Abstract
In a previous study we found that sumoylation of the DNA-binding protein heat shock factor 2 (HSF2) is up-regulated during mitosis, but the mechanism that mediates this regulation was unknown. Here we show that HSF2 interacts with the polycomb protein MEL-18, that this interaction decreases during mitosis, and that overexpression and RNA interference-mediated reduction of MEL-18 result in decreased and increased HSF2 sumoylation, respectively. Other results suggest that MEL-18 may also function to inhibit the sumoylation of other cellular proteins. The results also show that MEL-18 is able to interact with the small ubiquitin-like modifier (SUMO) ubiquitin carrier protein (E2) enzyme UBC9 and that MEL-18 inhibits the ability of UBC9 to transfer the SUMO protein to target proteins. Together, the results in this work suggest a mechanism in which MEL-18 bound to HSF2 inhibits its sumoylation by binding to and inhibiting the activity of UBC9 enzymes in the vicinity of HSF2. These results provide an explanation for how mitotic HSF2 sumoylation is regulated and suggest that MEL-18, in contrast to the sumoylation-stimulating activities of the polycomb protein PC2, actually functions like an anti-SUMO ubiquitin-protein isopeptide ligase (E3), interacting both with HSF2 and the SUMO E2 UBC9 but acting to inhibit UBC9 activity to decrease sumoylation of a target protein, in this case that of HSF2.
Covalent attachment of small ubiquitin-like modifier (SUMO)4 proteins to lysine residues in target proteins, or sumoylation, is an important regulator of protein functional properties (1–7). SUMO proteins are covalently attached to target lysine residues by the SUMO E2 enzyme UBC9, and these substrate lysines are typically found within the consensus sequence ψKXE (ψ represents hydrophobic amino acids) (8–11). SUMO E3 proteins have been identified that enhance the efficiency of sumoylation by interacting with both UBC9 (SUMO E2) and the target protein, thereby acting as bridging factors to increase the rate of the sumoylation reaction (12–14).
Previous studies in our and another laboratory revealed that a DNA-binding protein called heat shock factor 2 (HSF2) is a target of sumoylation in vivo (15, 16). Our prior work indicated that one of the functions of HSF2 is to bind to heat shock elements in the promoters of hsp70 and other heat shock protein genes during mitosis to mediate an epigenetic function called gene bookmarking on these promoters (17, 18). The results of this study also indicated that sumoylation of HSF2 is up-regulated during mitosis and is important for the interaction of this factor with a subunit of the condensin complex during the bookmarking process, suggesting that this modification is involved in regulating HSF2 bookmarking function (17). However, how the increase in HSF2 sumoylation during mitosis is regulated was not known.
MEL-18 is a member of the polycomb group of proteins that play a vital role in development and differentiation by controlling patterns of gene expression (19–23). One important development with respect to the functional roles of polycomb proteins was the discovery that at least one of them, PC2/CBX4, functions as a SUMO E3 to stimulate the sumoylation of specific target proteins (24, 25).
The results presented in this work now identify the existence of an interaction between HSF2 and the polycomb group protein MEL-18 and suggest that cell cycle-dependent interaction between MEL-18 and HSF2 functions as a mechanism for the previously observed up-regulation of HSF2 sumoylation during mitosis. The results also support the intriguing hypothesis that MEL-18, in contrast to the polycomb protein PC2/CBX4, in which SUMO E3 activity stimulates sumoylation of certain proteins, actually functions like an anti-SUMO E3 protein, interacting with both HSF2 and the SUMO E2 UBC9 but acting to inhibit UBC9 activity and thereby decreasing sumoylation of a target protein, in this case that of HSF2.
EXPERIMENTAL PROCEDURES
Cell Culture—HeLa ATCC cells and HEK 293T cells were grown at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Generation of Antibodies Against MEL-18—Affinity-purified goat polyclonal antibody to MEL-18 was prepared by Bethyl Laboratories (Montgomery, TX) and was raised against the synthetic peptide STSRGRKMTVNGAPVPPLT, which corresponds to the C-terminal sequence of the human MEL-18 polypeptide.
