surface hops are essentially diabatic, justifying our approach. In addition, both energy and momentum are conserved upon a transition, as the trajectory never leaves the diabatic energy surface. The theoretical background and algorithmic implementation of the diabatic surface hopping are detailed in the Supporting Information (Text S4).
Several theoretical studies on the photochemistry of the GFP chromophore have been conducted, applying both static ab initio [34]–[36] and DFT calculations [37], and dynamics simulations based on a semi-empirical Hamiltonian [38]. In addition, vertical excitation energies of asFP595 model chromophores in the gas phase and in a continuum dielectric were calculated by DFT and ab initio methods [39],[40], as well as in a minimal protein environment by means of DFT and CASSCF calculations within a QM/MM approach [41].
By identifying key residues in the cavity of the asFP595 chromophore, our nonadiabatic QM/MM molecular dynamics simulations elucidate how the protein surrounding governs the photoreactivity of this photoswitchable protein. Based on the simulations, we provide a new mechanism that qualitatively explains measured decay times and quantum yields, and that predicts the structures and protonation states of the photochemical intermediates and of the irreversibly fluorescent state. We also suggest excited state proton transfer (ESPT) to play an important mechanistic role. However, the detailed study of such ESPT processes is beyond the scope of this paper. Our predictions can be probed by, e.g., time-resolved Fourier transform infrared (FTIR) spectroscopy and x-ray crystallography.
Results/Discussion
Our results reveal that the excited state behavior of asFP595 is determined by the protonation pattern of the chromophore and some amino acids in the surrounding protein matrix rather than by the chromophore conformation (trans or cis). The latter, however, modulates the excited state properties by changing the hydrogen-bonded network in the chromophore cavity.
For both conformers, we identified three possible species that differ in protonation state, explaining the complex photochemical behavior of asFP595. First, the neutral chromophores Ntrans and Ncis undergo reversible trans-cis photoisomerization and thus account for the photoswitching between the dark off and fluorescent on states. Second, anionic chromophores Atrans and Acis lead to the observed ultra-fast radiationless deactivation. Third, fluorescence emission can in principle originate from both zwitterions Ztrans and Zcis. The protonation states are interchangeable via proton transfers. In the following we will describe the excited state behavior of all three protonation states.
trans-cis Isomerization of the Neutral Chromophore
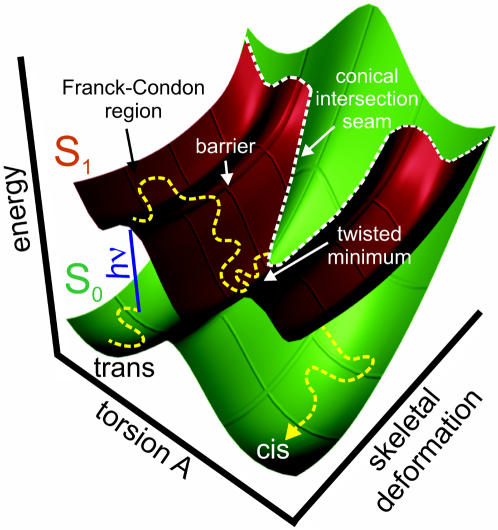
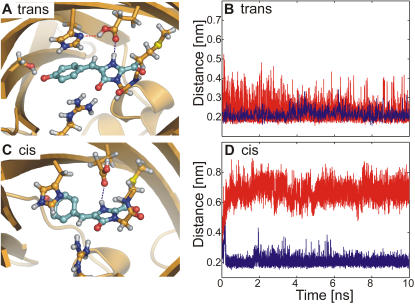
The five excited state simulations that were initiated from the ground state trajectory of the trans neutral chromophore Ntrans are listed in Table 1. Trans-to-cis photoisomerization of the chromophore was observed in one of these simulations (run b, Table 1; Video S2 in Supporting Information). Figure 2 shows a schematic representation of the S0 (green) and S1 (red) potential energy surfaces of the neutral chromophore, along with a photoisomerization MD trajectory (yellow dashed line). Two coordinates are shown, the isomerization coordinate and a skeletal deformation coordinate of the chromophore (see below). The dynamics can be separated into three distinct phases: (i) evolution on the electronic ground state S0, (ii) excitation and evolution on the excited state S1, and (iii) decay back to S0 at the surface crossing seam followed by subsequent relaxation on the ground state surface. The position of the surface crossing seam controls the passage of the trajectory from S1 to S0. The seam is accessed from a global twisted minimum on S1, which is separated by a small S1 barrier from a local planar minimum near the Franck-Condon (FC) region.
Table 1. Excited state lifetimes and final conformations from the MD simulations initiated in the neutral trans chromophore conformation.
| Starting in Ntrans | ||
| Run | S1 lifetime (ps) | Final conformation |
| a | 0.516 | trans |
| b | 0.475 | cis |
| c | 0.309 | trans |
| d | 0.224 | trans |
| e | 0.718 | trans |
Figure 2. Schematic representation of potential energy surfaces.
The excited (S1, red) and ground (S0, green) states of the neutral chromophore are shown along the trans-cis isomerization coordinate (torsion A) and a skeletal deformation coordinate of the chromophore. Radiationless decay occurs at the S1/S0 conical intersection (CI, dashed white line). In this representation, the CI occurs as an extended seam, because the torsion coordinate is from the (N-2)-dimensional intersection space, and the skeletal deformation coordinate is from the 2-dimensional branching pace. The dashed yellow line represents the path sampled in a QM/MM photoisomerization trajectory.
In our simulations, individual excited state (S1) lifetimes between 0.224 ps and 0.718 ps were observed (Table 1). A simple exponential fit to the observed lifetimes yielded a decay time of τ = 0.34 ps (σ+ = 0.21 ps, σ− = 0.13 ps, with σ being the statistical error). Given the low number of trajectories, the statistical error of our estimated lifetime may seem unexpectedly low, but results from a rigorous analysis assuming an underlying single exponential decay [42]. Recent femtosecond time-resolved pump/probe experiments by Schüttrigkeit and coworkers have yielded excited state decay time constants of 0.32 ps (78%), 2.6 ps (19%), and 12.1 ps (3%) as well as a fluorescence lifetime of 2.2 ns for asFP595 [43]. However, although the simulated decay times seem to be in good agreement to the experimental results, we believe that the results should not be directly compared. In previous work [24], we demonstrated that the Ntrans protonation state is hardly populated in asFP595 and therefore is unlikely to contribute to the observed excited state decay. Instead, the species that is predominantly responsible for the ultra-fast radiationless decay observed in the experiments is the anionic trans chromophore Atrans, as shown in detail below.
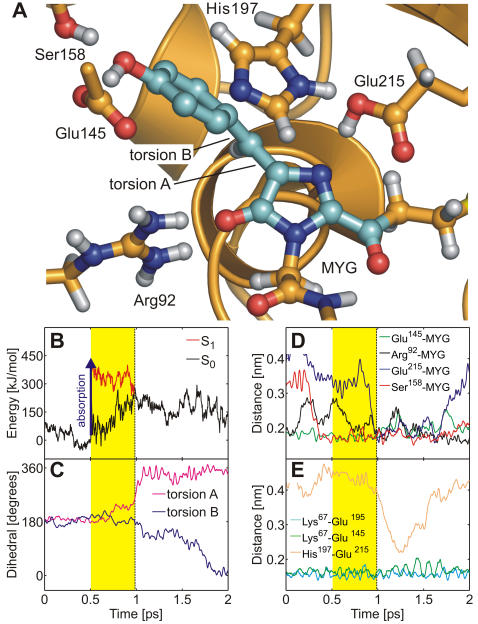
Figure 3A shows the snapshot from the isomerization trajectory (run b, Table 1) shortly before the surface crossing seam was encountered.
Figure 3. Trans-cis isomerization of the neutral chromophore.
(A) Chromophore (MYG) conical intersection geometry adopted during the MD simulation. MYG forms hydrogen bonds to Arg92, Glu145, Ser158, and Glu215. Color code as in Figure 1. (B) Ground (S0, black) and excited (S1, red) potential energy traces along the QM/MM molecular dynamics trajectory. Photon absorption (blue arrow) excites the chromophore into S1 (yellow area) until it decays back to S0 at the conical intersection seam (dashed line). (C) Time-evolution of the ring-bridging torsion angles A (magenta) and B (blue). (D, E) Change of the hydrogen bonding network in the chromophore cavity during trans-cis isomerization. The MYG-Arg92 (black), MYG-Glu145 (green), MYG-Ser158 (red), Lys67-Glu195 (cyan, residues not shown in (A)), and Lys67-Glu145 (green) hydrogen bonds were stable during isomerization. Additional hydrogen bonds between MYG and Glu215 (blue) as well as between His197 and Glu215 (orange) were transiently formed.