Plasmid Construction—The pEGFP-MEL-18 plasmid was generated by using PCR to amplify, from the plasmid pSG5-mel-18 cDNA, a coding fragment of MEL-18 having KpnI and BamHI sites at the ends using the following primers: 5′-GCG GGT ACC TCC ATG CAT CGG ACT ACA CGG-3′ and 5′-CGC GGA TCC AGA GGG TCC CTT TCC TCA AGG-3′. This PCR product was then cloned into the pEGFP-C1 vector at the KpnI and BamHI sites to form the pEGFP-MEL-18 plasmid. This plasmid was confirmed by DNA sequencing.
GST Pulldown Assays—For in vitro binding between MEL-18 and HSF2, GST-HSF2 was expressed in Escherichia coli from the pGEX-HSF2 plasmid, GST was expressed from the pGEX plasmid, and then lysates of these bacteria were incubated with glutathione-agarose beads for about 2 h at 4 °C with rotation followed by washing. The GST and GST-HSF2 bound to beads were then incubated with [35S]methionine-labeled MEL-18, created by a coupled in vitro transcription and translation system in rabbit reticulocyte lysates (TnT, Promega), overnight at 4 °C with rotation in Buffer A (20 mm HEPES (pH 7.9), 0.2 mm EDTA, 0.1 m KCl, and 20% (v/v) glycerol) in a total volume of 650 μl. After washing four times with Buffer D, the amount of [35S]methionine-labeled MEL-18 bound to GST or GST-HSF2 was determined by boiling the beads in SDS-PAGE buffer, followed by SDS-PAGE and autoradiography. For in vitro binding between MEL-18 and UBC9, similar amounts of purified GST or GST-UBC9 were bound to glutathione-agarose beads and then incubated with extracts of HeLa cells made using Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm dithiothreitol, and Complete protease inhibitor mixture (Roche Applied Science)). After washing six times with Nonidet P-40 buffer, bound protein complexes were separated by SDS-PAGE and analyzed by Western blotting using the anti-MEL-18 goat polyclonal antibodies (Bethyl Laboratories) raised as described above.
Immunoprecipitation Analysis—For co-immunoprecipitation experiments, asynchronous HeLa cells or HeLa cells blocked in mitosis (treated with 400 ng/ml nocodazole for 16 h) were extracted on ice with Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm dithiothreitol, and Complete protease inhibitor mixture) for 20 min. Lysates were then cleared by centrifugation at 13,000 rpm for 10 min at 4 °C. Supernatants were precleared by incubation with rabbit (control) IgG and protein G-Sepharose beads for 2 h at 4 °C with gentle rotation. Precleared extracts were then incubated with primary rabbit polyclonal HSF2 antibody or control IgG and 50% slurry of protein G-Sepharose for 4 h at 4 °C with rotation. After washing the beads six times for 5 min each at 4 °C with Nonidet P-40 buffer, bound proteins were released by boiling in SDS-PAGE sample dye and analyzed by Western blotting using the anti-MEL-18 goat polyclonal antibody or anti-HSF2 goat polyclonal antibody (Bethyl Laboratories). For immunoprecipitation analysis of HSF2 sumoylation, HEK 293T cells were transfected with pEGFP-MEL-18 or pEGFP-C1 along with Myc-SUMO-1 expression plasmid using Effectene transfection reagent according to the manufacturer's instructions (Qiagen). After 48 h, cell extracts were prepared in Nonidet P-40 lysis buffer and 10 mm N-ethylmaleimide before adding HSF2 polyclonal antibodies or nonspecific IgG to proceed as described above, followed by Western blot assay using anti-Myc monoclonal antibody (Invitrogen). For immunoprecipitation analysis of HSF1 sumoylation, HEK 293T cells were transiently transfected with pEGFP-MEL-18/pEGFP-C1 and Myc-SUMO-1 using Effectene transfection reagent according to the manufacturer's instructions. After 48 h, the cells were heat-treated at 42 °C for 1 h and then harvested. Cell extracts were prepared in Nonidet P-40 buffer before adding HSF1 polyclonal antibodies to proceed as above, followed by Western blotting using anti-Myc monoclonal antibody.
In Vitro Sumoylation Assay—Full-length HSF2 was translated in vitro in a rabbit reticulocyte lysate (TnT) and then subjected to in vitro SUMO-1 modification assay as described previously (26) in the presence or absence of purified recombinant GST-MEL-18 or GST.