After photon absorption (blue arrow in Figures 2 and 3B), the chromophore spontaneously rotated around torsion A (imidazolinone-twist), and the ring-bridging CH group pointed downwards (away from His197) by almost 90°. The time-evolution of the S0 and S1 potential energies and of the two ring-bridging torsion angles during trans-cis photoisomerization are shown in Figures 3B and 3C. After excitation to S1, the chromophore rapidly relaxed from the FC region into a nearby planar S1 minimum, as is evident from the decreasing S1 energy in panel b (red curve). The system stayed in this planar minimum for about 0.2 ps; subsequently the global twisted S1 minimum was reached through rotation around torsion A (Figures 2, 3B, and 3C). The system then oscillated around this minimum until the conical intersection seam was encountered. After the surface hop to S0, torsion B (hydroxyphenyl-twist) rotated after a short delay of about 0.5 ps. Previously, an ideal “hula-twist” isomerization mechanism of the zwitterionic chromophore was proposed based on a force field model [19]. This hula-twist involves a simultaneous rotation around both torsion angles A and B. In the QM/MM simulations of the neutral chromophore presented here, rotation around both torsions was also observed. However, twisting around torsion B was slower than around torsion A (Figure 3C), and the isomerization hence proceeded via the twisted conformer shown in Figure 3A, with perpendicular imidazolinone and hydroxyphenyl moieties. The hula-twist isomerization mechanism of the zwitterion at the QM/MM level will be discussed below.
During the initial equilibration of Ntrans, the hydrogen bonding network of the x-ray crystal structures of the anionic and zwitterionic chromophores (see above) changed to accommodate the non-native neutral chromophore. First, a stable hydrogen bond formed between the hydroxyphenyl OH group of MYG and Glu145 (Figure 3A). Second, the hydrogen bonds between the imidazolinone nitrogen and Glu215 as well as between His197 and Glu215 ruptured. Interestingly, these two hydrogen bonds were transiently re-established during the end of the second isomerization phase (blue and orange curves in Figures 3D and 3E, respectively) in which torsion B followed torsion A. In the twisted intermediate structure, the imidazolinone nitrogen atom was sterically more exposed as compared to the planar conformation, which facilitated the formation of the hydrogen bonds. During an extended 10 ns force field simulation, the His197-Glu215 and MYG-Glu215 hydrogen bonds repeatedly broke and re-formed at a timescale of several hundred picoseconds (data not shown), further underlining the flexibility of these two hydrogen bonds. Figures 3D and 3E furthermore show that during the isomerization, the hydrogen bonding network in the chromophore cavity was stable, as none of the hydrogen bonds that were established at the instant of photoexcitation ruptured. A similar stability was found in all simulations, irrespective of the chromophore conformation or protonation state. Thus, the hydrogen bonding network around the chromophore is flexible enough to allow for photoexcitation and even photoisomerization without being ruptured.
For the neutral cis chromophore Ncis, five excited state simulations were initiated from the ground state trajectory (Table 2). Only two of these trajectories returned to the ground state within 10 ps (Table 2, runs a and b), which was the maximum affordable trajectory length in terms of computation time. In one of these two simulations, a spontaneous cis-to-trans photoisomerization was observed (run a; Video S1 in Supporting Information). As expected, the isomerization pathway was similar to the reverse trans-to-cis pathway in that the conical intersection seam was accessed via rotation around torsion A, followed by a slightly delayed rotation around torsion B in S0 (see Figure S4 in Supporting Information). However, in contrast to the activation pathway, the ring-bridging CH group rotated upwards (i.e., towards His197). Thus, despite the anisotropic protein surrounding, both rotational orientations of the chromophore CH bridge are feasible. In the second simulation, the CI seam was also encountered after rotation around torsion A, but the chromophore returned to the initial cis conformation.
Table 2. Excited state lifetimes and final conformations from the MD simulations initiated in the neutral cis chromophore conformation.
| Starting in Ncis | ||
| Run | S1 lifetime (ps) | Final conformation |
| a | 0.374 | trans |
| b | 3.561 | cis |
| c* | 1.573 | trans |
| d* | 0.867 | cis |
| e* | 1.206 | trans |
*: In runs c, d, and e, the escape from the S1 minimum was accelerated by conformational flooding.
For the other three trajectories, the chromophore remained trapped in a planar S1 minimum conformation near the FC region throughout the simulation (data not shown in Table 2). The starting structures of these trajectories were used for three additional simulations in which the escape from the planar S1 minimum was accelerated by means of conformational flooding [42],[44] (Table 2, runs c–e). In these accelerated simulations, the flooding potential successfully induced the escape from the local S1 minimum, and the surface crossing seam was encountered in all cases. Isomerization was observed in two of these simulations. In total, cis-to-trans photoisomerization was seen in three out of five simulations initiated in the Ncis state. Although the number of trajectories is small, our simulations suggest that the probability for cis-to-trans photoisomerization is larger than for the reverse trans-to-cis process discussed before.
S1/S0 Conical Intersection Topology and S1 Minima
To further characterize the potential energy surfaces underlying the photochemical conversion processes of the neutral chromophore (Figure 2), we have optimized three S1 minima and a minimum energy S1/S0 conical intersection (MECI) in the gas phase (see Supporting Information, Figure S1, Tables S1 and S5). We found a local planar S1 minimum for the trans isomer 76 kJ/mol below the FC region. The structure corresponding to the global S1 minimum is twisted around torsion A by 85° and lies −131 kJ/mol relative to the FC region. The structure of the nearby MECI is twisted around torsion A by 81°. The MECI is energetically lower than the FC region by 62 kJ/mol, and the CI seam is therefore readily accessible. Twisting around torsion B instead of torsion A also leads to a local minimum on S1, whose energy is 28 kJ/mol below the FC energy.
The MD simulations reflect this surface topology. Immediately after excitation, the system relaxed from the FC region to the global S1 minimum by rotation around torsion A (Figure 3C). The system oscillated around this minimum until the conical intersection seam was encountered, with a subsequent surface hop back to S0. The gradients on S0 and S1 are almost parallel at the CI, which indicates that the CI is sloped. The gradient difference vector and the derivative coupling vector that span the branching space largely correspond to skeletal deformations of the imidazolinone moiety (see Supporting Information, Figure S1). Thus, as shown in Figure 2, the rotation coordinate around torsion A is parallel to the seam and does not lift the S1/S0 degeneracy. The seam is accessible anywhere along this torsional rotation coordinate, and therefore such torsional rotation is in principle not essential for the radiationless decay. The extended surface crossing seam parallel to the isomerization coordinate accounts for the low isomerization quantum yield seen in our simulations. In most of our MD simulations, the seam was encountered rather “early” along the torsional rotation coordinate (Figure 2), and the system thus returned to the ground state before overcoming the S0 barrier maximum. In these cases, relaxation on S0 after the surface hop led back to the starting conformation.
Role of the Protein Environment
To elucidate the influence of the protein environment on the photoisomerization process of the chromophore, we have re-calculated the S1 and S0 energies along two excited state trajectories (run b, Table 1 and run a, Table 2) in the gas phase. In these simulations, the chromophore followed the same trajectory as before, but did not interact with the rest of the system (protein and solvent surrounding). We have not attempted to further characterize the electrostatic influence of the surrounding by, e.g., pKa calculations.
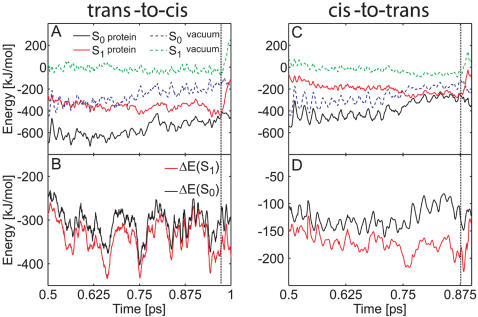
Figure 4A and 4C show the obtained energy traces. In the protein, both S1 and S0 are stabilized with respect to the gas phase. For the trans-to-cis isomerization process, the protein stabilized the energies of the S1 and S0 states on average by −339 kJ/mol and −307 kJ/mol, respectively. For the cis-to-trans process, the average stabilization energies were −173 kJ/mol and −126 kJ/mol, respectively. Thus, the protein (and solvent) environment favors S1 over S0 by about 30–50 kJ/mol. Figure 4B and 4D show the energy differences between the protein and the gas phase, ΔE = E(protein)−E(gas phase). The S1 stabilization was rather strong at the surface crossing seam (Figure 4). We found S1 to be stabilized stronger than S0 by 78 kJ/mol and 93 kJ/mol at the conical intersection in both MD simulations. In summary, the protein environment energetically stabilizes S1 more than S0, thereby enhancing fast radiationless decay.
Figure 4. Influence of the protein environment on the photoisomerization process of the neutral asFP595 chromophore.
(A, C) Ground and excited state energies along trans-to-cis (A) and cis-to-trans (C) isomerization trajectories (run b, Table 1 and run a, Table 2). The protein environment stabilizes S0 and S1 (black and red lines, respectively) relative to the gas phase (dashed blue and green lines, respectively). (B, D) Energy difference between the protein and the gas phase. ΔE(S0) = E(S0, protein)−E(S0, gas phase) is plotted in black, ΔE(S1) = E(S1, protein)−E(S1, gas phase) in red. The protein environment energetically stabilizes S1 more strongly than S0. The vertical dashed black line represents the surface crossing. The energy offset in (A) and (C) is 1.9699×106 kJ/mol.
Ultra-Fast Radiationless Deactivation of the Anionic Chromophore
In total, 20 simulations of the anionic chromophore protonation state were carried out, 10 of which were initiated in the trans conformation and the other 10 were initiated in the cis conformation. Ultra-fast radiationless deactivation was observed in all 20 trajectories (Table S6 in Supporting Information). However, trans-cis photoisomerization never occurred. A simple exponential fit to the S1 lifetimes of the trans anion yielded a decay time of τ = 0.45 ps (σ+ = 0.19 ps, σ− = 0.12 ps). Since Atrans is one of the two dominant protonation states in the off state besides Ztrans [24], we expect Atrans to significantly contribute to the experimentally observed decay. The measured decay time of 0.32 ps [43] agrees well with the decay time derived from the simulations. For Acis, an excited state decay time of τ = 1.81 ps (σ+ = 0.77 ps, σ− = 0.48 ps) was obtained, which is about four times longer as compared to the decay time of Atrans.
Figure 5A shows the conical intersection geometry adopted during a typical trajectory. In contrast to the neutral chromophore, the CI seam was accessed through a phenoxy-twist (rotation around torsion B, see Figure 5C), and the CH bridge remained in the imidazolinone plane. Shortly after excitation, rotation around torsion B drove the system towards the surface crossing seam (Figure 5B and 5C). Back on S0, the system returned to the initial configuration. The hydrogen bonding network in the chromophore cavity was very similar to the network observed in the x-ray crystal structures and remained stable during the excited state MD simulations.
Figure 5. Ultra-fast internal conversion mechanism of the trans anion.
(A) Snapshot at the conical intersection: the chromophore is twisted around torsion B, yet the hydrogen bonded network in the chromophore cavity remains intact. (B) Ground (S0, black) and excited (S1, red) potential energy traces along the QM/MM molecular dynamics trajectory. Photon absorption (green arrow) brings the chromophore into S1 (yellow area) until it decays back to S0 at the conical intersection seam (dashed line). (C) Time-evolution of the torsion angles A (magenta) and B (blue). (D) S0 and S1 energies along a representative excited state trajectory of Atrans. The protein environment strongly stabilizes S0 and S1 (black and red lines, respectively) relative to the gas phase (dashed blue and green lines, respectively). The energy offset is 1.9686×106 kJ/mol. (E) Energy difference ΔE between the protein and the gas phase for S0 (black) and S1 (red).
Since rotation around torsion B does not lead to trans-cis isomerization and rotation around torsion A did not occur, the quantum yield for the isomerization of the anion was zero in our simulations. However, due to the limited number of trajectories (20), we cannot rule out the trans-cis photoisomerization of the anion. Our results agree with recent MRPT2 computations of Olsen and coworkers on an anionic DsRed-like model chromophore in the gas phase [45], who have shown that the imidazolinone-twisted S1/S0 CI (i.e., twisted around torsion A), which leads to cis-trans isomerization, is disfavored by more than 150 kJ/mol as compared to the phenoxy-twisted CI. The latter CI is lower in energy than the planar S1 minimum by 9.2 and 48.1 kJ/mol at the MRPT2 and CASSCF levels, respectively [45]. In our calculations, this energy difference is about 30 kJ/mol (see Supporting Information, Table S2), in qualitative agreement with Olsen and coworkers. Due to the slightly different chromophores in DsRed and asFP595, a quantitative agreement cannot be expected.
The deviation from planarity of the trans chromophore observed in the crystal structures was speculated to enhance ultra-fast deactivation [24],[43]. Indeed, the difference between the S1 lifetimes of the cis and trans conformers seen in our simulations can be attributed to steric constraints imposed by the protein matrix. In the trans conformation the phenoxy-ring deviates from planarity by about 20°, whereas the cis chromophore is essentially planar [19],[24]. We observed in our simulations that only a slight additional twisting was required for the trans conformer to reach the surface crossing seam. Thus, the pre-twisting of the phenoxy-moiety due to the protein matrix facilitated fast internal conversion of the trans conformer.
S1/S0 Conical Intersection Topology and S1 Minima
As shown in Supporting Information (Figure S2, Table S2), we have optimized the S1/S0 MECI, a planar and two twisted S1 minima (imidazolinone-twist and phenoxy-twist) for an isolated anionic chromophore. The planar minimum lies 31 kJ/mol below the FC point. The structure of the global S1 minimum is twisted around torsion B by 269° and its energy lies −82 kJ/mol relative to the FC energy. The MECI structure is also twisted about torsion angle B by 269° and is energetically lower than the FC point by 61 kJ/mol, thus explaining the ultra-fast decay seen in our MD simulations. Twisting around torsion A leads to a local S1 minimum that is 46 kJ/mol below the FC point. The CI of the anion is sloped, and the gradient difference vector corresponds to a skeletal deformation of the imidazolinone ring, analogous to the neutral chromophore (see above). In contrast to the neutral chromophore, the derivative coupling vector involves rotation around torsion B. However, the amplitude of this vector is small. Thus, the two electronic states may remain close in energy along torsion B, allowing the system to decay at various phenoxy-twist angles.
Role of the Protein Environment
To study the influence of the protein matrix on the deactivation process of the anionic chromophore, we have re-evaluated the S0 and S1 energies along two representative excited state trajectories (trans and cis) with all interactions between the QM atoms of the chromophore and the MM surrounding switched off, as was done for the neutral chromophores (see above). As Figure 5D and 5E show, the protein (and solvent) environment stabilizes the chromophore with respect to the gas phase. The S0 and S1 states of the trans chromophore are strongly stabilized by −840 kJ/mol and −832 kJ/mol, respectively. The S0 and S1 states of the cis chromophore were stabilized relative to the gas phase by −407 kJ/mol and −433 kJ/mol, respectively, during a representative Acis trajectory (Figure S5 in Supporting Information). Similar to the neutral chromophore, the protein environment favors the trans conformation over cis. Before reaching the CI seam, the S0 and S1 states of the chromophore were stabilized to the same extent. At the CI, however, the protein environment lowered the energy of the S1 state more strongly than the energy of the S0 state by 26 kJ/mol and 20 kJ/mol for Atrans and Acis, respectively. This preferential stabilization of S1 enhanced the ultra-fast radiationless deactivation seen in our MD simulations.
Fluorescence Emission of the Zwitterionic Chromophores
Ten simulations were carried out for the zwitterion. Five simulations were started in the Ztrans conformation, and the other five simulations were initiated in the Zcis conformation. No decay back to the ground state was observed within a maximum trajectory length of 10 ps, neither for Ztrans nor for Zcis. The chromophore did not escape from a planar S1 minimum in the vicinity of the FC region in any of the excited state simulations. This suggests that Ztrans and Zcis could be the fluorescent species in asFP595, although the measured fluorescence lifetime of 2.2 ns [43] is still much longer than our maximal trajectory length (10 ps).