MEL-18 RNAi—MEL-18 short hairpin RNA (shRNA) was designed and cloned in the pSUPER-EGFP vector. The sequence of the shRNA used was 5′-CGACGCCACCACUAUCGUG-3′. pSUPER-shRNA-MEL-18 and pSUPER-scrambled (negative control) were transiently transfected into HeLa cells using Jet-PEI reagent according to the manufacturer's instructions (Bridge Bioscience). After 48 h, the cells were harvested. Cell extracts were prepared in Nonidet P-40 lysis buffer before adding HSF2 polyclonal antibodies to proceed as above, followed by Western blotting using anti-Myc monoclonal antibody (Invitrogen).
RESULTS
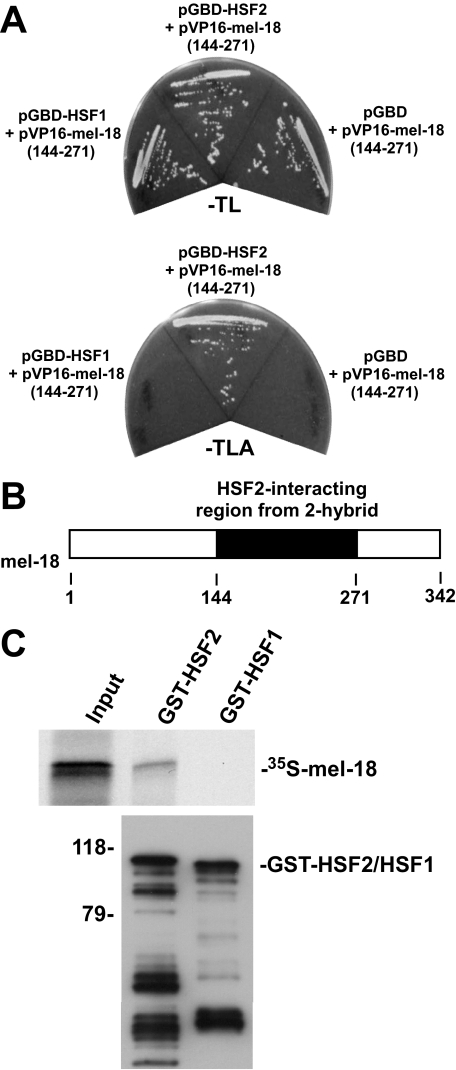
For further understanding of the regulation and function of HSF2 in cells, we performed a yeast two-hybrid screen using the HSF2 protein as a bait. One of the HSF2-interacting clones obtained from this screen represented a region of the polycomb group protein MEL-18 (Fig. 1A). This clone did not interact with a bait containing HSF1, a protein with high sequence relatedness to HSF2, indicating the specificity of this interaction. The location of the region in MEL-18 found in the interacting yeast two-hybrid clone, which comprises amino acids 144–271 of the protein, is shown in the schematic in Fig. 1B. As an independent test of the interaction between HSF2 and MEL-18 and to determine whether the interaction is direct, an in vitro binding experiment was performed in which 35S-labeled in vitro translated MEL-18 was incubated with GST-HSF2 or GST-HSF1 bound to glutathione-agarose beads. The results of this experiment demonstrate the ability of HSF2 to interact with MEL-18 and indicate that the interaction is direct (Fig. 1C).
FIGURE 1.
HSF2 interacts with MEL-18. A, yeast strain pJ694A transformed with pGBD-HSF1, pGBD-HSF2, or pGBD along with pVP16-MEL-18 (amino acids 144–271) was streaked on plates lacking tryptophan and leucine (–TL) or lacking tryptophan, leucine, and alanine (–TLA). B, the schematic depicts the location within MEL-18 of the segment (amino acids 144–271) identified as an HSF2-interacting region in the yeast two-hybrid assay. C, 35S-labeled in vitro translated MEL-18 was incubated with GST-HSF2 or GST-HSF1 that was bound to glutathione-agarose beads. After washing, the amount of bound 35S-labeled MEL-18 was determined by SDS-PAGE and autoradiography (upper panel). The amounts of GST-HSF2 and GST-HSF1 bound to the beads were determined by performing anti-GST Western blotting (lower panel).
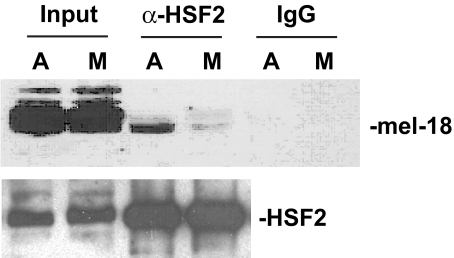
To determine whether endogenous HSF2 and MEL-18 proteins interact, immunoprecipitation analysis was performed. Because our previous studies revealed that HSF2 function is regulated in a mitosis-dependent manner (17), this analysis was performed using extracts of asynchronous cells as well as those of cells blocked in mitosis by nocodazole treatment. The results of this experiment indicate that endogenous HSF2 and MEL-18 proteins do associate and that lower levels of HSF2-MEL-18 complex are observed in extracts of mitotic cells compared with those of asynchronous cells (Fig. 2).