For Ztrans and for Zcis, we have carried out three additional simulations each, in which we applied the conformational flooding technique [42],[44],[46] to accelerate the escape from the S1 minimum in an unbiased manner (see Materials and Methods). The results of these flooding simulations are shown in the Supporting Information (Figure S6, Text S3). The flooding potential induced isomerization of the chromophore, which followed a hula-twist pathway, in agreement with our previous work [19]. From the flooding simulations, we obtained a lower bound for the excited state lifetime of the order of 1 ns. The qualitative agreement with the measured fluorescence decay time of 2.2 ns provides further support for the assignment of the zwitterion as the fluorescent species. The results thus obtained for the zwitterionic chromophore suggest that a hula-twist CI may be spontaneously accessed if the trajectories were extended to (significantly) longer times, i.e., nanoseconds. However, at the nanosecond timescale, fluorescence (and not isomerization) will be the predominant decay process. Note that in Ref. [19], the energy barrier for hula-twist isomerization of the zwitterion was underestimated, thus favoring this isomerization over fluorescence. In the next paragraph, we characterize the CI and show that the minimum energy crossing point for the zwitterionic chromophore has a high energy, thus hampering radiationless decay through hula-twist isomerization.
S1/S0 Conical Intersection Topology and Influence of the Protein Environment
To characterize the topology of the S1 and S0 potential energy surfaces and of the conical intersection that occurs between them, multiconfigurational calculations were carried out for the isolated zwitterionic chromophore, as was also done for the other protonation states (see above). We optimized a planar S1 minimum and a hula-twist S1/S0 MECI (see Supporting Information, Figure S3, Tables S3 and S4). In contrast to the anion and the neutral chromophore, no twisted S1 minima were found. The gradient difference vector and the derivative coupling vector at the MECI do not involve torsional rotation of either torsion A or torsion B, indicating that the CI seam lies parallel to the isomerization coordinate. The MECI lies 70 kJ/mol above the planar S1 minimum and 23.4 kJ/mol above the FC energy. Hence, in contrast to the anion and the neutral chromophore, no low-lying CI is present for the zwitterion, demonstrating that radiationless decay in the gas phase cannot occur in an unactivated manner. For the CI seam to become accessible, a significant stabilization of S1 relative to S0 by the protein environment would be required. However, as shown in Figure S6 in Supporting Information, the protein surrounding does not reduce the S1/S0 energy gap anywhere along the isomerization coordinate.
Deactivation of Ztrans through Proton Transfer
Our results suggest that the zwitterionic chromophore is potentially fluorescent, irrespective of the conformation. However, the x-ray analysis of the emitting species has shown that only the cis chromophore fluoresces, whereas the trans chromophore is dark [19]. A possible explanation for this discrepancy is the presence of an alternative deactivation channel that does not involve isomerization. This deactivation pathway would have to be more easily accessible for Ztrans than for Zcis. Only the latter would therefore be trapped in S1 and fluoresce.
The hydrogen bond between the NH group of the imidazolinone ring and Glu215 strongly suggests that the alternative decay involves an excited state proton transfer (ESPT). Such ESPT would quench the fluorescence, because the resulting anion rapidly deactivates, as shown above. However, by including only the chromophore into the QM subsystem, we have excluded the possibility of observing such ESPT in our QM/MM simulations.
To identify possible ESPT pathways, we have carried out extended force field MD simulations of both Ztrans and Zcis and analyzed the relevant hydrogen bonds. Figure 6A shows that, during the simulation of Ztrans, two stable hydrogen bonds were formed between the protonated OH group of Glu215 and His197 as well as between the NH proton of MYG and Glu215. These two hydrogen bonds allow for a proton transfer from Ztrans to the rapidly deactivating Atrans. The OH proton of Glu215 could transfer to the Nδ atom of His197, with a simultaneous or subsequent transfer of the NH proton of the imidazolinone moiety to Glu215. In contrast, during the force field simulation of Zcis, the MYG-Glu215 hydrogen bond remained intact, whereas the Glu215-His197 hydrogen bond broke after about 1 ns (Figure 6B). This differential behavior of Ztrans and Zcis was confirmed by two additional independent MD simulations (data not shown). Based on these results, we assume that only the trans zwitterion can be converted to the anion through a short proton wire. Therefore, an ultra-fast deactivation channel is available only for the trans zwitterion, and not for the fluorescent cis zwitterion. From the presence of the hydrogen bonding network in our force field trajectories, we do not obtain insights into the energetics of proton transfer. Studying these transfers along the identified pathways in asFP595, both in the ground and the excited state, is beyond the scope of the present work.
Figure 6. Hydrogen bonding network in the chromophore cavity during force field simulations of zwitterionic chromophores.
(A, C) Snapshots from MD simulations of Ztrans and Zcis, respectively. The blue dashed lines indicates the distance between the NH proton of MYG and Glu215, and the red dashed line that between the OH-group of Glu215 and the Nδ atom of His197. (B, D) Time-evolution of the two hydrogen bonds shown in (A) and (C) during representative force field MD simulations.
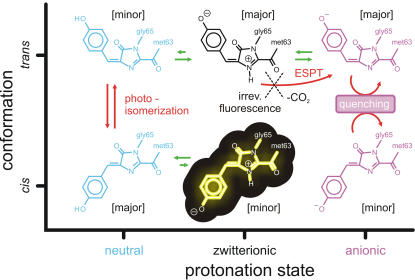
Having established that fluorescence can only originate from the zwitterionic chromophores, the structure of the irreversibly fluorescent state of asFP595 can now be predicted. We expect that intense irradiation over a prolonged period of time leads to a decarboxylation of the Glu215 side chain (Figure 7). Such process is also known to occur in GFP [47],[48] and DsRed [49]. A decarboxylated Glu215 can no longer take up the NH proton from the zwitterionic chromophores. The absence of an S1 ESPT deactivation channel leads to fluorescence. The finding that the irreversibly fluorescent state cannot be switched off by light (see Introduction) is corroborated by our observation that even in the flooding-induced isomerization trajectories, no radiationless decay back to S0 occurred (see Figure S6 in Supporting Information).
Figure 7. Scheme of the proposed decarboxylation of Glu215, which yields an irreversibly fluorescent zwitterion.
Switching Efficiency of asFP595
Figure 8 summarizes our proposed photoswitching mechanism. The proton distribution at the active site of asFP595 governs the photochemical conversion pathways of the chromophore in the protein matrix. Changes in the protonation state of the chromophore and several proximal amino acids lead to different photochemical states, which are all involved in the photoswitching process. These photochemical states are (i) the neutral chromophores Ntrans and Ncis, which can undergo trans-cis photoisomerization, (ii) the anionic chromophores Atrans and Acis, which rapidly undergo radiationless decay after excitation, and (iii) the potentially fluorescent zwitterions Ztrans and Zcis. The overall stability of the different protonation states is controlled by the isomeric state of the chromophore.
Figure 8. Scheme of the reversible photoswitching mechanism of asFP595 proposed in this work.
The fluorescent state Zcis is highlighted. The green arrows indicate ground state equilibria, whereas the red arrows indicate excited state processes. The major protonation states are the zwitterionic and the anionic chromophores in the trans conformation, and the neutral chromophore in the cis conformation, as indicated in the square brackets.
To switch from the non-fluorescent off to the fluorescent on state, the chromophore has to isomerize from trans to cis. As shown in Figure 8, this photoisomerization was only observed for the neutral form of the chromophore (Ntrans→ Ncis). However, the Ntrans state is only marginally populated [24], thus explaining the low quantum yield for switching asPF595 to the on state. Moreover, green light is used to switch on asFP595, whereas the absorption maximum of Ntrans is significantly blue-shifted. The use of blue light, however, would lead to an unfavorable Ncis→ Ntrans back-reaction due to the absorption of the blue light by Ncis.
The reverse cis-to-trans isomerization, i.e., on-to-off switching, requires the excitation of the neutral cis chromophore. Since Ncis is the predominant protonation state in the cis conformation, the efficiency for the on-to-off switching is high, as observed experimentally [19]. Fluorescence originates from Zcis, which is also hardly populated, like Ntrans [24]. Taken together, the low populations of the involved states give rise to the low overall fluorescence quantum yield of asFP595.
The insight obtained from our simulations can be exploited for a targeted improvement of asFP595 for applications as a fluorescence marker in optical microscopy. In particular, to improve the signal-to-noise ratio, a higher fluorescence quantum yield is desired. Our results suggest that one way to enhance fluorescence would be to increase the stability of Zcis, e.g., by introducing additional hydrogen bond donors near the phenoxy-group of the chromophore. Another possibility would be to implement an internal proton relay, similar to that in GFP. In GFP, a hydroxyphenyl-bound serine residue, a water molecule, and a glutamic acid form an internal proton wire that enhances the formation of the fluorescent Acis chromophore from the neutral chromophore via ESPT. GFP has a significantly higher fluorescence quantum yield as compared to asFP595 [50]–[54]. Although the fluorescent species in asFP595 and GFP are different, the similarity between the chromophores suggests that implementing a similar internal proton relay in asFP595 might increase its fluorescence quantum yield.
Note, however, that due to the competition between different reaction channels in asFP595, shifting the relative populations of the protonation states will also affect the photoswitchability. For example, increasing the population of the fluorescent species at the cost of the neutral species will decrease the back-isomerization efficiency. Thus, a compromise has to be found between increasing the fluorescence quantum yield on the one hand while maintaining the photoswitchability of asFP595 on the other hand.
Conclusions
Understanding the excited state dynamics of ultra-fast photoactivated processes in biomolecular systems such as the reversible photoswitching of the fluorescent protein asFP595 represents a major challenge, but is essential to unveil the underlying molecular mechanisms. In the present work we have demonstrated that by using an ab initio QM/MM excited state molecular dynamics strategy together with explicit surface hopping, it is not only possible to explain at the atomic level experimentally accessible quantities of asFP595, such as quantum yields and excited state lifetimes, but also to make predictions that are rigorously testable by experiment, such as the nature of the irreversibly fluorescent state or possible improvement of the fluorescence quantum of this protein.
We have revealed that the protonation pattern of the chromophore cavity determines the photochemical behavior of asFP595, and that photon absorption can lead to trans-cis isomerization of the neutral chromophore. Based on our results, we suggest that a reversibly switchable protein must fulfill three criteria. First, to enable switching, trans-cis photoisomerization is necessary. Second, this photoisomerization has to be coupled to proton transfer events, that is, the preferred protonation state is different for the two conformers. Third, only one of the two isomers fluoresces, while the other can undergo rapid radiationless decay.
Interestingly, other fluoroproteins contain a chromophoric moiety similar to asFP595, like, e.g., Dronpa [8],[55],[56], DsRed [49],[57], Kaede or KiKG [58],[59], eqFP611 [60]–[63], Rtms5 [64], and HcRed [65]. In these structures cis or trans conformations of the chromophore have been observed. Our simulations on asFP595 suggest that chromophore photoisomerization could also be possible in these fluoroproteins. In particular, the high structural similarity between asFP595 and the reversibly photoswitchable Dronpa protein suggests that the Dronpa chromophore can undergo trans-cis photoisomerization as well. Indeed, the crystal structure of the isomerized state of Dronpa was solved very recently [66] and confirms our prediction. Furthermore, recent experiments on Dronpa [67],[68] also provide strong support for our proposed protonation/deprotonation mechanism. The similarity between the chromophores in a variety of fluoroproteins suggests that during molecular evolution, the (p-hydroxybenzylidene)imidazolinone chromophoric moiety served as a template and that the photochromic properties - and thus the function - was fine-tuned by the protein environment.
Materials and Methods
To model the dynamics of the photoactivated asFP595 chromophore, we carried out excited state QM/MM [69] molecular dynamics (MD) simulations. Energies and forces of the excited (S1) and ground (S0) states were calculated on-the-fly at the CASSCF/3-21G level of theory with a reduced active space of 6 electrons in 6 orbitals. The studied processes start in S1 and end up in S0. The transitions (hops) between the two energy surfaces were modeled by surface selection at the conical intersection (CI) seam (see Supporting Information, Text S4 for details). This diabatic hopping procedure is known to potentially underestimate the surface hopping probability [70],[71]. However, our experience has shown that in large polyatomic systems, low-lying CI hyperlines are easily accessed due to their high dimensionality (N−2, where N is the number of internal degrees of freedom). We therefore expect that, for the case at hand, diabatic surface hopping is sufficiently accurate. For all MD simulations, Gromacs 3.3 [72] with an interface [70] to Gaussian03 [73] was used. Modifications were made to the one-electron integral routines of Gaussian03 to account for the polarization of the CASSCF wavefunctions due to the pointcharges on the MM atoms.
The reduced active space used in the MD simulations was validated using geometry optimizations of relevant excited state minima and minimum energy crossing points for the isolated chromophores. The full CASSCF active space for the π-system of the asFP595 chromophore would require 18 π electrons in 16 π orbitals, rendering geometry optimizations prohibitively expensive. To make the respective calculations feasible, we have restricted the number of excitations in the wavefunction by employing the RASSCF method (see Supporting Information, Text S1) [74]–[76]. We have characterized minima and conical intersections at the RASSCF(18,7+4+5)[2,2]/6-31G* level of theory. The final 6 electron, 6 orbital active space used in the CASSCF QM/MM MD simulations was selected from the RASSCF calculations such as to enable the simultaneous description of the electronic ground and first excited states. This approach allowed us to compute on-the-fly QM/MM trajectories using a CASSCF/3-21G wavefunction with an accuracy that is comparable to RASSCF/6-31G* at a drastically reduced computational cost.
All MD simulations were based on the crystal structure of a mutant of asFP595 [24]. The mutant has similar photochromic properties as the wild-type, but high-resolution crystal structures are available for both the trans and the cis conformations. For both trans and cis isomers, MD simulations were initiated for three different chromophore protonation states; anionic (A), neutral (N), and zwitterionic (Z). The simulations were performed in a rectangular periodic box of about 730 nm3. Each system contained about 21,500 TIP4P water molecules, including 340 crystallographic water molecules. After assigning the protonation pattern of the chromophore pocket, all other polar, aromatic, and aliphatic hydrogen atoms were added to the protein with the HB2MAK [77] routine of WHATIF [78]. To each of the systems, sodium and chloride ions were added at physiological concentration to compensate for the net positive charge of the protein. The actual number of ions varied with the total charge of the protein, which differed for the chosen protonation patterns of the chromophore cavity. The final systems comprised about 90,000 atoms.
Prior to the MD simulations, the systems were energy minimized (1000 steps steepest descent). Subsequently, force field based MD simulations were carried out. First, 500 ps MD simulations with harmonic position restraints on all protein heavy atoms (force constant 1000 kJ mol−1 nm−2) were carried out to equilibrate the solvent and the ions. Then, 500 ps free MD simulation were run at 300 K. All simulations were carried out using the OPLS all-atom force field [79]. Parameters for the chromophore were taken from [19].
The simulations were run at constant temperature and pressure by coupling to an external heat bath (τT = 0.1 ps, τp = 1 ps) [80]. In the force field simulations, LINCS [81] was used to constrain bond lengths, thus allowing a time step of 2 fs. SETTLE [82] was applied to constrain the internal degrees of freedom of the water molecules. A twin-range cut-off was used for the Lennard-Jones interactions. Interactions within 1.0 nm were updated at every step, whereas interactions between 1.0 nm and 1.6 nm were updated every ten steps. Coulomb interactions within 1.0 nm were computed at each step as well. Beyond this cut-off, the particle-mesh Ewald (PME) method [83] with a grid spacing of 0.12 nm was used.
The QM subsystem in the excited state QM/MM simulations comprised the chromophore; the rest of the system was described by the OPLS all-atom force field (Figure 1). The N−Cα bond of Gly65 and the Cα−Cβ bond of Met63, respectively, were replaced by a constraint, and the QM part was capped with hydrogen link atoms [84]. The forces on the link atoms were distributed over the two heavy atoms at the boundary according to the lever rule. The QM system experienced the Coulomb field of all MM atoms within a sphere of 1.6 nm, and Lennard-Jones interactions between QM and MM atoms were included. For the QM/MM simulations, a time step of 1 fs was used, and no constraints were applied in the QM subsystem. Prior to the excited state simulations, the systems were simulated in the ground state, first for 1 ps at the RHF/3-21G//OPLS level of theory and then for an additional 2.5 ps at the CASSCF(6,6)/3-21G//OPLS level. From the latter trajectory, frames at equal time intervals (Δt = 0.5 ps) were used as starting configurations for the excited state MD simulations.
To accelerate the escape from the S1 minima, additional excited state conformational flooding [42],[44] simulations were carried out for the Ncis, Zcis, and Ztrans systems, respectively. A Gaussian-shaped flooding potential Vfl was constructed from a principal component analysis (PCA) [85]–[87] of free QM/MM S1 simulations. For all systems, the covariance matrix of the motion of all QM atoms, except the exocyclic carbonyl group of the chromophore and the hydroxyphenyl-OH proton (for Ncis), was computed from two independent 10 ps S1 trajectories. All internal degrees of freedom of the chromophore were affected by the flooding potential to ensure that the escape from the initial minimum was accelerated in an unbiased manner. Unless stated differently, adaptive flooding [42],[46] with target destabilization free energies of 300 kJ/mol (Ztrans) or 100 kJ/mol (Ncis) and time constants of τ = 0.1 ps was applied. In all flooding simulations, Vfl was switched off as soon as the conical intersection seam was encountered to allow for an unperturbed relaxation on the ground state potential energy surface.