FIGURE 2.
Interaction between HSF2 and MEL-18 is decreased during mitosis. Extracts of asynchronous (A) or mitotic (M) HeLa cells were immunoprecipitated using anti-HSF2 antibodies or nonspecific IgG, and the immunoprecipitates were subjected to Western blotting using anti-MEL-18 antibodies. The amounts of HSF2 in the input and anti-HSF2 immunoprecipitate samples were measured by subjecting these samples to Western blotting using goat polyclonal anti-HSF2 antibodies.
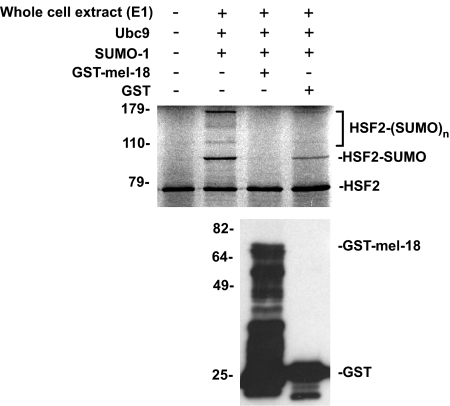
As described above in the Introduction, previous studies showed that the polycomb protein PC2/CBX4 functions as an E3 protein to stimulate sumoylation of specific target proteins (24, 25). Based on these findings, we sought to determine whether this polycomb protein, MEL-18, may play a role in regulating sumoylation of HSF2. Specifically, our previous finding of increased sumoylation of HSF2 during mitosis (17), coupled with our new results in Fig. 2 indicating that interaction between HSF2 and MEL-18 is decreased during mitosis, suggested the intriguing hypothesis that MEL-18 may actually function as a negative regulator of HSF2 sumoylation. This would be in contrast to the sumoylation stimulatory function of the polycomb protein PC2/CBX4. As a first test of this hypothesis, we determined whether adding purified recombinant GST-MEL-18 to an in vitro sumoylation assay would affect the SUMO modification of HSF2 in this system. As shown in Fig. 3, addition of purified GST-MEL-18 is associated with decreased sumoylation of HSF2 in the in vitro modification assay.
FIGURE 3.
Purified MEL-18 inhibits in vitro sumoylation of HSF2. 35S-Labeled in vitro translated HSF2 (first lane) was subjected to in vitro sumoylation in the absence of any additional purified proteins (second lane) or in the presence of purified GST-MEL-18 (third lane) or GST (fourth lane). Samples were then analyzed by SDS-PAGE and autoradiography (upper panel). The anti-GST Western blot (lower panel) shows the relative amounts of GST-MEL-18 or GST that were added to the reactions in the third and fourth lanes of the upper panel, respectively. E1, ubiquitin-activating enzyme.
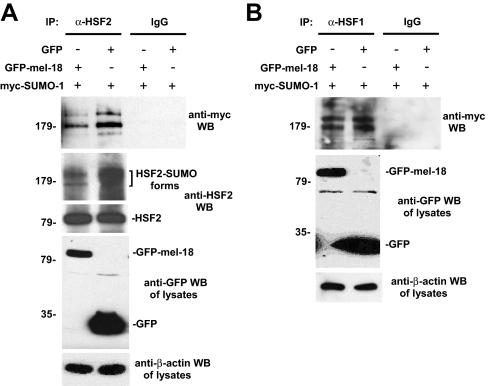
Next, we wanted to test whether MEL-18 can inhibit the sumoylation of HSF2 expressed in cells. To test this, GFP-MEL-18 or GFP expression constructs along with Myc-SUMO-1 expression plasmid were transfected into cells, and then extracts of the cells were subjected to immunoprecipitation with anti-HSF2 antibodies or nonspecific IgG (negative control), followed by anti-Myc Western blotting to detect the sumoylated forms of the HSF2 protein. The results of this experiment, shown in Fig. 4A, indicate that expression of GFP-MEL-18, but not GFP, is associated with decreased HSF2 sumoylation. To probe the specificity of this effect of GFP-MEL-18 in inhibiting HSF2 sumoylation, we performed a similar experiment to analyze the related HSF1 protein, which we and others have shown previously to be sumoylated in response to stress (27, 28). The results show that HSF1 sumoylation is not significantly affected by expression of GFP-MEL-18 (Fig. 4B), indicating the selectivity of the inhibitory effect of MEL-18 overexpression on sumoylation of HSF2.