In all our simulations of the anionic and zwitterionic chromophores, we considered both the cationic and neutral protonation states of the imidazole side chain of His197, which lies coplanar to the chromophore (Figure 1). Recent Poisson-Boltzmann electrostatics calculations have shown that slight structural fluctuations in the local environment change the preferred protonation of the His197 imidazole ring between cationic and neutral and that both protonation states are populated at room temperature [24]. Thus, we ran the same number of trajectories with a cationic and with a neutral His197. As shown in Supporting Information (Text S2), the His197 protonation state had no major influence on the decay mechanisms and lifetimes.
Supporting Information
Optimized ab initio geometries of Ntrans.
(0.53 MB MPG)
Optimized ab initio geometries of Atrans.
(0.48 MB DOC)
Optimized ab initio geometries of Ztrans.
(0.42 MB DOC)
Cis-to-trans isomerization of the neutral chromophore. (A) Chromophore (MYG) conical intersection geometry adopted during excited state QM/MM MD simulation. The CH bridge rotated upwards (i.e., towards His197). (B) Time-evolution of the ring-bridging dihedral angles A (magenta) and B (blue). (C) S0 (black) and S1 (red) potential energy traces along the QM/MM trajectory. Photon absorption brings the system into S1 (yellow area), until decay back to S0 occurs (dashed line) at the CI seam. (D,E) The hydrogen-bonding network in the chromophore cavity stayed stable during the isomerization of MYG.
(8.79 MB DOC)
Influence of the protein environment on the deactivation of the anionic cis chromophore Acis. (A) S0 and S1 energies along a representative trajectory (run h, Table S6). The protein environment stabilized S0 and S1 (black and red lines, respectively) relative to the gas phase (dashed blue and green lines, respectively). (B) Energy difference ΔE between the protein and the gas phase for S0 (black) and for S1 (red). The dashed line indicates the decay at the CI seam.
(1.30 MB TIF)
Isomerization of the cis zwitterion induced by conformational flooding. (A) Hula-twist structure adopted during isomerization trajectory (B) Ground (S0, black) and excited state (S1, red) potential energy traces along the trajectory. S0 and S1 come energetically close, but the surface crossing seam was not encountered. The surface hop was therefore imposed at the structure with the minimum energy gap (dashed black line). The time evolution of the flooding potential Vfl is shown in the inset. (C) Time evolution of the torsion angles A (magenta) and B (blue). (D) S0 and S1 energies (black and red lines, respectively) along the isomerization trajectory. The protein environment stabilizes S0 and S1 relative to the gas phase (dashed blue and green lines, respectively). The energy offset is 1.97·106 kJ/mol. (E) Energy difference ΔE between the protein and the gas phase for S0 (black) and S1 (red). S0 is stabilized slightly stronger than S1 along the whole isomerization pathway.
(7.52 MB TIF)
RASSCF(18,7+4+5)2,2/6-31G* results on Ntrans.
(0.03 MB TIF)
RASSCF(18,7+4+5)2,2/6-31G* results on Atrans.
(0.03 MB DOC)
RASSCF(18,7+4+5)2,2/6-31G* results on Ztrans.
(0.03 MB DOC)
CASSCF(6,6)/3-21G results on Ztrans.
(0.03 MB DOC)
Cartesian coordinates of optimized structures at RASSCF(18,7+4+5)2,2/6-31G* level.
(0.05 MB DOC)
Excited state lifetimes and conformations from the MD simulations of the anionic chromophores Atrans and Acis. In runs A–J, His197 was modelled as cationic, whereas in runs K-T, His197 was modelled as neutral (singly protonated at Nδ).
(0.04 MB DOC)
Ab initio calculations in the gas phase.
(0.03 MB DOC)
Influence of the π-stacked Histidine 197.
(0.02 MB DOC)
Conformational flooding simulations of the zwitterionic chromophores.
(0.02 MB DOC)
Diabatic Surface Hopping: Theory and Implementation.
(0.06 MB DOC)
cis-to-trans isomerization of neutral chromophore.
(24.36 MB DOC)
trans-to-cis isomerization of neutral chromophore.
(51.31 MB MPG)
Acknowledgments
We thank Ira Tremmel for carefully reading the manuscript and for valuable suggestions, and M. Andresen, C. Eggeling, S. Hell, S. Jakobs, A. Stiel, and M. Wahl for the crystal structures and stimulating discussions.
Footnotes
The authors have declared that no competing interests exist.
Support from the EU Nanomot project (grant 29084) is thankfully acknowledged. This work has been supported by EPSRC UK (grant GR/S94704/01). LVS thanks the Boehringer Ingelheim Fonds for a PhD fellowship.
References
- 1.Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol. 2003:S7–S14. [PubMed] [Google Scholar]
- 2.Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nat Cell Biol. 2003:S1–S7. [PubMed] [Google Scholar]
- 3.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 4.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 5.Chudakov DM, Belousov VV, Zarisky AG, Novoselov VV, Staroverov DB, et al. Kindling fluorescent proteins for precise in vivo photolabeling. Nature Biotechnol. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 6.Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, et al. Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J Biol Chem. 2000;275:25879–25882. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
- 7.Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilmann PG, Turcic K, Battad JM, Wilce MCJ, Devenish RJ, et al. The 1.7 Å crystal structure of Dronpa: A photoswitchable green fluorescent protein. J Mol Biol. 2006;364:213–224. doi: 10.1016/j.jmb.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 9.Dedecker P, Hotta J, Ando R, Miyawaki A, Engelborghs Y, et al. Fast and reversible photoswitching of the fluorescent protein Dronpa as evidenced by fluorescence correlation spectroscopy. Biophys J. 2006;91:L45–L47. doi: 10.1529/biophysj.106.089789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Crano JC, Guglielmetti RJ. Organic photochromic and thermochromic compounds: Main photochromic families, Vol. 1. New York: Plenum; 1999. [Google Scholar]
- 12.Hell SW, Jakobs S, Kastrup L. Imaging and writing at the nanoscale with focused visible light through saturable optical transitions. Appl Phys A-Mater Sci Process. 2003;77:859–860. [Google Scholar]
- 13.Hell SW. Toward fluorescence nanoscopy. Nature Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 14.Hell SW, Dyba M, Jakobs S. Concepts for nanoscale resolution in fluorescence microscopy. Curr Opin Neurobiol. 2004;14:599–609. doi: 10.1016/j.conb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Geisler C, Schonle A, von Middendorff C, Bock H, Eggeling C, et al. Resolution of lambda/10 in fluorescence microscopy using fast single molecule photo-switching. Appl Phys A-Mater Sci Process. 2007;88:223–226. [Google Scholar]
- 16.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 17.Sauer M. Reversible molecular photoswitches: A key technology for nanoscience and fluorescence imaging. Proc Natl Acad Sci USA. 2005;102:9433–9434. doi: 10.1073/pnas.0504264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. J Biol Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 19.Andresen M, Wahl MC, Stiel AC, Gräter F, Schäfer LV, et al. Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proc Natl Acad Sci USA. 2005;102:13070–13074. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmann PG, Petersen J, Devenish RJ, Prescott M, Rossjohn J. Variations on the GFP chromophore. J Biol Chem. 2005;280:2401–2404. doi: 10.1074/jbc.C400484200. [DOI] [PubMed] [Google Scholar]
- 22.Quillin ML, Anstrom DA, Shu XK, O'Leary S, Kallio K, et al. Kindling fluorescent protein from Anemonia sulcata: Dark-state structure at 1.38 Å resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 23.Yampolsky IV, Remington SJ, Martynov VI, Potapov VK, Lukyanov S, et al. Synthesis and properties of the chromophore of the asFP595 chromoprotein from Anemonia sulcata. Biochemistry. 2005;44:5788–5793. doi: 10.1021/bi0476432. [DOI] [PubMed] [Google Scholar]
- 24.Schäfer LV, Groenhof G, Klingen AR, Ullmann GM, Boggio-Pasqua M, et al. Photoswitching of the fluorescent protein asFP595: Mechanism, proton pathways, and absorption spectra. Angew Chemie Int Ed. 2007;46:530–536. doi: 10.1002/anie.200602315. [DOI] [PubMed] [Google Scholar]
- 25.Siegbahn P, Heiberg A, Roos B, Levy B. Comparison of the super-CL and the Newton-Raphson scheme in the complete active space SCF method. Phys Scr. 1980;21:323–327. [Google Scholar]
- 26.Siegbahn PEM, Almlof J, Heiberg A, Roos BO. The complete active space SCF (CASSCF) method in a Newton-Raphson formulation with application to the HNO molecule. J Chem Phys. 1981;74:2384–2396. [Google Scholar]
- 27.Lipkowitz KB, Boyd DB, Robb MA, Garavelli M, Olivucci M, et al. Reviews in computational chemistry. New York: Wiley; 2000. [Google Scholar]
- 28.Tully JC. Molecular dynamics with electronic transitions. J Chem Phys. 1990;93:1061–1071. [Google Scholar]
- 29.Ben-Nun M, Quenneville J, Martinez TJ. Ab initio multiple spawning: Photochemistry from first principles molecular dynamics. J Phys Chem A. 2000;104:5161–5175. [Google Scholar]
- 30.Martinez TJ. Insights for light-driven molecular devices from ab initio multiple spawning excited-state dynamics of organic and biological chromophores. Acc Chem Res. 2006;39:119–126. doi: 10.1021/ar040202q. [DOI] [PubMed] [Google Scholar]
- 31.Levine BG, Martinez TJ. Isomerization through conical intersections. Annu Rev Phys Chem. 2007;58:613–634. doi: 10.1146/annurev.physchem.57.032905.104612. [DOI] [PubMed] [Google Scholar]
- 32.Warshel A. Bicycle-pedal model for 1st step in vision process. Nature. 1976;260:679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- 33.Warshel A, Chu ZT. Nature of the surface crossing process in bacteriorhodopsin: Computer simulations of the quantum dynamics of the primary photochemical event. J Phys Chem B. 2001;105:9857–9871. [Google Scholar]
- 34.Martin ME, Negri F, Olivucci M. Origin, nature, and fate of the fluorescent state of the green fluorescent protein chromophore at the CASPT2//CASSCF resolution. J Am Chem Soc. 2004;126:5452–5264. doi: 10.1021/ja037278m. [DOI] [PubMed] [Google Scholar]
- 35.Altoe P, Bernardi F, Garavelli M, Orlandi G, Negri F. Solvent effects on the vibrational activity and photodynamics of the green fluorescent protein chromophore: A quantum-chemical study. J Am Chem Soc. 2005;127:11534–11535. doi: 10.1021/ja0451517. [DOI] [PubMed] [Google Scholar]
- 36.Sinicropi A, Andruniow T, Ferre N, Basosi R, Olivucci M. Properties of the emitting state of the green fluorescent protein resolved at the CASPT2//CASSCF/CHARMM level. J Am Chem Soc. 2005;127:11534–11535. doi: 10.1021/ja045269n. [DOI] [PubMed] [Google Scholar]
- 37.Laino T, Nifosi R, Tozzini V. Relationship between structure and optical properties in green fluorescent proteins: a quantum mechanical study of the chromophore environment. Chem Phys. 2004;298:17–28. [Google Scholar]
- 38.Toniolo A, Olsen S, Manohar L, Martinez TJ. Conical intersection dynamics in solution: the chromophore of green fluorescent protein. Faraday Discuss. 2004;127:149–163. doi: 10.1039/b401167h. [DOI] [PubMed] [Google Scholar]
- 39.Amat P, Granucci G, Buda F, Persico M, Tozzini V. The chromophore of asFP595: a theorectical study. J Phys Chem B. 2006;110:9348–9353. doi: 10.1021/jp057071a. [DOI] [PubMed] [Google Scholar]
- 40.Nemukhin AV, Topol IA, Burt SK. Electronic excitations of the chromophore from the fluorescent protein asFP595 in solutions. J Chem Theory Comput. 2006;2:292–299. doi: 10.1021/ct050243n. [DOI] [PubMed] [Google Scholar]
- 41.Grigorenko B, Savitsky A, Topol I, Burt S, Nemukhin A. Ground-state structures and vertical excitations for the kindling fluorescent protein asFP595. J Phys Chem B. 2006;110:18635–18640. doi: 10.1021/jp060124j. [DOI] [PubMed] [Google Scholar]
- 42.Lange OF, Schäfer LV, Grubmüller H. Flooding in GROMACS: Accelerated barrier crossings in molecular dynamics. J Comp Chem. 2006;27:1693–1702. doi: 10.1002/jcc.20473. [DOI] [PubMed] [Google Scholar]
- 43.Schüttrigkeit TA, von Feilitzsch T, Kompa CK, Lukyanov KA, Savitsky AP, et al. Femtosecond study of light-induced fluorescence increase of the dark chromoprotein asFP595. Chem Phys. 2006;323:149–160. [Google Scholar]
- 44.Grubmüller H. Predicting slow structural transitions in macromolecular systems: Conformational flooding. Phys Rev E. 1995;52:2893–2906. doi: 10.1103/physreve.52.2893. [DOI] [PubMed] [Google Scholar]
- 45.Olsen S, Smith SC. Radiationless decay of red fluorescent protein chromophore models via twisted intermolecular charge transfer states. J Am Chem Soc. 2007;129:2054–2065. doi: 10.1021/ja066430s. [DOI] [PubMed] [Google Scholar]
- 46.Müller EM, de Meijere A, Grubmüller H. Predicting unimolecular chemical reactions: Chemical flooding. J Chem Phys. 2002;116:897–905. [Google Scholar]
- 47.van Thor JJ, Gensch T, Hellingwerf KJ, Johnson LN. Phototransformation of green fluorescent protein with UV and visible light leads to decarboxylation of glutamate 222. Nat Struct Biol. 2002;9:37–41. doi: 10.1038/nsb739. [DOI] [PubMed] [Google Scholar]
- 48.Bell AF, Stoner-Ma D, Wachter RM, Tonge PJ. Light-driven decarboxylation of wild-type green fluorescent protein. J Am Chem Soc. 2003;125:6919–6926. doi: 10.1021/ja034588w. [DOI] [PubMed] [Google Scholar]
- 49.Habuchi S, Cotlet M, Gensch T, Bednarz T, Haber-Pohlmeier S, et al. Evidence for the isomerization and decarboxylation in the photoconversion of the red fluorescent protein DsRed. J Am Chem Soc. 2005;127:8977–8984. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 50.Helms V. Electronic excitations of biomolecules studied by quantum chemistry. Curr Opin Struct Biol. 2002;12:169–175. doi: 10.1016/s0959-440x(02)00305-6. [DOI] [PubMed] [Google Scholar]
- 51.Stoner-Ma D, Jaye AA, Matousek P, Towrie M, Meech SR, et al. Observation of excited-state proton transfer in green fluorescent protein using ultrafast vibrational spectroscopy. J Am Chem Soc. 2005;127:2864–2865. doi: 10.1021/ja042466d. [DOI] [PubMed] [Google Scholar]
- 52.Agmon N. Proton pathways in green fluorescence protein. Biophys J. 2005;88:2452–2461. doi: 10.1529/biophysj.104.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leiderman P, Huppert D, Agmon N. Transition in the temperature-dependence of GFP fluorescence: From proton wires to proton exit. Biophys J. 2006;90:1009–1018. doi: 10.1529/biophysj.105.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vendrell O, Gelabert R, Moreno M, Lluch JM. Potential energy landscape of the photoinduced multiple proton-transfer process in the green fluorescent protein: classical molecular dynamics and multiconfigurational electronic structure calculations. J Am Chem Soc. 2006;128:3564–3574. doi: 10.1021/ja0549998. [DOI] [PubMed] [Google Scholar]
- 55.Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, et al. 1.8 Å bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem J. 2006;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ando R, Flors C, Mizuno H, Hofkens J, Miyawaki A. Highlighted generation of fluorescence signals using simultaneous two-color irradiation on Dronpa mutants. Biophys J. 2007;92:L97–L99. doi: 10.1529/biophysj.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0 Å resolution. Proc Natl Acad Sci USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiedenmann J, Schenk A, Röcker C, Girod A, Spindler KD, et al. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from entacmaea quadricolor (anthozoa, actinaria). Proc Natl Acad Sci USA. 2002;99:11646–11651. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, et al. The 2.0 Å crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 62.Nienhaus K, Vallone B, Renzi F, Wiedenmann J, Nienhaus GU. Crystallization and preliminary x-ray diffraction analysis of the red fluorescent protein eqFP611. Acta Crystallogr Sect D-Biol Crystallogr. 2003;59:1253–1255. doi: 10.1107/s0907444903008837. [DOI] [PubMed] [Google Scholar]