FIGURE 4.
MEL-18 inhibits sumoylation of HSF2 in vivo. A, HEK 293 cells were transfected with GFP-MEL-18 or GFP expression constructs along with Myc-SUMO-1 expression plasmid, and then extracts of the cells were subjected to immunoprecipitation (IP) with anti-HSF2 antibodies (rabbit polyclonal) or nonspecific IgG (negative control), followed by anti-Myc Western blotting (WB) to detect the sumoylated forms of HSF2. Extracts of the cells were also subjected to Western blotting using goat polyclonal HSF2 antibodies. The cell lysates were subjected to anti-GFP Western blotting to analyze expression levels of GFP-MEL-18 and GFP and to anti-β-actin Western blotting as a loading control. B, an experiment similar to that described in A was performed, except that here the cells were subjected to a 42 °C heat treatment for 60 min prior to harvesting them for immunoprecipitation analysis (to allow stress-induced HSF1 sumoylation) and that anti-HSF1 antibodies were used for the immunoprecipitation step, so that sumoylated forms of HSF1 would be detected by the subsequent anti-Myc Western blotting (detecting Myc-SUMO-1).
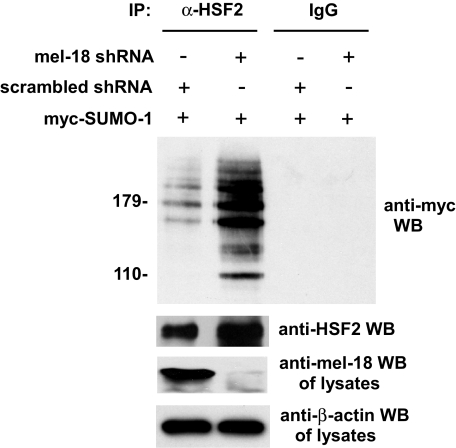
As a reverse, complementary approach for testing the hypothesis that MEL-18 may be a negative regulator of HSF2 sumoylation, we determined the effect of reducing cellular levels of MEL-18 on HSF2 sumoylation using the RNAi methodology. According to our hypothesis and the results in Figs. 3 and 4, we predicted that knockdown of MEL-18 should result in an increase in sumoylation of HSF2. To test this, cells were transfected with MEL-18 shRNA or scrambled shRNA along with Myc-SUMO-1 expression plasmid, and then extracts of the cells were subjected to immunoprecipitation with anti-HSF2 antibodies or nonspecific IgG (negative control), followed by anti-Myc Western blotting to detect the sumoylated forms of HSF2. As shown in Fig. 5, the results of this experiment indicate that, consistent with our hypothesis, a reduction in MEL-18 protein levels is associated with an increase in HSF2 sumoylation. These results, together with those shown in Figs. 3 and 4 above, support the hypothesis that MEL-18 functions as an inhibitor of HSF2 sumoylation.
FIGURE 5.
Knockdown of MEL-18 protein levels is associated with increased HSF2 sumoylation. HeLa cells were transfected with MEL-18 shRNA or scrambled shRNA along with Myc-SUMO-1 expression plasmid, and then extracts of the cells were subjected to immunoprecipitation (IP) with anti-HSF2 antibodies (rabbit polyclonal) or nonspecific IgG (negative control), followed by anti-Myc Western blotting (WB) to detect the sumoylated forms of HSF2. Extracts of the cells were also subjected to Western blotting using goat polyclonal HSF2 antibodies. The cell lysates were subjected to anti-MEL-18 Western blotting to confirm reduction of MEL-18 protein levels by MEL-18 shRNA treatment and to anti-β-actin Western blotting as a loading control.