- 63.Loos DC, Habuchi S, Flors C, Hotta JI, Wiedenmann JR, et al. Photoconversion in the red fluorescent protein from the sea anemone entacmaea quadricolor: Is cis-trans isomerization involved? J Am Chem Soc. 2006;128:6270–6271. doi: 10.1021/ja0545113. [DOI] [PubMed] [Google Scholar]
- 64.Prescott M, Ling M, Beddoe T, Oakley AJ, Dove S, et al. The 2.2 Å crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation. Structure. 2003;11:275–284. doi: 10.1016/s0969-2126(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 65.Wilmann PG, Petersen J, Pettikiriarachchi A, Buckle AM, Smith SC, et al. The 2.1 Å crystal structure of the far-red fluorescent protein HcRed: Inherent conformational flexibility of the chromophore. J Mol Biol. 2005;349:223–237. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 66.Andresen M, Stiel AC, Trowitzsch S, Weber G, Eggeling C, et al. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci USA. 2007;104:13005–13009. doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Habuchi S, Dedecker P, Hotta JI, Flors C, Ando R, et al. Photo-induced protonation/deprotonation in the GFP-like fluorescent protein Dronpa: mechanism responsible for the reversible photoswitching. Photochem Photobiol Sci. 2006;5:567–576. doi: 10.1039/b516339k. [DOI] [PubMed] [Google Scholar]
- 68.Fron E, Flors C, Schweitzer G, Habuchi S, Ando R, et al. Ultrafast excited-state dynamics of the photoswitchable protein Dronpa. J Am Chem Soc. 2007;129:4870–4871. doi: 10.1021/ja069365v. [DOI] [PubMed] [Google Scholar]
- 69.Warshel A, Levitt M. Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 70.Groenhof G, Bouxin-Cademartory M, Hess B, de Visser SP, Berendsen HJC, et al. Photoactivation of the Photoactive Yellow Protein: Why photon absorption triggers a trans-to-cis isomerization of the chromophore in the protein. J Am Chem Soc. 2004;126:4228–4233. doi: 10.1021/ja039557f. [DOI] [PubMed] [Google Scholar]
- 71.Groenhof G, Schäfer LV, Boggio-Pasqua M, Goette M, Grubmüller H, et al. Ultrafast deactivation of on excited cytosine-guanine base pair in DNA. J Am Chem Soc. 2007;129:6812–6819. doi: 10.1021/ja069176c. [DOI] [PubMed] [Google Scholar]
- 72.van der Spoel D, Hess B, Lindahl E, Groenhof G, Mark AE, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 73.Frisch MJ, et al. Gaussian 03, Revision B.04 Gaussian, Inc., Wallingford, CT, 2004. [Google Scholar]
- 74.Malmqvist PA, Rendell A, Roos BO. The restricted active space self-consistent-field method, implemented with a split graph unitary-group approach. J Phys Chem. 1990;94:5477–5482. [Google Scholar]
- 75.Olsen J, Roos BO, Jorgensen P, Jensen HJA. Determinant based configuration-interaction algorithms for complete and restricted configuration-interaction spaces. J Chem Phys. 1988;89:2185–2192. [Google Scholar]
- 76.Boggio-Pasqua M, Robb MA, Bearpark MJ. Photostability via a sloped conical intersection: A CASSCF and RASSCF study of pyracylene. J Phys Chem A. 2005;109:8849–8856. doi: 10.1021/jp053354r. [DOI] [PubMed] [Google Scholar]
- 77.Hooft RWW, Sander C, Vriend G. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins - Structure, Function, and Genetics. 1996;26:363–376. doi: 10.1002/(SICI)1097-0134(199612)26:4<363::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 78.Vriend G. WHAT IF: a molecular modelling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 79.Jorgensen WL, Tirado-Rives J. The OPLS potential functions for proteins. Energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 80.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 81.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. Lincs: A linear constraint solver for molecular simulations. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 82.Miyamoto S, Kollman PA. SETTLE: An analytical version of the SHAKE and RATTLE algorithms for rigid water models. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 83.Darden T, York D, Pedersen L. Particle Mesh Ewald - An Nlog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 84.Singh UC, Kollman PA. A combined ab initio quantum mechanical and molecular mechanical method for carrying out simulations on complex molecular systems: Applications to the CH3Cl + Cl- exchange reaction and gas phase protonation of polyethers. J Comput Chem. 1986;7:718–730. [Google Scholar]
- 85.Kitao A, Hirata F, Go N. The effects of solvent on the conformation and the collective motions of protein - normal mode analysis and molecular-dynamics simulations of melittin in water and in vacuum. Chem Phys. 1991;158:447–472. [Google Scholar]
- 86.Garcia AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 87.Amadei A, Linssen ABM, Berendsen HJC. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Optimized ab initio geometries of Ntrans.
(0.53 MB MPG)
Optimized ab initio geometries of Atrans.
(0.48 MB DOC)
Optimized ab initio geometries of Ztrans.
(0.42 MB DOC)
Cis-to-trans isomerization of the neutral chromophore. (A) Chromophore (MYG) conical intersection geometry adopted during excited state QM/MM MD simulation. The CH bridge rotated upwards (i.e., towards His197). (B) Time-evolution of the ring-bridging dihedral angles A (magenta) and B (blue). (C) S0 (black) and S1 (red) potential energy traces along the QM/MM trajectory. Photon absorption brings the system into S1 (yellow area), until decay back to S0 occurs (dashed line) at the CI seam. (D,E) The hydrogen-bonding network in the chromophore cavity stayed stable during the isomerization of MYG.
(8.79 MB DOC)
Influence of the protein environment on the deactivation of the anionic cis chromophore Acis. (A) S0 and S1 energies along a representative trajectory (run h, Table S6). The protein environment stabilized S0 and S1 (black and red lines, respectively) relative to the gas phase (dashed blue and green lines, respectively). (B) Energy difference ΔE between the protein and the gas phase for S0 (black) and for S1 (red). The dashed line indicates the decay at the CI seam.
(1.30 MB TIF)
Isomerization of the cis zwitterion induced by conformational flooding. (A) Hula-twist structure adopted during isomerization trajectory (B) Ground (S0, black) and excited state (S1, red) potential energy traces along the trajectory. S0 and S1 come energetically close, but the surface crossing seam was not encountered. The surface hop was therefore imposed at the structure with the minimum energy gap (dashed black line). The time evolution of the flooding potential Vfl is shown in the inset. (C) Time evolution of the torsion angles A (magenta) and B (blue). (D) S0 and S1 energies (black and red lines, respectively) along the isomerization trajectory. The protein environment stabilizes S0 and S1 relative to the gas phase (dashed blue and green lines, respectively). The energy offset is 1.97·106 kJ/mol. (E) Energy difference ΔE between the protein and the gas phase for S0 (black) and S1 (red). S0 is stabilized slightly stronger than S1 along the whole isomerization pathway.
(7.52 MB TIF)
RASSCF(18,7+4+5)2,2/6-31G* results on Ntrans.
(0.03 MB TIF)
RASSCF(18,7+4+5)2,2/6-31G* results on Atrans.
(0.03 MB DOC)
RASSCF(18,7+4+5)2,2/6-31G* results on Ztrans.
(0.03 MB DOC)
CASSCF(6,6)/3-21G results on Ztrans.
(0.03 MB DOC)
Cartesian coordinates of optimized structures at RASSCF(18,7+4+5)2,2/6-31G* level.
(0.05 MB DOC)
Excited state lifetimes and conformations from the MD simulations of the anionic chromophores Atrans and Acis. In runs A–J, His197 was modelled as cationic, whereas in runs K-T, His197 was modelled as neutral (singly protonated at Nδ).
(0.04 MB DOC)
Ab initio calculations in the gas phase.
(0.03 MB DOC)
Influence of the π-stacked Histidine 197.
(0.02 MB DOC)
Conformational flooding simulations of the zwitterionic chromophores.
(0.02 MB DOC)
Diabatic Surface Hopping: Theory and Implementation.
(0.06 MB DOC)
cis-to-trans isomerization of neutral chromophore.
(24.36 MB DOC)
trans-to-cis isomerization of neutral chromophore.
(51.31 MB MPG)