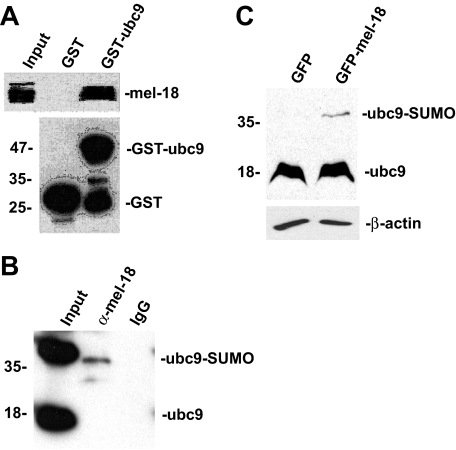
Next, we sought to probe the underlying mechanism by which MEL-18 inhibits the sumoylation of HSF2. A previous study showed that the polycomb protein PC2 mediates its effects on sumoylation of its target proteins by interacting with UBC9, the SUMO E2 enzyme (24). Based on this finding, we hypothesized that MEL-18 may also interact with UBC9 as part of its mechanism for inhibiting HSF2 sumoylation. To test this hypothesis, we performed an in vitro binding experiment to determine whether MEL-18 protein present in whole cell extracts can interact with purified recombinant GST-UBC9 bound to glutathione-agarose beads. In this experiment, after incubating the GST-UBC9 or GST bound to glutathione-agarose with HeLa cell extracts and washing the beads to remove unbound proteins, the amount of bound MEL-18 was determined by boiling the beads in SDS-PAGE buffer, followed by Western blotting using anti-MEL-18 antibodies. The results of this experiment indicate that purified recombinant UBC9 is able to interact with MEL-18 present in cell extracts (Fig. 6A).
FIGURE 6.
MEL-18 interacts with the SUMO E2 enzyme UBC9. A, GST or GST-UBC9 proteins bound to glutathione-agarose beads were incubated with extracts of HeLa cells, and then after washing the amount of bound MEL-18 was determined by Western blotting using anti-MEL-18 antibodies (upper panel). The anti-GST Western blot (lower panel) shows the relative amounts of GST-UBC9 or GST that were used in the binding reactions, respectively. B, extracts of HeLa cells were subjected to immunoprecipitation using anti-MEL-18 antibodies, followed by Western blotting of the immunoprecipitates using UBC9 antibodies. C, HEK 293 cells were transfected with the GFP or GFP-MEL-18 expression constructs, and then extracts of the transfected cells were subjected to Western blot assay using anti-UBC9 antibodies to detect the non-SUMO-conjugated (18-kDa) and SUMO-containing (39-kDa) forms of UBC9. The cell lysates were also subjected to anti-β-actin Western blotting as a loading control.
To test for interaction between endogenous MEL-18 and UBC9 proteins, we subjected cell extracts to immunoprecipitation using MEL-18 antibodies, followed by Western blotting of the immunoprecipitates using UBC9 antibodies. The results, shown in Fig. 6B, indicate that endogenous MEL-18 and UBC9 do interact. The results also suggest that there is more complex between MEL-18 and the form of UBC9 that is covalently charged with SUMO (39-kDa form) relative to the non-SUMO-carrying form of UBC9 (18-kDa form).
Based on this finding of interaction between MEL-18 and UBC9, we envisioned a mechanism in which MEL-18 bound to HSF2 inhibits its sumoylation by binding to and inhibiting the activity of UBC9 enzymes that approach HSF2. One way of inhibiting UBC9 activity would be to block its ability to transfer the SUMO group from its active site to the target protein. Therefore, as a means for testing this proposed mechanism, we determined whether increasing the level of MEL-18 in cells results in increased amounts of the form of UBC9 that has SUMO remaining covalently bound to it. GFP-MEL-18 or GFP alone was expressed in HEK 293 cells by transfection, and then extracts of the transfected cells were subjected to Western blotting with anti-UBC9 antibodies, which will detect both the 18-kDa non-SUMO-containing form of UBC9 and the 39-kDa SUMO-containing form of UBC9. The results of this experiment, shown in Fig. 6C, indicate that expression of GFP-MEL-18 results in increased levels of the SUMO-containing form of UBC9 compared with cells expressing the GFP-alone construct. This result supports the hypothesis that MEL-18 inhibits UBC9 activity by decreasing its ability to transfer SUMO groups to target proteins.
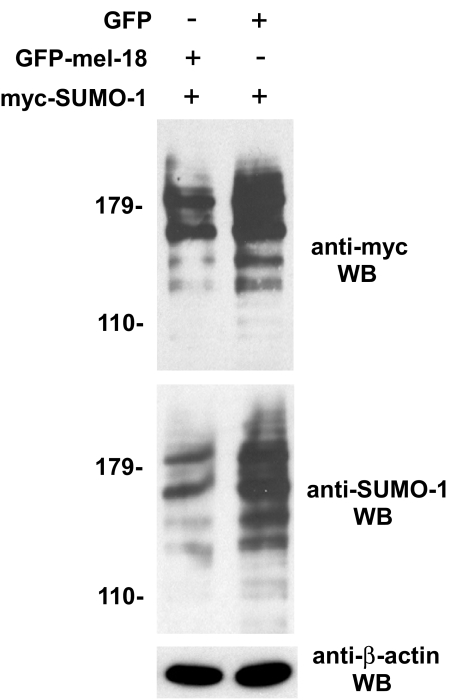
Based on the results shown in Fig. 6 indicating that MEL-18 interacts with UBC9 and may inhibit its ability to attach SUMO groups to proteins, we hypothesized that MEL-18 may inhibit the sumoylation of other proteins in addition to HSF2, perhaps even acting to down-regulate cellular sumoylation globally. To test this hypothesis, extracts of HEK 293 cells transfected with GFP-MEL-18 or GFP expression constructs, along with Myc-SUMO-1 expression plasmid from the experiment shown in Fig. 4, were subjected to Western blotting using anti-Myc and anti-SUMO-1 antibodies to detect sumoylated forms of cellular proteins. Consistent with this hypothesis, the results of this analysis indicate that increased expression of MEL-18 is associated with a detectable decrease in conjugation of SUMO-1 to cellular proteins (Fig. 7).
FIGURE 7.
MEL-18 inhibits general protein sumoylation in cells. Extracts of HEK 293 cells transfected with GFP-MEL-18 or GFP expression constructs, along with Myc-SUMO-1 expression plasmid from the experiment shown in Fig. 4, were subjected to Western blotting (WB) using anti-Myc and anti-SUMO-1 antibodies to detect sumoylated forms of cellular proteins and to anti-β-actin Western blotting as a loading control.
DISCUSSION
The results presented in this work support the hypothesis that MEL-18 bound to HSF2 inhibits its sumoylation by binding to and inhibiting the activity of SUMO E2 (UBC9) enzymes in the vicinity of the HSF2 protein. Furthermore, our results showing that the interaction between HSF2 and MEL-18 is decreased during mitosis would provide a mechanism to explain the previously observed finding that HSF2 sumoylation is increased during this stage of the cell cycle (17).
The results described in this work also suggest that MEL-18 actually functions like an anti-SUMO E3 protein because, like a traditional SUMO E3 protein, it interacts both with the SUMO E2 enzyme UBC9 and a sumoylation substrate protein (HSF2), but instead of stimulating SUMO modification of HSF2 it inhibits it. This is in contrast to the sumoylation-stimulating activities of another polycomb protein, PC2, a traditional SUMO E3, indicating that members of the polycomb group of proteins are involved in both the positive and negative regulation of protein sumoylation through their interactions with UBC9 and substrate proteins. The results also indicate that MEL-18 is able to detectably inhibit conjugation of SUMO-1 to proteins, suggesting that MEL-18 likely inhibits the sumoylation of other proteins in addition to HSF2, perhaps even acting to down-regulate sumoylation in cells globally. Future studies investigating whether additional polycomb proteins act as positive or negative regulators of the sumoylation of proteins in cells and identifying other SUMO substrate proteins in which modification is regulated by these polycomb proteins would likely provide important insights into both the regulation of protein sumoylation and the mechanisms by which polycomb proteins mediate their important biological functions.
Acknowledgments
We thank Guy Sauvageau for the pSG5-MEL-18 plasmid, Mike Matunis for the GST-UBC9 construct, and other members of the Sarge laboratory for insightful discussions.
This work was supported by National Institutes of Health Grants GM61053 and GM64606 (to K. D. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SUMO, small ubiquitin-like modifier; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; GFP, green fluorescent protein; EGFP, enhanced GFP; HSF, heat shock factor; RNAi, RNA interference; GST, glutathione S-transferase; shRNA, short hairpin RNA.
References
- 1.Dohmen, R. J. (2004) Biochim. Biophys. Acta 1695 113–131 [DOI] [PubMed] [Google Scholar]
- 2.Johnson, E. S. (2004) Annu. Rev. Biochem. 73 355–382 [DOI] [PubMed] [Google Scholar]
- 3.Hay, R. T. (2005) Mol. Cell 18 1–12 [DOI] [PubMed] [Google Scholar]
- 4.Gill, G. (2005) Curr. Opin. Genet. Dev. 15 536–541 [DOI] [PubMed] [Google Scholar]
- 5.Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006) Annu. Rev. Cell Dev. Biol. 22 159–180 [DOI] [PubMed] [Google Scholar]
- 6.Bossis, G., and Melchior, F. (2006) Cell Div. 29 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay, D., and Dasso, M. (2007) Trends Biochem. Sci. 32 286–295 [DOI] [PubMed] [Google Scholar]
- 8.Desterro, J. M., Thomson, J., and Hay, R. T. (1997) FEBS Lett. 417 297–300 [DOI] [PubMed] [Google Scholar]
- 9.Johnson, E. S., and Blobel, G. (1997) J. Biol. Chem. 272 26799–26802 [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez, M. S., Dargemont, C., and Hay, R. T. (2001) J. Biol. Chem. 276 12654–12659 [DOI] [PubMed] [Google Scholar]
- 11.Sampson, D. A., Wang, M., and Matunis, M. J. (2001) J. Biol. Chem. 276 21664–21669 [DOI] [PubMed] [Google Scholar]
- 12.Johnson E. S., and Gupta, A. A. (2001) Cell 106 735–744 [DOI] [PubMed] [Google Scholar]
- 13.Kahyo, T., Nishida, T., and Yasuda, H. (2001) Mol. Cell 8 713–718 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi, Y., Toh-e, A., and Kikuchi, Y. (2001) Gene (Amst.) 275 223–231 [DOI] [PubMed] [Google Scholar]
- 15.Goodson, M. L., Hong, Y., Rogers, R., Matunis, M. J., Park-Sarge, O. K., and Sarge, K. D. (2001) J. Biol. Chem. 276 18513–18518 [DOI] [PubMed] [Google Scholar]
- 16.Anckar, J., Hietakangas, V., Denessiouk, K., Thiele, D. J., Johnson, M. S., and Sistonen, L. (2006) Mol. Cell. Biol. 26 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing, H., Wilkerson, D. C., Mayhew, C. N., Lubert, E. J., Skaggs, H. S., Goodson, M. L., Hong, Y., Park-Sarge, O. K., and Sarge, K. D. (2005) Science 307 421–423 [DOI] [PubMed] [Google Scholar]
- 18.Sarge, K. D., and Park-Sarge, O. K. (2005) Trends Biochem. Sci. 30 605–610 [DOI] [PubMed] [Google Scholar]
- 19.Breiling, A., Sessa, L., and Orlando, V. (2007) Int. Rev. Cytol. 258 83–136 [DOI] [PubMed] [Google Scholar]
- 20.Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B., and Cavalli, G. (2007) Cell 128 735–745 [DOI] [PubMed] [Google Scholar]
- 21.Ringrose, L., and Paro, R. (2007) Development (Camb.) 134 223–232 [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, Y. B., and Pirrotta, V. (2007) Nat. Rev. Genet. 8 9–22 [DOI] [PubMed] [Google Scholar]
- 23.Sparmann, A., and van Lohuizen, M. (2006) Nat. Rev. Cancer 6 846–856 [DOI] [PubMed] [Google Scholar]
- 24.Kagey, M. H., Melhuish, T. A., and Wotton, D. (2003) Cell 113 127–137 [DOI] [PubMed] [Google Scholar]
- 25.Li, B., Zhou, J., Liu, P., Hu, J., Jin, H., Shimono, Y., Takahashi, M., and Xu, G. (2007) Biochem. J. 405 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilgarth, R. S., and Sarge, K. D. (2005) Methods Mol. Biol. 301 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong, Y., Rogers, R., Matunis, M. J., Mayhew, C. N., Goodson, M. L., Park-Sarge, O. K., and Sarge, K. D. (2001) J. Biol. Chem. 276 40263–40267 [DOI] [PubMed] [Google Scholar]
- 28.Hietakangas, V., Ahlskog, J. K., Jakobsson, A. M., Hellesuo, M., Sahlberg, N. M., Holmberg, C. I., Mikhailov, A., Palvimo, J. J., Pirkkala, L., and Sistonen, L. (2003) Mol. Cell. Biol. 23 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]